Abstract
Eukaryotic transcriptional activators usually recognize short DNA motifs, which are not only located within promoter regions, but also scattered throughout the genome. Assuming that the function of activators at non-promoter regions is wasteful and perhaps harmful, one can ask whether such binding is somehow prevented or if transcription is blocked at a downstream step. Here, we show that the yeast transcriptional activator Gcn4 is associated in vivo with several non-promoter euchromatic sites. This association results in the recruitment of the SAGA (Spt3/Ada/Gcn5/acetyltransferase) complex and the consequent activity of the Gcn5 histone acetyltransferase. The functional recruitment of the Swi/Snf nucleosome-remodelling complex was also evident at sites located in positioned nucleosomes. We show that this assemblage of coactivator complexes is not productive because of the absence of core promoter elements, other than the TATA box, that are required for stable mediator recruitment.
Introduction
A long-standing question in the field of transcriptional activation is what prevents the potentially hazardous functioning of DNA-binding transcriptional activators outside promoter regions. This question arises from the fact that such proteins recognize and bind short DNA motifs that are found throughout the genome. For example, the yeast Gcn4 activator recognizes the consensus sequence RTGACTCAY, which can be identified at 166 sites in the yeast genome, of which 119 are located in regions outside promoters. There are two possible answers to this question: either the accessibility of activators is somehow restricted to promoter sites, or their function is blocked if binding occurs.
The accessibility of transcriptional regulators to non-promoter regions has been explored by both specific and whole-genome approaches. In one report, examination of accessibility at a small number of sites suggested that binding of activators might be favoured in promoter regions (Mai et al., 2000). By contrast, Gal4 occupancy was detected at consensus sites located within the open reading frame (ORF) of the ACC1 gene (Li & Johnston, 2001). In yeast, among studies analysing occupancy on a genome-wide scale, two have examined the recruitment of transcriptional regulators (Sbf, Mbf and Rap1) to both intergenic and coding regions (Iyer et al., 2001; Lieb et al., 2001). Although a preference for binding at promoter regions was again shown, recruitment was evident at several sites located within coding regions, and this fact was not commented on. Similarly, binding of Drosophila homeobox proteins at sites located in euchromatic coding regions has also been reported (Carr & Biggin, 1999). Finally, a study aiming to identify the genomic targets of human c-MYC reported the detection of binding at non-promoter sites, although this occurred at a much lower level than in yeast (Fernandez et al., 2003).
Given the results above, it seems that binding of transcriptional activators at non-promoter sites is not always precluded. What prevents functional consequences of such recruitment from occurring? We addressed this question in yeast, taking advantage of the extensive knowledge of the function of the Gcn4 transcriptional activator.
Results
We first asked whether Gcn4 can occupy intragenic regions containing the consensus Gcn4-response element (GCRE) in vivo. We selected such regions in genes that are either constitutively expressed or repressed under the growth conditions used. In addition, and to avoid background signal, we ensured that the selected regions did not contain any other, even degenerate, GCREs located within at least 10 kb from the consensus element. As well as these euchromatic sites, we also monitored occupancy at a consensus GCRE that is located close to the telomere of chromosome VI. As shown in Fig. 1A, a dose-dependent occupancy of Gcn4 was detected in the ORFs of the PHO8 (repressed) and FRE6 and POL2 (active) genes. By contrast, occupancy of Gcn4 was not detected at the telomeric site. For FRE6 and POL2, in which the GCRE is located well within the coding region, the extent of recruitment was similar to that measured in the GCRE-containing TRP3 promoter. Gcn4 occupancy of the PHO8 gene was extremely high, probably due to the location of the GCRE element at the start of the coding region, near to a naturally accessible promoter and, as shown below, at the linker region between two positioned nucleosomes. Nevertheless, this high level of recruitment prompted us to investigate any potential functional significance. We found that, irrespective of its level of expression, Gcn4 was not required for either repression or activation of the PHO8 gene, nor was it used for the expression of the KRE2 gene, which is located 900 bp upstream of PHO8 (Fig. 2).
