Abstract
Most chromatin in interphase nuclei is part of condensed chromatin domains. Previous work has indicated that transcription takes place predominantly at the surface of chromatin domains, that is, in the perichromatin region. It is possible that genes inside chromatin domains are silenced due to inaccessibility to macromolecular components of the transcription machinery. We have tested the accessibility of chromatin domains in nuclei of living cells with proteins and dextrans of different molecular sizes. Our results show that chromatin domains are readily accessible to large macromolecules, including proteins with a molecular weight of several hundred kilodaltons. Therefore, the silencing of genes that are incorporated into such domains is not due to the physical inaccessibility of condensed chromatin domains to transcription factors.
Introduction
There is growing evidence that the architecture of the interphase nucleus is an important element in the epigenetic regulation of gene expression (Francastel et al., 2000). Using a variety of techniques, it has been shown that transcriptionally active genes are predominantly excluded from condensed chromatin domains (Cmarko et al., 1999; Fakan, 1994; Verschure et al., 1999). Little or no transcriptional activity was detected inside chromatin domains, which suggests that transcription units inside chromatin domains are inactive. Similarly, most (but not all) genes associated with pericentromeric heterochromatin are silenced (Brown et al., 1999; Cockell & Gasser, 1999; Fisher & Merkenschlager, 2002). It has been suggested that the transcription machinery has no access to genes inside chromatin domains because these structures are too dense. In support of this, transcriptional activation correlates with considerable chromatin decondensation, which may be necessary for the access of transcription factors to genes (for example, see Tumbar et al., 1999).
Previous studies have shown that several large components involved in transcription initiation, replication and repair move rapidly inside the cell nucleus (Houtsmuller et al., 1999; Phair & Misteli, 2000). However, these studies do not address the relationship between unconstrained movement and the distribution of condensed chromatin. Here, we test the accessibility of condensed chromatin domains in nuclei of living cells with proteins and dextrans of various molecular sizes. The results indicate that chromatin domains are relatively open structures that are readily accessible to transcription, replication and repair factors.
Results and Discussion
Spatial distribution of 3-, 10- and 70-kDa dextrans
Fluorescently labelled dextrans of various molecular sizes were microinjected into the nuclei of living HeLa cells that express green fluorescent protein (GFP)-tagged histone H2B, which enables visualization of the chromatin (Kanda et al., 1998). Dual-colour, three-dimensional confocal images were obtained, allowing comparison of the spatial distribution of dextran molecules and GFP-tagged chromatin. We used uncharged dextrans because previous studies have shown that fluorescently labelled dextrans diffuse freely in the nucleus and behave like inert macromolecules (Seksek et al., 1997). Figure 1 shows that 3- and 10-kDa dextrans are evenly distributed throughout the nucleoplasm, except for in the nucleoli, which partially exclude both dextrans. The line scans show that in nucleoli the 3-kDa dextran signal (Fig. 1Ad,Bd) and the 10-kDa dextran signal (Fig. 1Cd,Dd) are reduced by 20% and 40%, respectively. Peripheral heterochromatin (close to the nuclear envelope) and perinucleolar heterochromatin (around the nucleoli) are completely accessible (regions are marked with arrows in the overlay images (Fig. 1Ac,Bc,Cc,Dc); corresponding to asterisks in the line scans (Fig. 1Ad,Bd,Cd,Dd). The results show that 3- and 10-kDa dextran molecules have access to all compartments in the nucleus.
Figure 1.
