Abstract
We present the first experimental evidence for the existence of an exosome-like protein complex in Archaea. In Eukarya, the exosome is essential for many pathways of RNA processing and degradation. Co-immunoprecipitation with antibodies directed against the previously predicted Sulfolobus solfataricus orthologue of the exosome subunit ribosomal-RNA-processing protein 41 (Rrp41) led to the purification of a 250-kDa protein complex from S. solfataricus. Approximately half of the complex cosediments with ribosomal subunits. It comprises four previously predicted orthologues of the core exosome subunits from yeast (Rrp41, Rrp42, Rrp4 and Csl4 (cep1 synthetic lethality 4; an RNA-binding protein and exosome subunit)), whereas other predicted subunits were not found. Surprisingly, the archaeal homologue of the bacterial DNA primase DnaG was tightly associated with the complex. This suggests an RNA-related function for the archaeal DnaG-like proteins. Comparison of experimental data from different organisms shows that the minimal core of the exosome consists of at least one phosphate-dependent ribonuclease PH homologue, and of Rrp4 and Csl4. Such a protein complex was probably present in the last common ancestor of Archaea and Eukarya.
Introduction
Aspects of RNA metabolism in bacteria and eukaryotes are well studied, but there is still only limited information about RNA processing in Archaea. RNA maturation and degradation are highly ordered processes that are phylogenetically well conserved (Anantharaman et al., 2002). In bacteria, as well as in Eukarya, large protein complexes participate in RNA maturation and decay. In bacteria, the endoribonuclease RNase E organizes a protein complex called the degradosome (Py et al., 1996; Jäger et al., 2001). Eukaryotic cells have a conserved RNA processing and degrading protein complex called the exosome, which comprises several 3′ to 5′ exoribonucleases, RNA-binding proteins, RNA helicases and associated protein factors (Mitchell et al., 1997; Allmang et al., 1999a). It has a central function in the maturation of ribosomal RNA, small nucleolar RNA (snoRNA) and small nuclear RNA (snRNA), as well as in messenger RNA decay (Allmang et al., 1999b; Jacobs et al., 1998). Six of the exosome core subunits have homology to the Escherichia coli phosphate-dependent ribonuclease PH (RNase PH), which is a phosphorolytic ribonuclease; these are ribosomal-RNA-processing protein 41 (Rrp41), Rrp42, Rrp43, Rrp45, Rrp46 and Mtr3. The other core subunits are the hydrolytic RNases Rrp4, Rrp40, and Rrp44, and Csl4 (cep1 synthetic lethality 4; an RNA-binding protein and exosome subunit; Aloy et al., 2002). Yeast, plant and human cells contain large exosomes (300–400 kDa), the compositions of which are similar, but not identical, to each other (Mitchell et al., 1997; Allmang et al., 1999a; Chekanova et al., 2000). In Trypanosoma brucei, which diverged early in eukaryotic evolution, only a subset of the orthologues of exosome subunits are found in a complex. It has been suggested that the exosome was present in primitive eukaryotes and became increasingly complex during subsequent evolution (Estevez et al., 2001).
By comparing the gene order in completely sequenced archaeal genomes, complemented by sequence-profile analysis, the existence of the archaeal counterpart of the eukaryotic exosome was proposed. In most archaeal genomes, including that of the hyperthermophile Sulfolobus solfataricus, orthologues of the exosome core subunits Rrp4, Rrp41 and Rrp42 were found to be encoded by adjacent open reading frames (ORFs), which are a central part of the so-called exosomal superoperon. In addition, other potential subunits of the archaeal exosome were predicted to be encoded by this superoperon and by smaller, partially conserved operons (Koonin et al., 2001).
To learn more about RNA processing and degradation in Archaea, we decided to investigate the predicted exosome of S. solfataricus. We found that only four out of of the ten proteins that have been proposed to represent subunits of the archaeal exosome are part of an exosome-like protein complex. This complex coprecipitates with other, Archaea-specific proteins, which may have unexpected functions in RNA metabolism. Our findings provide a foundation for future investigations of the machinery for RNA turnover in Archaea. In addition, they contribute to the understanding of the development of protein complexes during evolution.