Figure 1.
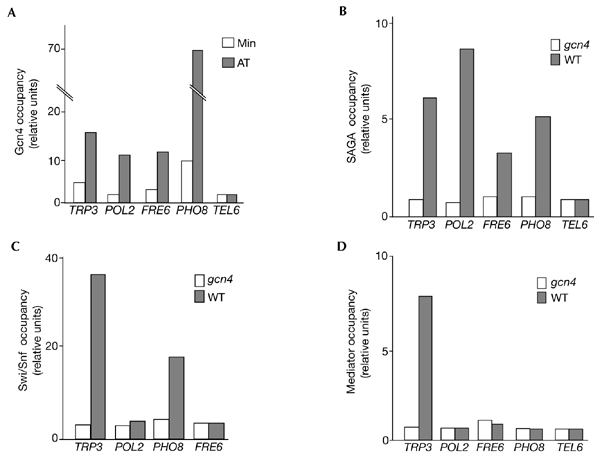
Recruitment of the Gcn4 transcriptional activator and coactivators at non-promoter chromosomal regions. (A) In vivo occupancy by Gcn4 of the TRP3 promoter and target sites located at the indicated regions and after growth in minimal (Min) or histidine starvation (AT) media. Levels shown on the y axis reflect the enrichment of the DNA relative to a region of the PHO5 open reading frame after correction for the ratios of amplification achieved using input DNA. (B) Occupancy of the same sites by the SAGA (Spt3/Ada/Gcn5/acetyltransferase) complex, monitored using a MYC-tagged Spt3 component, in relation to the presence (WT) or absence (gcn4) of Gcn4. (C) Occupancy of Swi/Snf in relation to the presence or absence of Gcn4, probed through a MYC-tagged Snf2 component. (D) Occupancy of the mediator complex in relation to the presence or absence of Gcn4, probed using a MYC-tagged Srb4 component. In the experiments presented in (B–D), strains were grown in conditions of histidine starvation.
Figure 2.
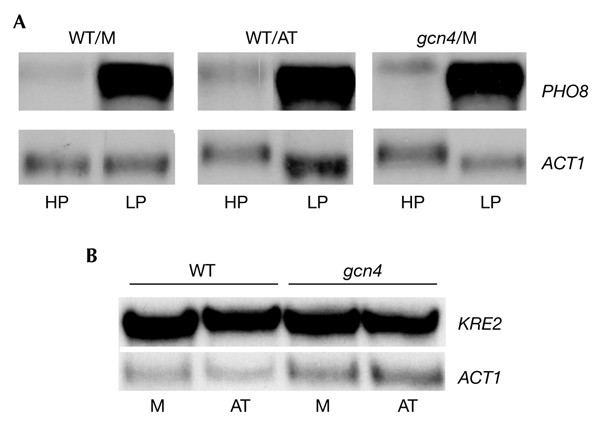
The high level of occupancy of the PHO8 site by Gcn4 does not contribute to the regulation of PHO8 expression, or that of the upstream KRE2 gene. (A) The messenger RNA levels of the PHO8 gene from cells grown in either high- or low-phosphate media (HP and LP, respectively) are not affected by the presence of either low (WT/M) or high (WT/AT) levels of Gcn4, or by the absence (gcn4) of this protein. (B) mRNA levels of the KRE2 gene in wild-type (WT) and gcn4 strains grown in either minimal (M) or histidine starvation (AT) media. Actin mRNA (ACT1) was used as loading control in both experiments.