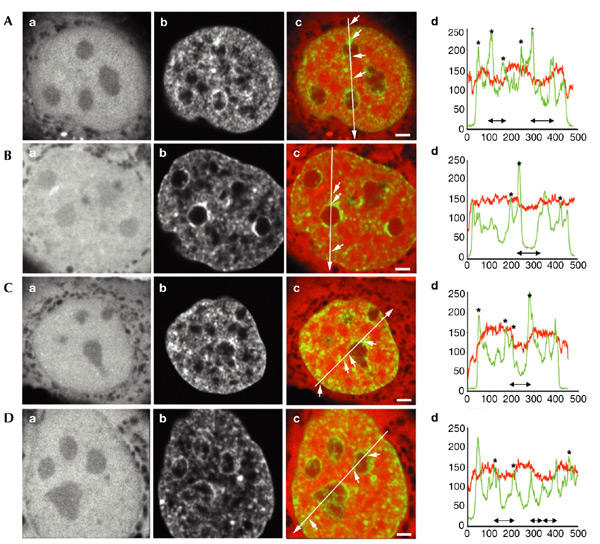
Spatial distribution of 3- and 10-kDa dextrans in relation to chromatin domains. Fluorescently labelled dextrans with molecular weights of 3 and 10 kDa were microinjected into nuclei of living HeLa cells. (A,B) Typical nuclei microinjected with 3-kDa dextran. (C,D) Nuclei microinjected with 10-kDa dextran. (Aa,Ba,Ca,Da) Distribution of microinjected fluorescently labelled dextrans. (Ab,Bb,Cb,Db) Histone-H2B–green-fluorescent-protein (GFP) signal. (Ac,Bc,Cc,Dc) Overlay of dextrans in red and chromatin in green. (Ad,Bd,Cd,Dd) Line scans showing the local intensity distribution of fluorescently labelled dextrans (red) and GFP-tagged chromatin (green). In the overlay images, the positions and directions of the line scans are indicated by long arrows. Small arrows point to several positions on the lines that correspond to asterisks in the line scans. The positions of the nucleoli are shown by double-headed black arrows below the line scans. Three-dimensional images were recorded; images shown are individual mid-nuclear optical sections. Scale bars in (Ac,Bc,Cc), 2 μm; scale bar in (Dc), 1.5 μm.
The 70-kDa dextran shows a more complex distribution (Fig. 2). Some nuclear domains with high chromatin concentrations are less accessible to these dextran molecules (Fig. 2Ad,Bd, regions marked with arrows in the overlay; corresponding to asterisks in the line scans in Fig. 2Ae,Be). This is seen for perinuclear heterochromatin (for example, Fig. 2Ad, first arrow on line 2 (in the direction of the arrow); corresponding to the first asterisk in Fig. 2Aeii), for perinucleolar heterochromatin (for example, Fig. 2Bd, first arrow on line 2; corresponding to the first asterisk in Fig. 2Beii), and for some chromatin-dense domains elsewhere in the nucleoplasm, which probably represents pericentromeric heterochromatin (for example, Fig. 2Ad, second arrow on line 2; corresponding to the second asterisk in Fig. 2Aeii). A significant fraction of the chromatin-dense domains, however, did not show a reduction in local dextran concentration, indicating that these domains are fully accessible (for example, Fig. 2Bd, third arrow on line 2; corresponding to the third asterisk in Fig. 2Beii). Nucleoli exclude 70-kDa dextran molecules to a level that is 25% of that in the nucleoplasm. In addition, in various interchromatin regions, that contain little or no chromatin, a significant local decrease in intensity of the dextran signal was seen (for example, Fig. 2Ad, first arrow on line 3; corresponding to the first asterisk in Fig. 2Aeiii). These results show that different types of condensed chromatin domains exist that can be distinguished by their accessibility to high-molecular-weight dextrans, and that several non-chromatin subnuclear domains exist that also are poorly accessible to 70-kDa dextrans.
Figure 2.