Results
Sulfolobus solfataricus Rrp41 is found in a complex
S. solfataricus Rrp41, the putative exosome subunit, was purified as a His6-tagged protein under denaturing conditions and used for antibody production. Fractionation of a whole-cell extract was performed through a 10–30% glycerol gradient at 150 mM NaCl, and the presence of S. solfataricus Rrp41 was monitored by western blot analysis. S. solfataricus Rrp41 was not detected in monomeric form, but only in fractions with estimated molecular weights of 270–400 kDa, and in the pellet (data not shown). Fractionation of the cell extract through a 10–60% glycerol gradient at 150 mM NaCl followed by western blot analysis revealed two peaks (Fig. 1A). The first peak (Fig. 1A, fractions 5–7) corresponds to fractions with experimentally determined sedimentation coefficients that are similar to that of catalase. We assume that this peak represents an exosome-like protein complex of ∼250 kDa. The second peak, which contains an even higher amount of S. solfataricus Rrp41 (Fig. 1A, lanes 10–15) corresponds to fractions that contain ribosomal subunits (Fig. 1B, lanes 11–15). To exclude the possibility that S. solfataricus Rrp41 (and probably the S. solfataricus Rrp41-containing complex) binds non-specifically to ribosomal subunits, we performed 5–20% glycerol-gradient centrifugation under high-salt conditions (500 mM NaCl). A large amount of S. solfataricus Rrp41 was found in the pellet (Fig. 2, lane P). This suggests that the association with ribosomal subunits is specific. In addition, we detected the free S. solfataricus Rrp41-containing complex, which has a sedimentation coefficient similar to that of catalase (Fig. 2, lanes 6–9).
Figure 1.
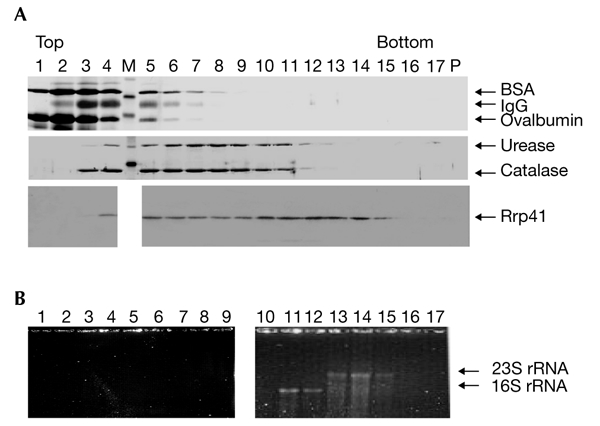
Sulfolobus solfataricus Rrp41 is a complex-bound protein. (A) Sulfolobus solfataricus lysate and marker proteins were fractionated in parallel through 10–60% glycerol gradients at 150 mM NaCl. Aliquots of each fraction were resolved by SDS–polyacrylamide gel electrophoresis. Rrp41 was detected by western blot analysis and marker proteins were detected by silver staining. Lanes 1–17 contain glycerol-gradient fractions. The proteins detected are indicated by arrows. (B) Analysis of the RNA content of the phenol/chloroform-extracted glycerol-gradient fractions in an ethidium-bromide-stained formaldehyde–agarose gel. The two panels are from the same gel. M, protein marker; P, pellet; Rrp41, ribosomal-RNA-processing protein 41.
Figure 2.
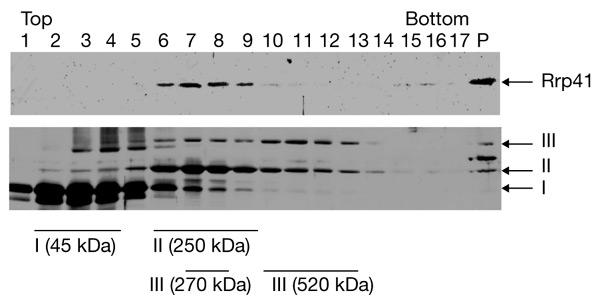
Sulfolobus solfataricus Rrp41 cosediments with ribosomal subunits under high-salt conditions. Sulfolobus solfataricus lysate and marker proteins were fractionated in parallel through 5–20% glycerol gradients in the presence of 500 mM NaCl. Aliquots of each fraction were resolved by SDS–polyacrylamide gel electrophoresis. Rrp41 was detected by western blot analysis and marker proteins were detected by silver staining. Lanes 1–17 contain glycerol-gradient fractions. The proteins detected are indicated by arrows. The peak fractions of each marker are indicated under the lower panel. P, pellet; Rrp41, ribosomal-RNA-processing protein 41. I, Ovalbumin; II, catalase; III, urease.