Several transcriptional complexes are recruited by direct interactions with the Gcn4 activation domain (Natarajan et al., 1999; Brown et al., 2001; Neely et al., 2002). We therefore asked whether such complexes are recruited in addition to the activator at sites containing GCRE targets. We found that the SAGA (Spt3/Ada/Gcn5/acetyltransferase) complex was recruited in a Gcn4-dependent manner at all sites tested (Fig. 1B). Swi/Snf was recruited only at the PHO8 site (Fig. 1C), whereas the mediator complex did not stably occupy any of the sites (Fig. 1D). Moreover, the recruited SAGA and Swi/Snf complexes were functional with regard to their known activities. As shown in Fig. 3A, Gcn5-dependent histone H3 acetylation was detected in nucleosomes located in the region of the GCRE, and this acetylation was increased further when the amount of Gcn4 was elevated. Similarly, the positioned nucleosomes around the PHO8 site were remodelled in a Gcn4- and Snf2-dependent manner (Fig. 3B, left panel). In addition, we tested one of the sites that did not support Swi/Snf recruitment. Interestingly, no positioning or remodelling was detected for the POL2 site (Fig. 3B, right panel). Thus, the differences observed in Gcn5-dependent acetylation and Swi/Snf recruitment between the various non-promoter sites might be due to differences in chromatin organization, that is, positioned versus stochastic nucleosomes (see Discussion). We conclude that chromatin-modifying complexes, such as SAGA and Swi/Snf, can be recruited by Gcn4 at non-promoter sites. By contrast, stable recruitment of the mediator complex was not detected at any site, and this presumably blocks the assembly of pre-initiation complexes.
Figure 3.
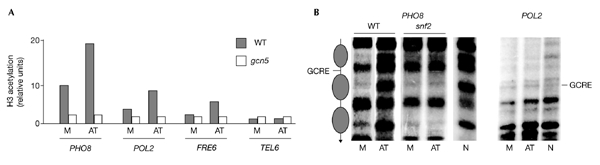
Functional consequences of coactivator recruitment. (A) Gcn5-dependent acetylation of nucleosomal histones located at the regions indicated was measured at low (M) or high (AT) Gcn4 concentrations by chromatin immunoprecipitation using antibodies raised against a diacetylated form of histone H3. Numbers indicate the enrichment of the DNA relative to a telomeric region (TEL) and corrected for the level of amplification achieved using input DNA. TEL (Vogelauer et al., 2000) was used as a control as it is heterochromatic and therefore under-acetylated. (B) The diagram in the left panel shows the mapped positions of nucleosomes around the Gcn4-response element (GCRE) site of the PHO8 gene. Gcn4-dependent remodelling of the nucleosomes positioned on either side of the GCRE located in the PHO8 open reading frame is shown in the left panel. Remodelling was monitored in wild-type (WT) or snf2 strains by determining the sensitivity to micrococcal nuclease and indirect end-labelling. M and AT indicate cells with low and high amounts of Gcn4, respectively. N is the micrococcal-nuclease cleavage pattern of naked DNA. By contrast, similar experiments for the POL2 site (right panel) showed the absence of positioned nucleosomes, as indicated by the identical patterns of micrococcal nuclease cleavages in nuclear (M and AT) or naked (N) DNA.
How generally applicable are these observations? Short of performing a whole-genome study, we attempted to generalize by engineering a non-promoter site by the systematic reduction of a promoter, an approach that aimed towards the definition of elements contributing to or preventing the formation of the above assemblages. Our starting point was a PHO5 promoter (Almer et al., 1986), in which a GCRE site was introduced at the linker region between nucleosomes −1 and −2 (Fig. 4A, top left panel). The use of this synthetic promoter has advantages over natural ones, as no factors other than Gcn4 and a single TATA element are required for its transcriptional activation. In addition, its nucleosomal organization provides an opportunity for the functional monitoring of chromatin remodelling complexes. It should be noted that under the conditions used, both the upstream activating sequence 1 (UAS1) and UAS2 elements were irrelevant, as all the results were identical in a pho4Δ strain. Occupancy of Gcn4 on this synthetic promoter resulted in the recruitment of the SAGA and mediator complexes (Fig. 4A, right panel), as well as that of Swi/Snf, as revealed by its requirement for remodelling of the −2 nucleosome (Fig. 4A, bottom left panel). Thus, all three complexes tested were recruited in a Gcn4-dependent manner to this promoter.
Figure 4.
