Abstract
Depletion of the mitochondrial matrix protein frataxin is the molecular cause of the neurodegenerative disease Friedreich ataxia. The function of frataxin is unclear, although recent studies have suggested a function of frataxin (yeast Yfh1) in iron/sulphur (Fe/S) protein biogenesis. Here, we show that Yfh1 specifically binds to the central Fe/S-cluster (ISC)-assembly complex, which is composed of the scaffold protein Isu1 and the cysteine desulphurase Nfs1. Association between Yfh1 and Isu1/Nfs1 was markedly increased by ferrous iron, but did not depend on ISCs on Isu1. Functional analyses in vivo showed an involvement of Yfh1 in de novo ISC synthesis on Isu1. Our data demonstrate a crucial function of Yfh1 in Fe/S protein biogenesis by defining its function in an early step of this essential process. The iron-dependent binding of Yfh1 to Isu1/Nfs1 suggests a role of frataxin/Yfh1 in iron loading of the Isu scaffold proteins.
Introduction
The neurodegenerative disorder Friedreich ataxia (FRDA) is caused by a deficiency of the mitochondrial matrix protein frataxin (Babcock et al., 1997; Foury & Cazzalini, 1997; Wilson & Roof, 1997). Although the cellular function of frataxin and its yeast orthologue Yfh1 is still a matter of debate (for a review, see Puccio & Koenig, 2002; Wilson, 2003), a role in cellular iron homeostasis seems likely. A deficiency of frataxin/Yfh1 leads to an accumulation of iron in mitochondria (Babcock et al., 1997; Foury & Cazzalini, 1997; Puccio et al., 2001). In vitro studies suggest that frataxin may bind iron and form large aggregates that are reminiscent of the iron-storage protein ferritin (Adamec et al., 2000; Cavadini et al., 2002). However, the physiological relevance of the multimeric forms of frataxin has been questioned (Adinolfi et al., 2002). Another general phenotype of frataxin-deficient cells is a defect in mitochondrial Fe/S proteins (Rötig et al., 1997; Foury, 1999; Chen et al., 2002; Lutz et al., 2001). Recently, frataxin was found to be required for de novo biogenesis of cellular Fe/S proteins in vivo (Mühlenhoff et al., 2002a), which demonstrated a direct function of frataxin in this essential process.
Biosynthesis of Fe/S proteins in eukaryotes is mediated by a set of highly conserved components that are located in mitochondria (for a review, see Mühlenhoff & Lill, 2000; Craig & Marszalek, 2002; Gerber & Lill, 2002). Central components of this Fe/S cluster (ISC)-assembly machinery in yeast are the cysteine desulphurase Nfs1, which provides sulphur, and the homologous protein pair Isu1/Isu2, which bind ferrous iron and function as scaffolds for de novo synthesis of the nascent ISC (Frazzon & Dean, 2003). In addition, the electron-transfer system that is comprised of the ferredoxin Yah1 and the ferredoxin reductase Arh1 is essential and may reduce elemental sulphur (S0) to sulphide. Furthermore, the DnaK-like and DnaJ-like chaperones Ssq1 and Jac1, respectively, interact with Isu1/2 and are required after ISC assembly on Isu1/2 (Dutkiewicz et al., 2003; Mühlenhoff et al., 2003).
To elucidate the molecular function of frataxin, we attempted to identify its cellular interaction partners. We report a direct interaction between yeast Yfh1 and the central ISC-assembly complex Isu1/Nfs1 in mitochondrial extracts. Interaction was strongly dependent on the presence of physiological concentrations of ferrous iron. Furthermore, Yfh1 is crucially involved in the de novo synthesis of an ISC on the Isu1 scaffold, where it may support the iron loading of Isu1.
Results
Yfh1 binds to the core ISC-assembly complex Isu1/Nfs1
To identify the interaction partners of yeast frataxin (Yfh1), we first used the glutathione-S-transferase (GST)-affinity system. We constructed a yeast expression vector that allows the synthesis of the fusion protein pSu9GST–Yfh1. This carries the amino-terminal mitochondrial targeting sequence of the ATP synthase subunit 9 (pSu9) and GST at the N terminus of Yfh1, which replaces the Yfh1 pre-sequence. As a control, the fusion protein pSu9GST was used. After synthesis in Saccharomyces cerevisiae, the mature forms of both fusion proteins were located exclusively in mitochondria (data not shown). GST–Yfh1 was functional, as it restored the growth defect of a Gal–YFH1 mutant in which endogenous Yfh1 is depleted (data not shown; Mühlenhoff et al., 2002a).