Analysis of the Sulfolobus solfataricus Rrp41 complex
Glycerol-gradient fractions (10–60% gradient) that contained the ribosome-free S. solfataricus Rrp41 complex were pooled and immunoprecipitation was performed using antibodies directed against His6-tagged S. solfataricus Rrp41. Six other proteins coprecipitated with Rrp41 (Fig. 3, lanes E1 and E2). Five of them also coprecipitated directly from cell-free lysate (Fig. 3, lane E3). The identity of S. solfataricus Rrp41 was confirmed by western blot analysis (Fig. 3, lane W). No other proteins cross-react with the antibodies. All polypeptides were unambiguously identified by mass spectrometry. Table 1 summarizes the sequence coverage of the proteins identified. To confirm these results, an additional tandem mass-spectrometric analysis was performed on one or two peptides of the peptide-mass fingerprints. As expected, the proposed subunits of the archaeal exosome, the S. solfataricus orthologues of Rrp4 and Rrp42, were part of the complex. The polypeptide of ∼20 kDa was identified as the orthologue of the core subunit of the eukaryotic exosome, Csl4. Csl4 from S. solfataricus was omitted in the work of Koonin et al. (2001). Using the National Center for Biotechnology Information database (NCBI Tools for Data Mining; http://www.ncbi.nlm.nih.gov/About/tools/index.html), protein–protein BLAST analysis and comparison to the Clusters of Orthologous Groups (COGs) database (http://www.ncbi.nlm.nih.gov/COG/), we found that the gene encoding S. solfataricus Csl4 is located in the same conserved genomic context as in other Archaea (data not shown; Koonin et al., 2001). In addition to orthologues of eukaryotic exosome subunits, the S. solfataricus DnaG homologue, the 60-kDa chaperonin (Cpn) of S. solfataricus, and the cell-division cycle 48 (Cdc48) homologue were coprecipitated (Table 1; Fig. 3).
Figure 3.
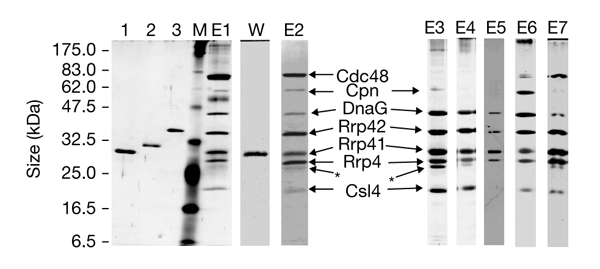
Composition of the Sulfolobus solfataricus exosome. Gels from SDS–polyacrylamide gel electrophoresis and a western blot, showing the proteins that copurify with Sulfolobus solfataricus ribosomal-RNA-processing protein 41 (Rrp41) during immunoprecipitation. At the left side of the panel, the size of the marker polypeptides is shown. The coprecipitated proteins are indicated by arrows. The asterisks indicate the Rrp4 degradation product. Lane 1, His6-tagged Rrp4; lane 2, His6-tagged Rrp41; lane 3, His6-tagged Rrp42; lane M, protein marker; lane W, western hybridization of lane E1 with antibodies against Rrp41; lanes E1–E7, independent purifications of the exosome (first elution fractions); lanes E1 and E2, coprecipitation from 10–60% glycerol-gradient fractions; lanes E3 and E4, coprecipitation directly from cell-free extract; lane E4, treatment with ribonucleases was performed before elution; E5, coprecipitation after partial purification on anion- and cation-exchange columns; lane E6, coprecipitation after anion-exchange and size-exclusion chromatography steps; lane E7, coprecipitation from 5–20% glycerol-gradient fractions 6–9 (see Fig. 2). Lane E2 shows Coomassie-blue-stained proteins. The proteins in lanes 1–3, M, E1 and E3–E7 are silver stained.
Table 1.