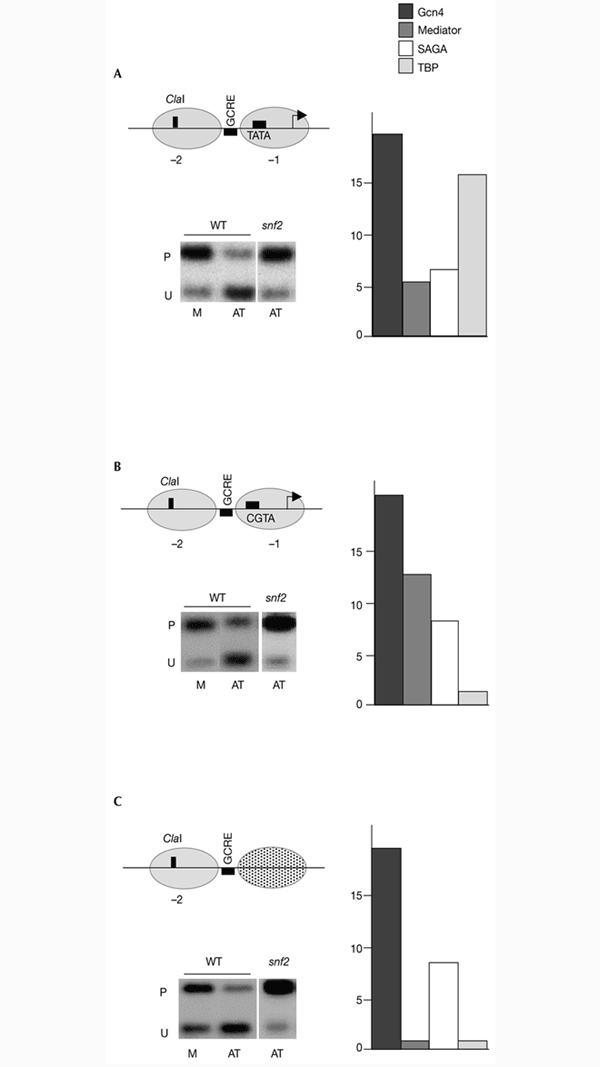
An engineered non-promoter site defines the requirements for coactivator recruitment. (A) The top left panel shows a representation of the PHO5 promoter with a Gcn4-response element (GCRE) site introduced into the linker between nucleosomes −2 and −1. Also indicated are the ClaI cleavage site used for monitoring nucleosome remodelling, the TATA box and the transcription initiation site (arrow). The right panel shows that in cells growing under histidine limitation (high levels of Gcn4), the wild-type promoter is occupied by Gcn4, the mediator complex (Srb4), SAGA (Spt3) and TATA-binding protein (TBP). The bottom left panel shows that Snf2 is required for remodelling of nucleosome −2. Remodelling was assayed by determining the accessibility to ClaI of cells containing low (M) or high (AT) amounts of Gcn4 in wild-type (WT) or snf2 strains. P indicates the protected DNA; U indicates one fragment that results from cleavage by the enzyme used and secondary digestions. (B) Mutating the TATA element (top left panel) affects only the recruitment of TBP. (C) The absence of all promoter elements and downstream open reading frame sequences (top left panel) allows the recruitment of SAGA.
It is reasonable to assume that the presence of TATA elements at a functional distance from the non-promoter sites is highly unlikely. On the basis of this, we investigated whether the recruitments described above are dependent on the presence of this element. This was done by introducing a two-base alteration that changed the TATA core to CGTA (Fig. 4B, top left panel). Although TATA-binding protein (TBP) occupancy was completely abolished, SAGA, the mediator complex and a functional Swi/Snf complex were recruited to this mutated promoter (Fig. 4B, right and bottom left panels). We conclude that the recruitment of coactivator complexes does not depend on the presence of TATA elements. Assuming that the presence of transcription start sites near to non-promoter targets is also unlikely, we transferred the region containing nucleosome −2 and the GCRE to the polylinker region of a yeast centromeric plasmid (Fig. 4C, top left panel) and monitored Gcn4-dependent recruitment. This region retained the ability to position nucleosomes (Fig. 4C, bottom left panel). Interestingly, occupancy by Gcn4 resulted in SAGA and Swi/Snf recruitment, but also in failure of the mediator complex to be positioned in this region (Fig. 4C, right and bottom left panels). Thus, an engineered non-promoter target confirmed that occupancy by Gcn4 allows the recruitment of SAGA and Swi/Snf but not of the mediator complex. We hypothesize that stable occupancy of the latter was not possible due to the lack of core promoter regions other than the TATA element.