Mitochondria that harboured the GST proteins were isolated and lysed in detergent-containing buffer, and the extracts were subjected to GST-affinity purification. The components associated with the purified GST proteins were identified by immunostaining. The two predominant proteins that precipitated with GST–Yfh1 were Isu1 and Nfs1 (Fig. 1A). No significant amounts of other components of the ISC-assembly machinery, such as Ssq1, Yah1, Grx5, Jac1, or of other mitochondrial proteins, such as aconitase, were detected (Fig. 1A; and data not shown). In particular, native Yfh1 did not co-purify with GST–Yfh1, indicating that the Yfh1 species that was isolated in our assay system functions as a monomer (see below). Binding of Isu1 and Nfs1 to GST–Yfh1 was highly specific, as these and other mitochondrial proteins were not found to be associated with GST (Fig. 1B). Furthermore, the amount of co-purification of Isu1 increased substantially when Isu1 was overproduced, indicating the specificity of the interaction (Fig. 1A).
Figure 1.
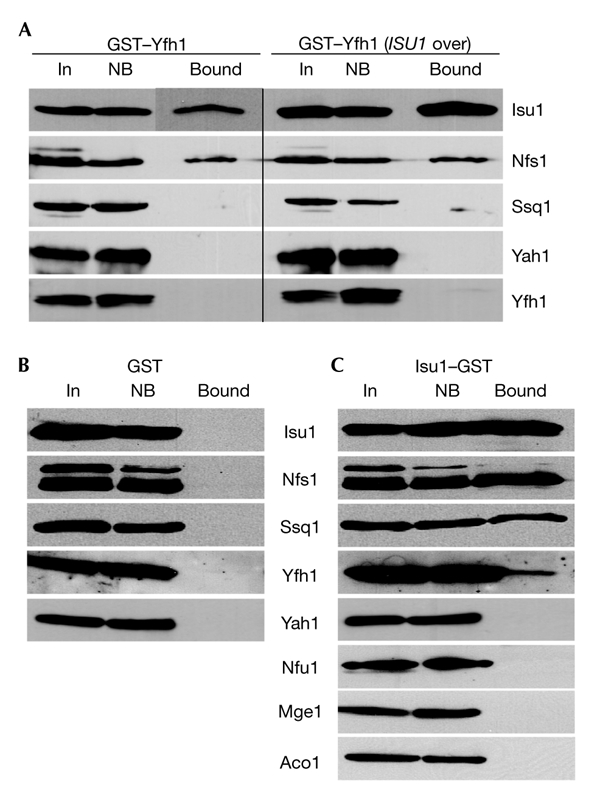
Yfh1 binds to the Isu1/Nfs1 complex. The fusion proteins pSu9GST–Yfh1, pSu9GST and Isu1–GST (glutathione-S-transferase) were produced in wild-type yeast cells grown in the presence of galactose. Mitochondria were isolated, lysed in detergent-containing buffer A, and GST-affinity purification was performed. Fractions were analysed for the presence of specific proteins by immunostaining (A–C). Results obtained using cells that overexpressed ISU1 are shown in the right panel of (A) (ISU1 over.). Bound, GST-affinity purified protein; In, input mitochondrial extract (10% of total); NB, non-bound fraction (10% of total).
Yfh1 is co-purified with Isu1–GST
To verify the specificity of the protein–protein interactions analysed above, we constructed an expression vector for the synthesis of an Isu1 fusion protein (Isu1–GST) that has the GST domain at its carboxyl terminus. Isu1–GST was imported into mitochondria and fully restored the growth defects of the promoter-exchange mutant Gal–ISU1/Δisu2 on depletion of Isu1 (see below), which indicates that the Isu1 domain was functional (data not shown). Wild-type mitochondria that harboured Isu1–GST were subjected to GST-affinity purification. In addition to Isu1–GST, purified fractions contained the ISC proteins Nfs1, Ssq1 and Yfh1, thus supporting the data described above (Fig. 1C). Moreover, untagged Isu1 was co-purified, indicating that both Isu1 proteins interact, a finding that is consistent with that for the dimeric form of purified yeast Isu1 (data not shown). Our data also show that similar ISC protein complexes are formed in the yeast and bacterial ISC systems (Frazzon & Dean, 2003). The interactions detected were highly specific, as no other proteins of the mitochondrial ISC-assembly machinery or of other pathways were detected in the purified fractions (Fig. 1C; and data not shown).