Sulfolobus solfataricus proteins identified by mass spectrometry
| Coprecipitated proteins | NCBI entry number | Amino-acid coverage (%) | Number of peptides confirmed by MS/MS |
|---|---|---|---|
| Rrp41 | 15897636 | 25 | 2 |
| Rrp42 | 15897635 | 23 | 2 |
| Rrp4 | 15897637 | 38 | 1 |
| Csl4* | 15897237 | 24 | 2 |
| DnaG | 15897045 | 31 | 2 |
| Cpn-α | 1174646 | 26 | 2 |
| Cpn-β | 135647 | 32 | 2 |
| Cdc48‡ | 15897351 | 15 | 2 |
*NCBI 15897237 shows 37% identity and 54% similarity to the Aeropyrum pernix 0445 protein, which is an orthologue of yeast Csl4 (cep1 synthetic lethality 4) and is a predicted subunit of the archaeal exosome (Koonin et al., 2001). NCBI 1174646 and 135647 correspond to the α- and β-subunits, respectively, of the 60-kDa chaperonin (thermophilic factor 55 (TF55) or Cpn; Knapp et al., 1994; Cerchia et al., 2000) of the closely related Sulfolobus shibatae, and show 90% and 95% identity, respectively, to the corresponding S. solfataricus polypeptides.
‡NCBI 15897351 was identified as a cell-division cycle 48 (Cdc48) homologue by BLAST analysis. NCBI, National Center for Biotechnology Information. MS/MS, tandem mass spectrometry; Rrp, ribosomal-RNA-processing proteins.
Control experiments were performed to check whether the coprecipitated proteins were in a complex. S. solfataricus Cpn interacted weakly with pre-immune serum (data not shown). None of the other proteins was precipitated when pre-immune serum was used (data not shown). In addition, to ensure that the proteins did not coprecipitate because of their interaction with an RNA substrate, RNase treatment was performed before elution. The four orthologues of core subunits of the eukaryotic exosome and the DnaG homologue remained in a complex (Fig. 3, lane E4).
To provide more evidence for the specific association of the coprecipitated proteins, the Rrp41 complex was enriched on an anion-exchange column. Half of the S. solfataricus Rrp41-containing fraction was applied to a cation-exchange column and immunoprecipitation was performed. S. solfataricus DnaG remained associated with S. solfataricus Rrp4, Rrp41 and Rrp42 in the exosome-like complex during the chromatography steps (Fig. 3A, lane E5). Only a small amount of protein was seen, and we cannot exclude the possibility that S. solfataricus Csl4 was also present in the complex. The second half of the S. solfataricus Rrp41-containing anion-exchange fraction was applied onto a size-exclusion chromatography column. All seven proteins were coprecipitated, although only a small amount of the Cdc48 homologue was present (Fig. 3, compare lanes E2 and E6). In addition, co-immunoprecipitation was performed using the fractions that contained the 250-kDa complex in the glycerol gradient at 500 mM NaCl (Fig. 2). Again, all seven proteins were coprecipitated, although Cpn was at the limit of detection (Fig. 3A, lane E7). Our data suggest that the Rrp4, Rrp41, Rrp42 and Csl4 orthologues, as well as the DnaG homologue, are tightly associated in a complex, and only part of the complex pool interacts with the Cdc48 homologue or Cpn.
Discussion
We have, for the first time, isolated and biochemically characterized an archaeal protein complex that is orthologous to the eukaryotic exosome. Its composition differs from the predictions made on the basis of genome-sequence comparisons (Koonin et al., 2001). In S. solfataricus, the complex comprises four previously predicted exosomal subunits (the orthologues of the yeast proteins Rrp4, Rrp41, Rrp42 and Csl4) and, unexpectedly, the DnaG homologue. In addition, the chaperonin Cpn, the Cdc48 homologue and ribosomal subunits were found to be associated with the complex.