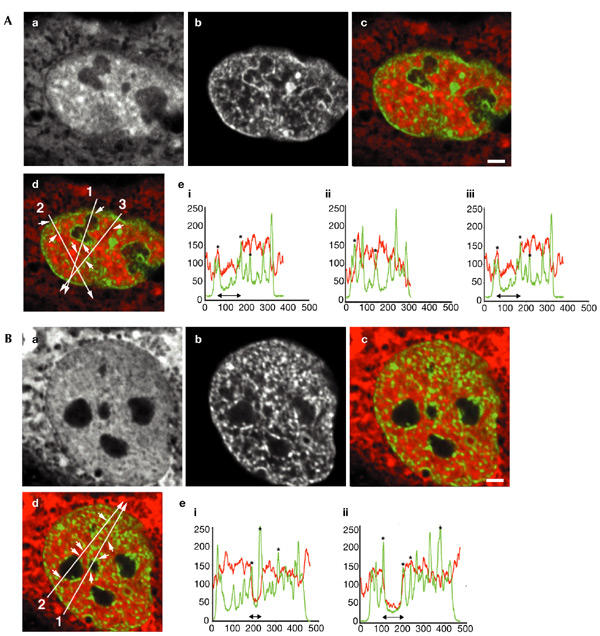
Spatial distribution of 70-kDa dextran in relation to chromatin domains. Fluorescently labelled dextran with a molecular weight of 70 kDa was microinjected into nuclei of living HeLa cells. (A,B) Typical nuclei, showing (Aa,Ba) the distribution of microinjected fluorescently labelled dextrans, (Ab,Bb) the histone-H2B–green-fluorescent-protein (GFP) signal, (Ac,Ad,Bc,Bd) overlays of the dextran signal in red and GFP-tagged chromatin in green, (Ae,Be) line scans through the two nuclei shown in (Ac,Ad,Bc,Bd). In the overlay pictures in (Ad,Bd), the line-scan positions are indicated by long arrows. Small arrows point to several positions on the line that are represented by asterisks in the line scans. The positions of the nucleoli are represented by black double-headed arrows in the line scans (Ae,Be). Three-dimensional images were recorded; images show individual mid-nuclear optical sections. Scale bars, 2 μm.
Spatial distribution of GFP-tagged proteins
In addition to inert dextrans, we analysed the accessibility of chromatin to two large, endogenous protein complexes, the transcription/repair factor TFIIH–GFP and GFP–RNA-polymerase-II, in human fibroblasts and CHO-K1 cells, respectively (Hoogstraten et al., 2002; Kimura et al., 2002). Interestingly, TFIIH–GFP is homogeneously distributed in the nucleoplasm (Fig. 3A,B), including peripheral and perinucleolar heterochromatin and other nuclear domains with a high local chromatin concentration (marked with arrows in the overlay images in Fig. 3Ac,Bc; corresponding to asterisks in Fig. 3Ad,Bd, respectively). In addition, TFIIH–GFP accumulates in nucleoli, consistent with its recently described role in RNA polymerase I transcription (Iben et al., 2002; Hoogstraten et al., 2002). In addition, the multisubunit complex GFP–RNA-polymerase-II is diffusely distributed throughout the nucleoplasm and cytoplasm (Kimura et al., 2002), and is excluded from nucleoli to ∼55% of that in the cytoplasm (Fig. 3C,D). Consistent with the dextran data, peripheral and perinucleolar heterochromatin and other nuclear domains with a high local chromatin concentration are completely accessible to GFP-tagged RNA polymerase II (marked with arrows in the overlay images in Fig. 3Cc,Dc; corresponding to asterisks in Fig. 3Cd,Dd, respectively).
Figure 3.