Binding of Yfh1 to Isu1–GST is concentration dependent
The amount of Yfh1 that is co-isolated with Isu1–GST should decrease on overexpression of the ISU1 gene, due to competition of native Isu1 with its tagged version. Similarly, the yield of Yfh1 and Isu1–GST complex formation should increase on overproduction of native Yfh1. Variation of Isu1 and Yfh1 levels was achieved by using the promoter-exchange mutant Gal–ISU1/Δisu2, which carries a disrupted ISU2 gene and ISU1 under the control of the galactose-inducible and glucose-repressible GAL1-10 promoter (Fig. 2A). These cells were transformed with a plasmid that carries the YFH1 gene downstream of the doxycycline-repressible TetO2 promoter and a plasmid that encodes Isu1–GST. On downregulation of Isu1, a significant amount of Yfh1 was co-isolated with Isu1–GST by GST-affinity purification (Fig. 2C). This amount was considerably higher than that seen in wild-type cells (Fig. 1C) and increased further on overproduction of Yfh1. This result indicates that the binding equilibrium between Yfh1 and the two Isu1 proteins was shifted towards the formation of the Yfh1/Isu1–GST complex. Conversely, on overproduction of Isu1 by the growth of Gal–ISU1/Δisu2 cells in the presence of galactose, no significant amount of Yfh1 was co-purified with Isu1–GST, even after overproduction of Yfh1 (Fig. 2B). These findings show that Isu1–GST efficiently competed with native Isu1 for binding to Yfh1, Nfs1 and Ssq1, and therefore demonstrate further the specificity of these interactions.
Figure 2.
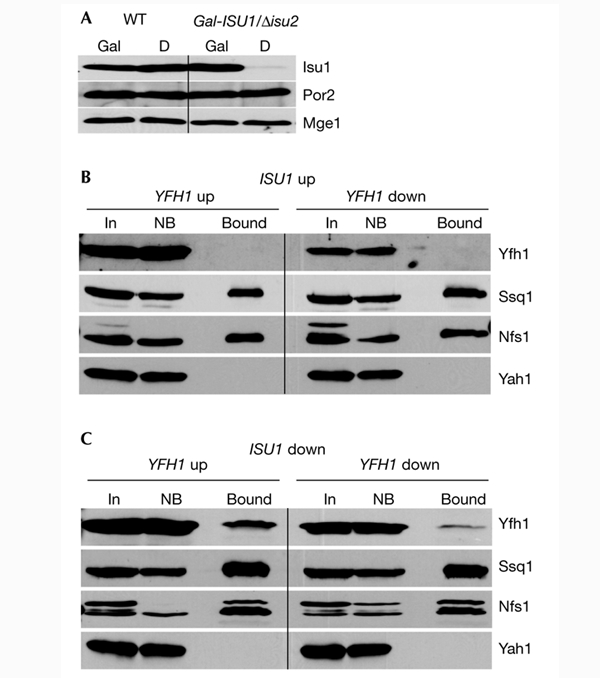
The interaction between Isu1–GST and Yfh1 is dependent on the concentrations of Isu1 and Yfh1. (A) Wild-type (WT) and Gal–ISU1/Δisu2 cells were grown on synthetic minimal media supplemented with galactose (Gal) or glucose (D). Mitochondria were isolated and analysed by immunostaining for Isu1, porin (Por2) and Mge1. (B,C) Isu1–GST was overproduced in Gal–ISU1/Δisu2 cells containing a plasmid (pCM182–YFH1) that carries YFH1 under the control of the TetO2 promoter. In this strain, levels of native Isu1 (without GST) are downregulated by growth in the presence of glucose, and the overexpression of YFH1 is blocked by the addition of 5 μg ml−1 doxycycline. Mitochondria were isolated from cells cultivated on synthetic minimal media supplemented with galactose (ISU1 up) or glucose (ISU1 down) in the absence (YFH1 up) or presence (YFH1 down) of doxycycline, as indicated. Further analysis was carried out as described for Fig. 1. Bound, glutathione-S-transferase (GST)-affinity purified protein; In, input mitochondrial extract (10% of total); NB, non-bound fraction (10% of total).
Co-immunoprecipitation of Yfh1 with Isu1/Nfs1
As an independent method to study protein–protein interactions, we used co-immunoprecipitation. Specific anti-Yfh1 antisera quantitatively immunoprecipitated Yfh1 from mitochondrial extracts (Fig. 3A). The anti-Yfh1 immunoprecipitate contained significant amounts of Isu1 and Ssq1, but not of Yah1 and non-ISC proteins, such as Heat shock protein 60 (Hsp60; Fig. 3A; and data not shown). No ISC proteins or other mitochondrial proteins were detected in the antibody-bound fraction using pre-immune serum (Fig. 3A, left panel). These findings establish a specific interaction in vivo between the native forms of Yfh1 and Isu1/Nfs1.