The tight association of S. solfataricus DnaG with the exosome-like complex suggests that this protein is involved in RNA metabolism rather than in DNA metabolism. In bacteria, DnaG synthesizes the RNA primer during DNA replication. The bacterial primase is different from that of eukaryotes. Archaeal genomes encode apparent orthologues of the two subunits of the eukaryotic primase and also encode DnaG homologues (Aravind et al., 1998). The data available suggest that the DNA replication machinery in Archaea is a simplified version of the eukaryotic machinery (for a review, see Böhlke et al., 2002). The archaeal DnaG homologues form a unique, conserved group of proteins with a domain organization that differs from that of the bacterial primase. Their annotation is based on the Toprim domain, which is a conserved catalytic domain in topoisomerases, DnaG-type primases, OLD (overcoming lysogenization defect) family nucleases, and recombination R (RecR) proteins (Aravind et al., 1998). At the amino terminus, they have a conserved motif (motif VI) that is unique to superfamily II helicases (and is involved in DNA or RNA binding) and this family of archaeal DnaG-like proteins. At the carboxyl terminus, they have a highly conserved motif of unknown function, which is unique to the archaeal DnaG homologues (Aravind et al., 1998). The domain composition of S. solfataricus DnaG, its coprecipitation with the Rrp41 complex, and the lack of any other helicase in the complex suggests a possible function for S. solfataricus DnaG as an RNA helicase.
As well as its protein-folding ability (Cerchia et al., 2000), S. solfataricus Cpn also has specific RNA-binding and endoribonucleolytic properties, and participates in 16S rRNA maturation (Ruggero et al., 1998). Therefore, its coprecipitation with the S. solfataricus Rrp41 complex might reflect a functional interaction with the same substrates as those with which the complex interacts, such as 16S rRNA precursors, rather than non-specific binding to denatured proteins.
The interaction of the Cdc48 homologue with the complex seems to be strong (Fig. 3, lanes E6 and E7), although not all complex preparations contained this protein (Fig. 3, compare lanes E2 and E3). However, it has been noted that yeast Cdc48 has a chaperone-like activity and can be a source of non-specific binding in biochemical interaction experiments (Thoms, 2002).
The cosedimentation of the S. solfataricus Rrp41 complex with ribosomal subunits suggests that it might function in rRNA maturation. The function of the eukaryotic exosome in rRNA processing is well established (Allmang et al., 1999b). The significance of the cosedimentation of Cpn, the Cdc48 homolgue and ribosomal subunits with the exosome-like complex remains to be elucidated.
The S. solfataricus proteins Rrp41, Rrp42 and Rrp4 are present in apparently equimolar amounts in the 250-kDa protein complex, a smaller amount of S. solfataricus Csl4 is copurified, and the amount of S. solfataricus DnaG coprecipitated varies (Fig. 3). Similar to the yeast exosome (Aloy et al., 2002), a ring of six RNase PH homologues might consist of three S. solfataricus Rrp41 and three S. solfataricus Rrp42 polypeptides. Three S. solfataricus Rrp4 molecules might also be present in the complex as counterparts of the yeast hydrolytic RNases Rrp4, Rrp40 and Rrp44. The tenth core subunit of the S. solfataricus complex is Csl4, the counterpart of yeast Csl4. In addition, at least one S. solfataricus DnaG polypeptide seems to be present in the exosome-like complex. The combined molecular weight of the 11 polypeptides (∼300 kDa) is consistent with the experimentally determined size of the complex, which is ∼250 kDa. Cpn and the Cdc48 homologue probably interact with only part of the complex pool.
Counterparts of yeast Rrp4 and Csl4, as well as an RNase PH homologue (Rrp45), are also present in the small exosome of T. brucei (Estevez et al., 2001), suggesting an essential role of these proteins in the assembly and function of the protein complex. Our data imply that an exosome-like protein complex was present before the onset of the eukaryotic lineage. We propose that the 'minimal' exosome core consists of at least one RNase PH homologue and of Rrp4- and Csl4-like proteins.
Methods
Construction and purification of recombinant proteins.
The ORFs 6015740, 6015742 and 6015744, which encode S. solfataricus Rrp4, Rrp41 and Rrp42 (orthologues of the yeast exosome subunits Rrp4, Rrp41 and Rrp42; Koonin et al., 2001) were amplified (from the second to the last codon) and cloned into the pQE30 His6-tag vector. Overexpression in E. coli M15 (REP4) cells and purification using nickel–nitriloacetic-acid (Ni-NTA) agarose were performed in accordance with the manufacturer's instructions (Qiagen).
Purification of S. solfataricus Rrp41 complexes.