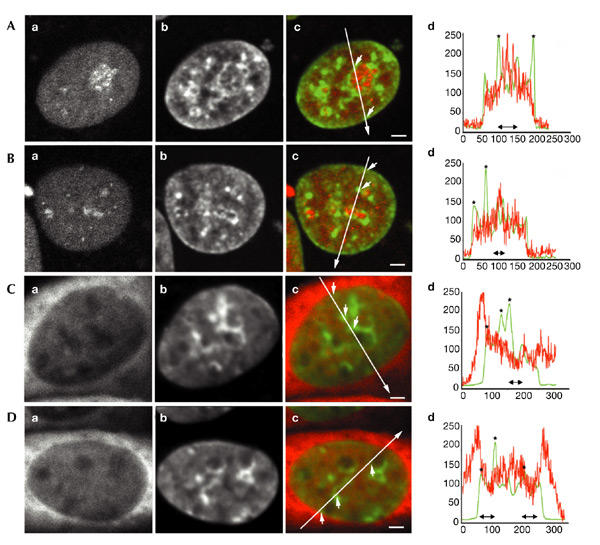
Spatial distribution of GFP-tagged TFIIH and RNA polymerase II in relation to chromatin domains. Chromatin from human SV40 (simian virus 40)-transformed fibroblasts that express GFP-tagged TFIIH, and from CHO-K1 cells that express GFP-tagged RNA polymerase II, were stained with Hoechst. (A,B) Typical nuclei; TFIIH–GFP distribution in relation to condensed chromatin is shown. (C,D) Typical nuclei; RNA-polymerase–II-GFP distribution in relation to condensed chromatin is shown. (Aa,Ba,Ca,Da) Distribution of GFP-tagged protein. (Ab,Bb,Cb,Db) Hoechst-labelled condensed chromatin. (Ac,Bc,Cc,Dc) Overlay of the GFP-tagged protein signal in red and the Hoechst-labelled chromatin in green. (Ad,Bd,Cd,Dd) Line scans through the nuclei shown in Ac,Bc,Cc and Dc, respectively. In the overlay images, the positions and directions of the line scans are indicated by long arrows. Small arrows point to several positions on the lines that correspond to asterisks in the line scans. The positions of the nucleoli are shown by double-headed black arrows below the line scans. Three-dimensional images were recorded; images shown are individual, mid-nuclear optical sections. Scale bars, 1.5 mm. GFP, green fluorescent protein.
Accessibility of chromatin domains
Several studies have indicated that many nuclear proteins diffuse freely throughout the nucleus (for example, Houtsmuller et al., 1999; Phair & Misteli, 2000). However, none of these studies have analysed the relationships between protein mobility and large-scale chromatin organization. The behaviour of inert macromolecules in the nucleus is predominantly determined by their hydrodynamic properties (Lukacs et al., 2000; Seksek et al., 1997). This behaviour depends on their radius of gyration (RG). RG values for 3-, 10- and 70-kDa dextrans are 4, 6 and 10 nm, respectively (Lukacs et al., 2000; Seksek et al., 1997). For spherical proteins, RG = 0.775R, where 2R is the diameter of the protein molecule (Van Holde, 1971). Starting with this information, simple calculations show that RG values of 4, 6 and 10 nm correspond to spherical proteins with molecular weights of ∼400, ∼1,400 and ∼6,600 kDa, respectively. For non-spherical proteins, for example, those that can be described as a prolate ellipsoid with an axial ratio of 3, RG = 2.2R (2R is the short axis of the ellipsoid), the corresponding molecular weights are 55, 180 and 850 kDa, respectively. Clearly, these numbers are only indicative of the size range of protein molecules that have hydrodynamic properties similar to the dextrans that we used in this study. The calculations indicate that protein molecules with molecular dimensions in the size range of components of the transcription machinery can diffuse freely inside condensed chromatin domains. In agreement with this, our results show that the GFP-tagged endogenous proteins TFIIH and RNA polymerase II (with molecular weights of ∼550 and ∼600 kDa, respectively), have access to all chromatin domains.
The results obtained for 70-kDa dextran, which were calculated to correspond to proteins with molecular weights between ∼850 kDa and 6,600 kDa (depending on their shape), allow us to put limits to the sizes of molecules or complexes that have access to condensed chromatin domains. The 70-kDa dextrans show limited access to some domains with high chromatin concentration, that is, perinuclear and perinucleolar heterochromatin and (peri)centromeric heterochromatin. Consistent with these results, Politz et al. (1999) have shown that endogenous heterogeneous nuclear ribonucleoprotein (hnRNP) particles (large poly(A) RNA–protein complexes) can move freely in the interchromatin space, but are excluded from chromatin domains and nucleoli. hnRNP particles are much larger than most protein components in the cell, which explains their exclusion from condensed chromatin domains.