Figure 3.
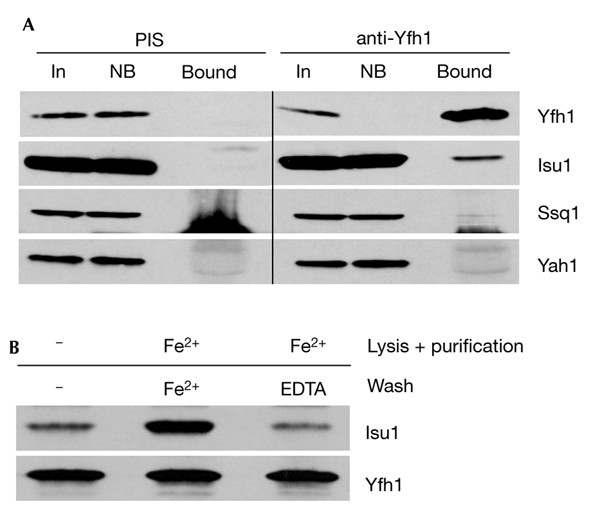
Co-immunoprecipitation of Yfh1 and the Isu1/Nfs1 complex. (A) Isolated wild-type mitochondria that overexpressed Isu1 (p426–Isu1) were lysed in detergent-containing buffer A and an immunoprecipitation was performed using a specific antiserum raised against purified Yfh1 and a pre-immune serum (PIS). The immunoprecipitates were analysed by SDS-polyacrylamide gel electrophoresis, followed by immunostaining for the indicated proteins. The staining below Ssq1 is due to the heavy chains of IgG, which preclude the analysis of Nfs1 by this method. (B) The lysis, purification and washing steps were carried out in the absence or presence of 50 μM Fe2+/1 mM ascorbate (Fe2+) and 1 mM EDTA, as indicated. Bound, glutathione-S-transferae (GST)-affinity purified protein; In, input mitochondrial extract (10% of total); NB, non-bound fraction (10% of total).
Iron reversibly increases the binding of Yfh1 to Isu1/Nfs1
As Isu1 binds iron in a labile manner in vitro (Agar et al., 2000; Nuth et al., 2002), we tested the potential influence of reduced iron on the physical interaction between Yfh1 and Isu1/Nfs1. Maintenance of physiological concentrations of ferrous iron (50 μM) during detergent lysis of mitochondria and throughout the GST purification procedure induced a substantial increase in the yield of Isu1/Nfs1 that co-purified with GST–Yfh1 (Fig. 4A). When the glutathione beads were treated with EDTA or other metal chelators during the washing steps, the yield of co-isolated Isu1/Nfs1 decreased almost to the levels found in the absence of added iron, indicating the reversible character of binding. Highly similar results were obtained using the co-immunoprecipitation method with anti-Yfh1 antibodies (Fig. 3B). The effect is specific for iron, as 50 μM Mg2+, Ca2+ or Mn2+ ions did not influence complex formation (Fig. 4B). Similarly, low-molecular-mass compounds that are involved in Fe/S protein biogenesis (cysteine, NADH and ATP; Mühlenhoff et al., 2002b) had no effect (data not shown). In summary, reversible complex formation between Yfh1 and Isu1/Nfs1 is strongly dependent on ferrous iron. In contrast to previous in vitro studies, ferrous iron did not induce oligomerization of Yfh1 in our in organello assay system, as we could not observe any co-purification of wild-type Yfh1 with GST–Yfh1 (Fig. 4A; Cavadini et al., 2002).
Figure 4.