S. solfataricus strain P2 was grown as previously described (Evguenieva-Hackenberg et al., 2002). Cells (1.7 g) were harvested at an optical density (measured at a wavelength of 600 nm) of 0.5, resuspended in 6 ml of TMN buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% Nonidet P-40 (NP-40), 1 mM PMSF (phenylmethylsulphonyl fluoride; Allmang et al., 1999a) and sonicated. Cell debris was removed and 600 μl of the supernatant was layered on an 11-ml 10–60% (w/v) glycerol gradient containing TMN buffer. Alternatively, a 5–20% glycerol gradient containing 500 mM NaCl was used. Centrifugation was performed in a TH-641 rotor at 4 °C for 21.5 h at 26,600 r.p.m. (121,262g), and 630-μl fractions were collected. For calibration purposes, ovalbumin (45 kDa), BSA (66 kDa), IgG (150 kDa), catalase (250 kDa) and urease (a trimer of 270 kDa or a hexamer of 520 kDa) were centrifuged under identical conditions. The antibodies against His6–Rrp41 (raised in rabbits by Eurogentec) were directly coupled to protein-A–sepharose beads (Harlow & Lane, 1988) and incubated with glycerol-gradient fractions, cell lysate or protein fractions obtained after chromatographic enrichment of the Rrp41 complex. After washing with TMN buffer, proteins were eluted with 50 μl 0.1 M glycine, pH 1.8, and analysed by SDS–polyacrylamide gel electrophoresis. As a control, RNase A and RNase T treatment were performed before elution (Jäger et al., 2001).
Chromatography.
The FPLC (fast-performance liquid chromatography) System (Pharmacia) was used. S. solfataricus cells (7 g) were resuspended in buffer QA (10 mM Tris, pH 7.5, 5 mM MgCl2, 10% glycerol, 0.1 % NP-40, 1 mM PMSF) and sonicated. After ultracentrifugation at 100,000g, the supernatant was applied to an anion-exchange column (BioRad Econo-Pac Q-cartridge) equilibrated in buffer QA. Bound proteins were eluted with buffer QB (10 mM Tris, pH 7.5, 5 mM MgCl2, 10% glycerol, 2M NaCl). Fractions containing Rrp41 were eluted at NaCl concentrations between 300 and 600 mM. These fractions were pooled, diluted tenfold in buffer SA (10 mM HEPES, pH 7.5, 5 mM MgCl2, 10% glycerol, 0.1% NP-40) and applied to a cation-exchange column (BioRad Econo-Pac S-cartridge) equilibrated with buffer SA. Elution was performed with buffer SB (10 mM HEPES, pH 7.5, 5 mM MgCl2, 10% glycerol, 2M NaCl). The Rrp41-containing fraction was eluted at 400–500 mM NaCl. It was then diluted sevenfold in buffer TMN and used for co-immunoprecipitation. Size-exclusion chromatography was performed using the HiLoad Superdex 200 column (Pharmacia) and TMN buffer. The presence of Rrp41 was monitored by western blot analysis.
Protein identification.
Protein bands were excised from Coomassie-blue-stained SDS gels and were destained with alternating washing steps using 50 mM ammonium bicarbonate buffer and acetonitrile. Trypsin was added (at an enzyme:substrate ratio of ∼1:10) and incubation at 36 °C overnight was carried out. To elute peptides, gel pieces were incubated with acetonitrile and then with 10% formic acid. This step was then repeated once. Supernatants were collected and combined, and mass-spectrometric analysis was performed using a 4700 Proteomics Analyser (Applied Biosystems) equipped with an Nd-YAG (neodymium–yttrium aluminium garnet) laser that produces pulsed power at 355 nm at pulse rates of 200 Hz. A 0.5-μl aliquot of the sample was mixed on the target with 0.5 μl of the matrix solution (5 mg ml−1 α-cyano-4-hydroxycinnamic acid dissolved in 50% acetonitrile, 0.1% trifluoroacetic acid) and dried at 18 °C. Mass analysis was performed using positive reflector mode with a deflection cut-off range with a mass/charge ratio (m/z) of 800. One thousand laser shots were accumulated to produce a single spectrum. Subsequently, high-energy MALDI–TOF (matrix-assisted laser desorption/ionization time-of-flight)/TOF collision-induced dissociation (CID) spectra were recorded on selected ions from the same sample spot. The collision energy was 1 kV. Air was used as the collision gas, and the pressure, which was measured by the ionization gauge in the high-pressure region near the collision cell, was 2 × 10−8 Torr for tandem mass spectrometry (MS/MS) without collision gas and 1 × 10−6 Torr with collision gas. The peptide-mass fingerprint and the tandem mass spectra were submitted to a search of the NCBI protein NR data base using MASCOT (http://www.matrixscience.com).