The difference in nucleolar accessibility of GFP–TFIIH compared with GFP–RNA-polymerase-II and the dextran molecules is an intriguing observation. GFP–TFIIH accumulates in the nucleolus, probably because of binding to nucleolar transcription sites (Iben et al., 2002; Hoogstraten et al., 2002), whereas GFP–RNA-polymerase-II and the dextrans have limited access. These findings are consistent with the fact that observed spatial distributions of nuclear proteins are determined by their accessibility to nuclear compartments and their affinity for nuclear binding sites.
Large-scale chromatin structure and accessibility
The observation that condensed chromatin domains are freely accessible to large macromolecules has important implications for understanding large-scale chromatin structure. Apparently, the higher-order structure of the folded nucleosomal fibre is relatively open. Protein complexes in the size range of a nucleosome, and those that are larger, can readily enter condensed chromatin domains. This may in part be explained by the constrained diffusion of chromatin in vivo, resulting in 'breathing' of the structure, thereby giving access to large protein complexes (Chubb et al., 2002; Gasser, 2002; Marshall et al., 1997; Vazquez et al., 2001).
Chromatin function and large-scale organization
Our results suggest that it is unlikely that genes embedded in dense chromatin are inactive due to exclusion of the transcription machinery from the chromatin domain. This is consistent with the observation that some genes that are transcribed by RNA polymerase II are active in a heterochromatin environment (Hilliker et al., 1980; Lu et al., 2000). In addition, DNA repair factors seem to have rapid access to condensed chromatin domains (Houtsmuller et al., 1999; Essers et al., 2002). Despite the apparent accessibility of dense chromatin domains, many transcriptionally active loci are excluded from condensed chromatin (Cmarko et al., 1999; Fakan, 1994; Verschure et al., 1999). Similarly, DNA replication occurs predominantly near the surface of condensed chromatin domains (Fakan & Hancock, 1974; Jaunin & Fakan, 2002; Jaunin et al., 2000). Our data suggest that principles other than simple steric exclusion are responsible for the silencing of loci inside condensed chromatin. It is likely that the accessibility of DNA sequence elements inside condensed chromatin domains is controlled by the recruitment of histone-modifying enzymes and chromatin-remodelling factors. The question remains as to whether the observed positioning of transcriptionally active loci and factors involved in gene transcription outside condensed chromatin domains (for example, in the perichromatin space) is a prerequisite for gene expression, or is somehow the consequence of that process (Singer & Green, 1997; Chubb & Bickmore, 2003).
Methods
Cell culture.
HeLa cells that express histone H2B–GFP (Kanda et al., 1998; provided by G. Wahl) were grown in DMEM that contained 10% FCS, with selection using 0.35 mg ml−1 G418 (Sigma). CHO-K1 cells that stably express the largest catalytic subunit of RNA polymerase II, with a GFP tag (provided by H. Kimura; Kimura et al., 2002), were grown in F12 medium that contained 10% FCS. Human SV40 (simian virus 40)-immortalized XPCS2BA fibroblasts without a functional XPB gene (which encodes one of the core subunits of the general transcription factor TFIIH), but that stably express enhanced-GFP–XPB (Hoogstraten et al., 2002), were used to analyse the behaviour of TFIIH. Cells were grown in F10/DMEM that contained 10% FCS. Cells expressing GFP-tagged proteins were grown at 37 °C in a 5% CO2 atmosphere on glass-bottom microwell dishes that were coated with poly-D-lysine (Mattek) to 50–70% confluency. To stain the chromatin in cells expressing GFP-tagged TFIIH and RNA polymerase II, cells were incubated for 1 h in 100 μg ml−1 Hoechst 33258.