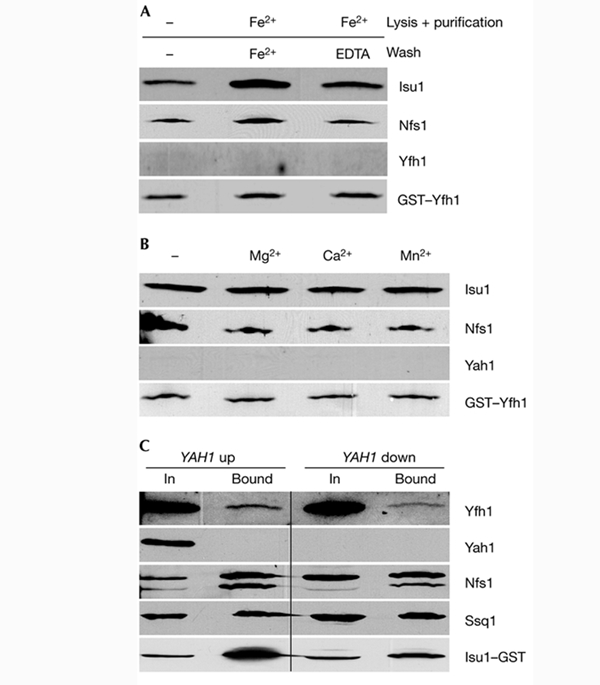
Binding of Isu1 to Yfh1 is increased at higher iron concentrations, but does not require an Fe/S cluster on Isu1. (A,B) Mitochondria from wild-type cells that overproduce glutathione-S-transferease (GST)–Yfh1 were used for GST-affinity purification, as described for Fig. 1. Treatment of samples in (A) was as described for Fig. 3B; samples in (B) were incubated in the presence of 50 μM of metal ions, as indicated. (C) Gal–YAH1 cells that overexpress Isu1–GST were grown on synthetic minimal media supplemented with galactose (YAH up) or glucose (YAH down). Mitochondria were isolated, Isu1–GST was purified and bound proteins were analysed as described for Fig. 1. Bound, GST-affinity purified protein; In, input mitochondrial extract (10% of total).
Yfh1 binds to the Fe/S cluster-free apoform of Isu1
Does Isu1 require a transiently bound ISC for its association with Yfh1? To determine this, we synthesized Isu1–GST in the promoter-exchange mutant Gal-YAH1, which carries a regulatable YAH1 gene (Lange et al., 2000). In vivo analyses have shown that depletion of the ferredoxin Yah1 results in severe defects in ISC synthesis on the Isu1 scaffold, that is Isu1 remains in the apoform in conditions of Yah1 depletion (Mühlenhoff et al., 2003). In Yah1-depleted Gal–YAH1 mitochondria, Yfh1 was co-isolated with Isu1–GST by GST-affinity chromatography (Fig. 4C). The amount of purified Yfh1 relative to the input protein was similar to that in wild-type cells (Fig. 1C), showing that Yfh1 bound efficiently to the ISC-free apoform of Isu1. For unknown reasons, the level of affinity-purified Isu1–GST was lower in conditions of Yah1 depletion; that is, co-isolation of Yfh1 relative to purified Isu1–GST increased on depletion of Yah1. We conclude that the interaction between Yfh1 and Isu1/Nfs1 does not require the presence of an ISC on Isu1. It was not possible to test whether Yfh1 also associates with the ISC-containing holoform of Isu1, as the cluster is only transiently bound and the steady-state levels of the Isu1 holoprotein in vivo may be low.
Yfh1 is crucial for Fe/S cluster synthesis on Isu1
To obtain an insight into the functional significance of the interaction between Isu1/Nfs1 and Yfh1, we analysed whether Yfh1 is crucial for ISC synthesis on the Isu1 scaffold. Yfh1 was depleted using Gal–YFH1 cells that carry a regulatable YFH1 gene (Mühlenhoff et al., 2002a). To monitor Isu1-bound ISCs, Isu1 was overexpressed in Gal–YFH1 and wild-type cells. After growth in galactose- or glucose-containing media, cells were radiolabelled with 55Fe, cell extracts were prepared and Isu1 was immunoprecipitated. In wild-type cells or Gal–YFH1 cells cultivated in the presence of galactose, a significant amount of radioactive 55Fe was co-immunoprecipitated with Isu1 (Fig. 5), indicating the formation of an ISC on Isu1 (Mühlenhoff et al., 2003). The iron that was associated with the immunobeads was specific, as levels of radioactivity close to background levels (<0.3 pmol of 55Fe per gram of cells) were detected without overproduction of Isu1, or with preimmune serum. Conversely, on depletion of Yfh1 by growth in the presence of glucose, the amount of 55Fe associated with Isu1 decreased fourfold. As the levels of overexpressed Isu1 and other mitochondrial proteins did not decrease detectably (Fig. 5, inset; Mühlenhoff et al., 2002a), the data strongly indicate a crucial requirement of Yfh1 in de novo ISC assembly on Isu1.
Figure 5.