RNA methods.
Fractions of glycerol gradients were extracted with phenol/chloroform and ethanol precipitated. The presence of RNA in the fractions was tested for by denaturing formaldehyde-gel electrophoresis and ethidium-bromide staining. As a marker for the migration of rRNA in the gel, total RNA isolated from S. solfataricus was loaded.
Acknowledgments
We thank F. Klein for help with immunoprecipitations, and S. Hundt, A. Balzer and S. Mair for help with cloning experiments. This work was supported by the Justus-Liebig-Universität, Giessen and Fonds der Chemischen Industrie.
References
- Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D. & Mitchell P. ( 1999a) The yeast exosome and human PM–Scl are related complexes of 3′ to 5′ exonucleases. Genes Dev., 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E. & Tollervey D. ( 1999b) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloy P., Ciccarelli F.D., Leutwein C., Gavin A.C., Superti-Furga G., Bork P., Bottcher B. & Russell R.B. ( 2002) A complex prediction: three-dimensional model of the yeast exosome. EMBO Rep., 3, 628–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V., Koonin E.V. & Aravind L. ( 2002) Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res., 30, 1427–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Leipe D.D. & Koonin E.V. ( 1998) Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res., 26, 4205–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhlke K., Pisani F.M., Rossi M. & Antranikian G. ( 2002) Archaeal DNA replication: spotlight on a rapidly moving field. Extremophiles, 6, 1–14. [DOI] [PubMed] [Google Scholar]
- Cerchia L., Rossi M. & Guagliardi A. ( 2000) An archaeal chaperonin-based reactor for renaturation of denatured proteins. Extremophiles, 4, 1–7. [DOI] [PubMed] [Google Scholar]
- Chekanova J.A., Shaw R.J., Wills M.A. & Belostotsky D.A. ( 2000) Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J. Biol. Chem., 275, 33158–33166. [DOI] [PubMed] [Google Scholar]
- Estevez A.M., Kempf T. & Clayton C. ( 2001) The exosome of Trypanosoma brucei. EMBO J., 20, 3831–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evguenieva-Hackenberg E., Schiltz E. & Klug G. ( 2002) Dehydrogenases from all three domains of life cleave RNA. J. Biol. Chem., 277, 46145–46150. [DOI] [PubMed] [Google Scholar]
- Harlow E. & Lane D. ( 1988) Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA. [Google Scholar]
- Jacobs J.S., Anderson A.R. & Parker R.P. ( 1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S., Fuhrmann O., Heck C., Hebermehl M., Schiltz E., Rauhut R. & Klug G. ( 2001) An mRNA degrading complex in Rhodobacter capsulatus. Nucleic Acids Res., 29, 4581–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S., Schmidt-Krey I., Hebert H., Bergman T., Jornvall H. & Ladenstein R. ( 1994) The molecular chaperonin TF55 from the thermophilic archaeon Sulfolobus solfataricus. A biochemical and structural characterization. J. Mol. Biol., 242, 397–407. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Wolf Y.I. & Aravind L. ( 2001) Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res., 11, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Shevchenko A., Mann M. & Tollervey D. ( 1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′–5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- Py B., Higgins C.F., Krisch H.M. & Carpousis A.J. ( 1996) A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature, 381, 169–172. [DOI] [PubMed] [Google Scholar]
- Ruggero D., Ciammaruconi A. & Londei P. ( 1998) The chaperonin of the archaeon Sulfolobus solfataricus is an RNA-binding protein that participates in ribosomal RNA processing. EMBO J., 17, 3471–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S. ( 2002) Cdc48 can distinguish between native and non-native proteins in the absence of cofactors. FEBS Lett., 520, 107–110. [DOI] [PubMed] [Google Scholar]