Microinjection of fluorescently labelled dextrans.
The nuclei of living cells were microinjected with a solution of 10 mg ml−1 dextrans in PBS pH 7.4. The following dextrans were used for microinjection: a 3-kDa dextran labelled with Texas Red; 10- and 70-kDa dextrans labelled with tetramethylrhodamine (Molecular Probes). A volume of approximately 5% of the nuclear volume was microinjected into the cell nucleus (Narashige microinjection system). After microinjection, cells were cultured at 37 °C for at least 2 h before analysis.
Confocal laser-scanning microscopy.
All experiments were performed in duplicate. For each experiment, ten nuclei were visualized and at least five nuclei were imaged. Images were recorded using a Zeiss LSM 510 confocal laser-scanning microscope (Zeiss) equipped with a 100×/1.23 NA oil-immersion objective. We used an Ar laser at 488 nm and a He/Ne laser at 543 nm to excite green and red fluorochromes simultaneously. Fluorescence was detected with a 505–530-nm bandpass filter (green) and a 560-nm longpass filter (red). Pairs of images were collected simultaneously. Three-dimensional images were scanned as 512 × 512 × 32 voxel images (sampling rate, 49 nm lateral and 208 nm axial).
Image processing.
Image analysis was performed using the Huygens System 2 software package (Scientific Volume Imaging). For semi-quantitative analysis of the spatial relationship between the relative distributions of components in dual-labelled cells, we made line scans. The signal intensities of the two labels were plotted along a line through the nucleus. The highest intensity inside the nucleus of each of two labels was set at 100% and the lowest intensity (outside the nucleus) was set at zero.
Acknowledgments
This work was supported in part by the European Union BIOMED II programme and by the biological branch of the Dutch National Research Council (NWO) programme.
References
- Brown K.E., Baxter J., Graf D., Merkenschlager M. & Fisher A.G. ( 1999) Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell, 3, 207–217. [DOI] [PubMed] [Google Scholar]
- Chubb J.R. & Bickmore W.A. ( 2003) Considering nuclear comparmentalization in the light of nuclear compartments. Cell, 112, 403–406. [DOI] [PubMed] [Google Scholar]
- Chubb J.R., Boyle S., Perry P. & Bickmore W.A. ( 2002) Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol., 12, 439–445. [DOI] [PubMed] [Google Scholar]
- Cmarko D., Verschure P.J., Martin T.E., Dahmus M.E., Krause S., Fu X.D., Van Driel R. & Fakan S. ( 1999) Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell, 10, 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M. & Gasser S.M. ( 1999) Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev., 9, 199–205. [DOI] [PubMed] [Google Scholar]
- Essers J., Houtsmuller A.B., van Veelen L., Paulusma C., Nigg A.L., Pastink A., Vermeulen W., Hoeijmakers J.H. & Kanaar R. ( 2002) Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J., 21, 2030–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S. ( 1994) Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol., 4, 86–90. [DOI] [PubMed] [Google Scholar]
- Fakan S. & Hancock R. ( 1974) Localization of newly-synthesized DNA in a mammalian cell as visualized by high resolution autoradiography. Exp. Cell Res., 83, 95–102. [DOI] [PubMed] [Google Scholar]
- Fisher A.G. & Merkenschlager M. ( 2002) Gene silencing, cell fate and nuclear organisation. Curr. Opin. Genet. Dev., 12, 193–197. [DOI] [PubMed] [Google Scholar]
- Francastel C., Schubeler D., Martin D.I.K. & Groudine M. ( 2000) Nuclear compartmentalization and gene activity. Nature Rev. Mol. Cell Biol., 1, 137–143. [DOI] [PubMed] [Google Scholar]