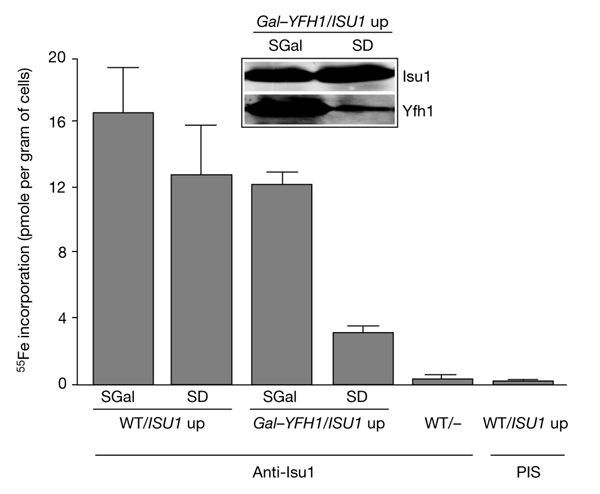
Yfh1 is required for de novo Fe/S-cluster synthesis on Isu1. Wild-type (WT) and Gal–YFH1 cells that overexpress ISU1 (ISU1 up) were incubated in an iron-poor medium supplemented with galactose (SGal) or glucose (SD). Cells were radiolabelled with 55Fe and lysed mechanically, and Isu1 was immunoprecipitated from the cell lysates using Isu1-specific antibodies. The amount of radioactive 55Fe associated with the immunobeads was quantified by liquid scintillation counting. The inset (immunostaining) shows the levels of Isu1 and Yfh1 in Gal–YFH1 cells. PIS, immunoprecipitation with pre-immune serum.
Discussion
In this study, we have defined the molecular function of yeast frataxin in the complex process of Fe/S-protein biogenesis. We identified binding partners of Yfh1 in mitochondria, defined the conditions for interaction and performed functional studies to detect the site of action of Yfh1 in Fe/S protein biogenesis. Several lines of evidence indicate that Yfh1 specifically binds to the core ISC-assembly complex Isu1/Nfs1. Yfh1 was affinity purified with an Isu1–GST fusion protein, and the Isu1/Nfs1 complex was also found to associate with GST–Yfh1. The specificity of the interaction was demonstrated by the fact that the extent of binding to the GST fusion proteins was decreased by increasing the amounts of the corresponding native proteins. These findings were supported using co-immunoprecipitation as a second, independent method to demonstrate the association between native Yfh1 and Isu1/Nfs1. As we used isolated mitochondria for our analyses, the results reported here might reflect the situation in vivo. In support of this, ISU1 gene expression is increased in yfh1 mutant cells, which indicates a genetic link between these genes (Foury & Talibi, 2001).
Binding between Yfh1 and Isu1/Nfs1 was significantly higher when physiological concentrations of ferrous iron were maintained during complex isolation. However, formation of the complex did not require an ISC on Isu1. These observations are consistent with our functional studies (Fig. 5), which indicate a crucial involvement of Yfh1 in de novo ISC synthesis on Isu1. Other components required for this early step in Fe/S protein biogenesis are Nfs1 and Yah1/Arh1, whereas the chaperones Ssq1 and Jac1 are dispensable and may function later in biogenesis (Mühlenhoff et al., 2003). What is the specific contribution of Yfh1 to the synthesis of an ISC on Isu1? Frataxin might be involved in iron loading of Isu1, sulphur transfer from Nfs1 to Isu1, or the combination of iron and sulphur to a cluster on the Isu1 scaffold. On the basis of the iron-stimulated binding of Yfh1 to Isu1/Nfs1, the most likely possibility is that frataxin supports the iron loading of Isu1. This suggestion, which is derived from our in organello and in vivo studies, is supported by elegant in vitro work that was published during the revision of our manuscript. Yoon & Cowan (2003) have demonstrated direct iron transfer from frataxin to the human ISU protein. The authors also reported stable binding of 6–7 iron ions per frataxin molecule. However, under more physiological conditions, and with mitochondrial extracts, no stable binding of iron to frataxin/Yfh1 was seen (Adinolfi et al., 2002; Mühlenhoff et al., 2002a). Further studies under conditions that are close to the physiological situation are needed to resolve this apparent discrepancy between in vivo and in vitro analyses and to clarify the issue of stable versus labile iron-binding to Isu proteins. Our investigation and the work of Yoon & Cowan (2003) now open the way to elucidate the functions of Yfh1 in iron transfer to Isu1 and in ISC synthesis at a molecular level.
The results presented here unequivocally support suggestions of a primary function of frataxin in Fe/S protein biogenesis and define the site of action of the protein. This function explains the mitochondrial iron accumulation in frataxin-deficient cells, as an increased mitochondrial iron concentration is a well-documented secondary phenotype of cells that are defective in Fe/S protein biogenesis (Lill & Kispal, 2000; Chen et al., 2002). As frataxin is conserved from bacteria to humans, our data are of relevance to the neurodegenerative disorder Friedreich ataxia, which is associated with defects in Fe/S proteins and mitochondrial iron accumulation in affected cells and in mouse models (Rötig et al., 1997; Puccio et al., 2001).
Methods
Yeast strains and cell growth.
The Saccharomyces cerevisiae strains used were as follows: W303-1A (MATa, ura3-1, ade2-1, trp1-1, his3-11,15, leu2-3,112), which was used as a wild-type strain; Gal–ISU1/Δisu2 (W303-1A; isu2::HIS3; nucleotides −477 to −16 of the ISU1 gene were replaced by the GAL1-10/LEU2 cassette); Gal–YFH1 (Mühlenhoff et al., 2002a). Cells were grown as described previously (Mühlenhoff et al., 2002a).
Plasmid constructs.
The GST open reading frame was amplified by PCR from the vector pGEX-2TK (Amersham). For pSu9GST, codons 1–69 of the mitochondrial targeting sequence of the ATPase subunit 9 from Neurospora crassa were fused to codons 1–220 of the GST gene from Schistosoma japonicum. For Isu1–GST, full-length ISU1 from S. cerevisiae (codons 1–163) was fused to codons 1–220 of GST. For pSu9GST–Yfh1, pSu9 (codons 1–69) and GST (codons 1–233) were fused to YFH1 from S. cerevisiae (codons 52–174). Constructs were inserted into the ARS/CEN vector p416MET25 (Mumberg et al., 1995). The fusion constructs were verified by DNA sequencing.
Affinity purification of GST fusion proteins and co-immunoprecipitations.
Mitochondria (250 μg of protein) were lysed for 3 min on ice in 200 μl buffer A (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.2% Tween 20, 10 μM leupeptin, 10 μM pepstatin, 2 mM PMSF (phenylmethylsulphonylfluoride), 50 μM pyridoxal phosphate, and 1 mM ascorbic acid). Membrane debris was removed by centrifugation for 10 min at 15,000 r.p.m. and the supernatant was incubated with 30 μl of Glutathione-Sepharose 4B beads (Amersham) for 1 h on a rotary shaker at 4 °C. Beads were collected by centrifugation for 5 min at 3,000 r.p.m. and washed three times with 300 μl of buffer A. Bound proteins were subjected to SDS–polyacrylamide gel electrophoresis and identified by immunostaining. Co-immunoprecipitation was performed under the same conditions using specific antiserum against Yfh1. All other methods have been described previously (Mühlenhoff et al., 2002a).
Acknowledgments
We thank H. Herrmann for providing us with antibodies against Ssq1. This study was supported by grants from the Sonderforschungsbereiche 286 and 593, Deutsche Forschungsgemeinschaft, Fonds der Chemischen Industrie, Deutsches Humangenomprojekt (HITOP) and the European Commission (QLG1-CT-2001-00966).
References
- Adamec J., Rusnak F., Owen W.G., Naylor S., Benson L.M., Gacy A.M. & Isaya G. ( 2000) Iron-dependent self-assembly of recombinant yeast frataxin: implications for Friedreich ataxia. Am. J. Hum. Genet., 67, 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adinolfi S., Trifuoggi M., Politou A.S., Martin S. & Pastore A. ( 2002) A structural approach to understanding the iron-binding properties of phylogenetically different frataxins. Hum. Mol. Genet., 11, 1865–1877. [DOI] [PubMed] [Google Scholar]
- Agar J.N., Yuvaniyama P., Jack R.F., Cash V.L., Smith A.D., Dean D.R. & Johnson M.K. ( 2000) Modular organization and identification of a mononuclear iron-binding site within the NifU protein. J. Biol. Inorg. Chem., 5, 167–177. [DOI] [PubMed] [Google Scholar]
- Babcock M., De Silva D., Oaks R., Davis-Kaplan S., Jiralerspong S., Montermini L., Pandolfo M. & Kaplan J. ( 1997) Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science, 276, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Cavadini P., O'Neill H.A., Benada O. & Isaya G. ( 2002) Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum. Mol. Genet., 11, 217–227. [DOI] [PubMed] [Google Scholar]
- Chen O.S., Hemenway S. & Kaplan J. ( 2002) Inhibition of Fe–S cluster biosynthesis decreases mitochondrial iron export: evidence that Yfh1p affects Fe-S cluster synthesis. Proc. Natl Acad. Sci. USA, 99, 12321–12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E.A. & Marszalek J. ( 2002) A specialized mitochondrial molecular chaperone system: a role in formation of Fe/S centers. Cell. Mol. Life Sci., 59, 1658–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkiewicz R., Schilke B., Kneiszner H., Walter W., Craig E.A. & Marszalek J. ( 2003) Ssq1, a mitochondrial Hsp70 involved in iron–sulfur (Fe/S) center biogenesis: similarities to and differences from its bacterial counterpart. J. Biol. Chem. (in the press). [DOI] [PubMed] [Google Scholar]