- Gasser S.M. ( 2002) Nuclear architecture—visualizing chromatin dynamics in interphase nuclei. Science, 296, 1412–1416. [DOI] [PubMed] [Google Scholar]
- Hilliker A.J., Appels R. & Schalet A. ( 1980) The genetic analysis of D. melanogaster heterochromatin. Cell, 21, 607–619. [DOI] [PubMed] [Google Scholar]
- Hoogstraten D., Nigg A.L., Heath H., Mullenders H.F., Van Driel R., Hoeijmakers J.H.J., Vermeulen W. & Houtsmuller A.B. ( 2002) Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell, 10, 1163–1174. [DOI] [PubMed] [Google Scholar]
- Houtsmuller A.B., Rademakers S., Nigg A.L., Hoogstraten D., Hoeijmakers J.H.J. & Vermeulen W. ( 1999) Action of DNA repair endonuclease ERCC1/XPF in living cells. Science, 284, 958–961. [DOI] [PubMed] [Google Scholar]
- Iben S., Tschochner H., Bier M., Hoogstraten D., Hozak P., Egly J.M. & Grummt I. ( 2002) TFIIH plays an essential role in RNA polymerase I transcription. Cell, 109, 297–306. [DOI] [PubMed] [Google Scholar]
- Jaunin F. & Fakan S. ( 2002) DNA replication and nuclear architecture. J. Cell. Biochem., 85, 1–9. [PubMed] [Google Scholar]
- Jaunin F., Visser A.E., Cmarko D., Aten J.A. & Fakan S. ( 2000) Fine structural in situ analysis of nascent DNA movement following DNA replication. Exp. Cell Res., 260, 313–323. [DOI] [PubMed] [Google Scholar]
- Kanda T., Sullivan K.F. & Wahl G.M. ( 1998) Histone–GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol., 8, 377–385. [DOI] [PubMed] [Google Scholar]
- Kimura H., Sugaya K. & Cook P.R. ( 2002) The transcription cycle of RNA polymerase II in living cells. J. Cell Biol., 159, 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B.Y., Emtage P.C.R., Duyf B.J., Hilliker A.J. & Eissenberg J.C. ( 2000) Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics, 155, 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs G.L., Haggie P., Seksek O., Lechardeur D., Freedman N. & Verkman A.S. ( 2000) Size-dependent DNA mobility in cytoplasm and nucleus. J. Biol. Chem., 275, 1625–1629. [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Straight A., Marko J.F., Swedlow J., Dernburg A., Belmont A., Murray A.W., Agard D.A. & Sedat J.W. ( 1997) Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol., 7, 930–939. [DOI] [PubMed] [Google Scholar]
- Phair R.D. & Misteli T. ( 2000) High mobility of proteins in the mammalian cell nucleus. Nature, 404, 604–609. [DOI] [PubMed] [Google Scholar]
- Politz J.C., Tuft R.A., Pederson T. & Singer R.H. ( 1999) Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr. Biol., 9, 285–291. [DOI] [PubMed] [Google Scholar]
- Seksek O., Biwersi J. & Verkman A.S. ( 1997) Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J. Cell Biol., 138, 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer R.H. & Green M.R. ( 1997) Compartmentalization of eukaryotic gene expression. Cell, 91, 291–294. [DOI] [PubMed] [Google Scholar]
- Tumbar T., Sudlow G. & Belmont A.S. ( 1999) Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol., 145, 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holde K.E. ( 1971) In Physical Biochemistry (eds Hager, L. & Wold, F.), 180–202. Prentice-Hall, Inc., Englewood Cliffs, New Jersey, USA. [Google Scholar]
- Vazquez J., Belmont A.S. & Sedat J.W. ( 2001) Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr. Biol., 11, 1227–1239. [DOI] [PubMed] [Google Scholar]
- Verschure P.J., van der Kraan I., Manders E.M. & van Driel R. ( 1999) Spatial relationship between transcription sites and chromosome territories. J. Cell Biol., 147, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]