- Foury F. ( 1999) Low iron concentration and aconitase deficiency in a yeast frataxin homologue deficient strain. FEBS Lett., 456, 281–284. [DOI] [PubMed] [Google Scholar]
- Foury F. & Cazzalini O. ( 1997) Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS Lett., 411, 373–377. [DOI] [PubMed] [Google Scholar]
- Foury F. & Talibi D. ( 2001) Mitochondrial control of iron homeostasis. A genome wide analysis of gene expression in a yeast frataxin-deficient strain. J. Biol. Chem., 276, 7762–7768. [DOI] [PubMed] [Google Scholar]
- Frazzon J. & Dean D.R. ( 2003) Formation of iron-sulphur clusters in bacteria—an emerging field in bioinorganic chemistry. Curr. Opin. Chem. Biol., 6, 166–173. [DOI] [PubMed] [Google Scholar]
- Gerber J. & Lill R. ( 2002) Biogenesis of iron–sulfur proteins in eukaryotes: components, mechanism and pathology. Mitochondrion, 2, 71–86. [DOI] [PubMed] [Google Scholar]
- Lange H., Kispal G., Kaut A. & Lill R. ( 2000) A mitochondrial ferredoxin is essential for biogenesis of intra- and extra-mitochondrial Fe/S proteins. Proc. Natl Acad. Sci. USA, 97, 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R. & Kispal G. ( 2000) Maturation of cellular Fe/S proteins: the essential function of mitochondria. Trends Biochem. Sci., 25, 352–356. [DOI] [PubMed] [Google Scholar]
- Lutz T., Westermann B., Neupert W. & Herrmann J.M. ( 2001) The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J. Mol. Biol., 307, 815–825. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U. & Lill R. ( 2000) Biogenesis of iron–sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Biochim. Biophys. Acta, 1459, 370–382. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U., Richhardt N., Ristow M., Kispal G. & Lill R. ( 2002a) The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum. Mol. Genet., 11, 2025–2036. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U., Richhardt N., Gerber J. & Lill R. ( 2002b) Characterization of iron–sulfur protein assembly in isolated mitochondria. A requirement for ATP, NADH and reduced iron. J. Biol. Chem., 277, 29810–29816. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U., Richhardt N., Gerber J. & Lill R. ( 2003) Components involved in assembly and dislocation of iron–sulfur clusters on the scaffold protein Isu1p. EMBO J., in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Müller R. & Funk M. ( 1995) Yeast vectors for controlled expression of heterologous proteins in different genetic backgrounds. Gene, 156, 119–122. [DOI] [PubMed] [Google Scholar]
- Nuth M., Yoon T. & Cowan J.A. ( 2002) Iron–sulfur cluster biosynthesis: characterization of iron nucleation sites for assembly of the [2Fe–2S]2+ cluster core in IscU proteins. J. Am. Chem. Soc., 124, 8774–8775. [DOI] [PubMed] [Google Scholar]
- Puccio H. & Koenig M. ( 2002) Friedreich ataxia: a paradigm for mitochondrial diseases. Curr. Opin. Genet. Dev., 12, 272–277. [DOI] [PubMed] [Google Scholar]
- Puccio H., Simon D., Cossee M., Criqui-Filipe P., Tiziano F., Melki J., Hindelang C., Matyas R., Rustin P. & Koenig M. ( 2001) Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe–S enzyme deficiency followed by intramitochondrial iron deposits. Nature Genet., 27, 181–186. [DOI] [PubMed] [Google Scholar]
- Rötig A., de Lonlay P., Chretien D., Foury F., Koenig M., Sidi D., Munnich A. & Rustin P. ( 1997) Aconitase and mitochondrial iron–sulphur protein deficiency in Friedreich ataxia. Nature Genet., 17, 215–217. [DOI] [PubMed] [Google Scholar]
- Wilson R.B. ( 2003) Frataxin and frataxin deficiency in Friedreich's ataxia. J. Neurol. Sci., 207, 103–105. [DOI] [PubMed] [Google Scholar]
- Wilson R.B. & Roof D.M. ( 1997) Respiratory deficiency due to loss of mitochondrial DNA in yeast lacking the frataxin homologue. Nature Genet., 16, 352–357. [DOI] [PubMed] [Google Scholar]
- Yoon T. & Cowan J.A. ( 2003) Iron–sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in Isu-type proteins. J. Am. Chem. Soc., 125, 6078–6084. [DOI] [PubMed] [Google Scholar]