Discussion
In this study, we examined the occupancy by Gcn4 of a set of DNA targets located outside promoter regions. Our first conclusion is that Gcn4 can occupy such binding sites located within euchromatic ORF coding regions. By contrast, recruitment was prohibited at a site located close to a telomere, presumably due to the spreading of heterochromatin. As stated previously, whole-genome studies in yeast have indicated that ∼20% of potential non-promoter sites are occupied by specific activators (Iyer et al., 2001; Lieb et al., 2001). In this study, although it was limited in the number of sites examined, we saw a bias towards recruitment, which probably reflects the fact that sites containing the optimum target were selected. Nevertheless, these results confirm that activators can occupy non-promoter targets, and provide tools to investigate the functional consequences of such recruitment.
A second key finding of this study is that, given this occupancy, the safeguard against unwanted transcriptional events is not the prohibition of functional recruitment of nucleosome-modifying complexes such as SAGA and Swi/Snf, but the lack of stable recruitment of the mediator complex. We came to this conclusion not only by examining recruitment at natural non-promoter sites, but also from the properties of an engineered site that was derived by sequential promoter subtractions. Thus, SAGA was always recruited, a result consistent with its reported interaction with the activation domains of Gcn4 and other proteins (Brown et al., 2001; Bryant & Ptashne, 2003), which is sufficient for recruitment independently of TBP and other core promoter elements (Bhaumik & Green, 2001; Larschan & Winston, 2001). Conversely, Swi/Snf recruitment correlated strongly with the presence of organized positioned nucleosomes. This could be explained by the reported weaker interaction of this complex with Gcn4 (Natarajan et al., 1999) and evidence from several studies that have shown that positioned and acetylated nucleosomes contribute to the stable recruitment of Swi/Snf (Hassan et al., 2001, 2002). Alternatively, and given the fact that FRE6 and POL2, but not PHO8, are expressed under the growth conditions used, it is likely that the presence of an elongating form of RNA polymerase II selectively destabilizes Swi/Snf. Finally, stable recruitment of the mediator complex was not evident in all instances. Given that this complex is required for the transcription of 93% of all yeast genes (Holstege et al., 1998), this should interrupt the further assembly of the transcription pre-initiation complex. Our results show that this complex failed to be stably recruited not because of the absence of nearby TATA elements, but rather because of the lack of other regions that probably define a transcription initiation site. We speculate that such regions might stabilize components of the pre-initiation complex, which in turn dock the mediator complex in place to facilitate successive rounds of transcription initiation/re-initiation (Yudkovsky et al., 2000). Such functional interplay of promoter elements should involve an aspect of directionality, as suggested by the results with the GCRE at the PHO8 ORF, which is located nearby, but to the left of this gene's transcription initiation site.
It is worth noting that although after activator binding at non-promoter sites transcription is ultimately prevented, the functional recruitment of chromatin-related coactivator complexes is not. Gcn5-dependent acetylation was clearly evident at all non-promoter sites, and such acetylation events might provide a simple mechanistic explanation for at least part of what is called global 'untargeted' Gcn5-dependent histone acetylation in yeast (Vogelauer et al., 2000). Consistent with this idea was the lack of such global effects on telomeres, where binding of transcription factors such as Gcn4 is precluded. If this assumption is correct then it is possible that the functional recruitment of Swi/Snf at PHO8, and presumably other loci, might be part of the as yet undescribed global nucleosome remodelling. It is interesting that the evolution of mechanisms of activated transcription necessitated tolerance to, or even benefited from, such global effects on chromatin.
Methods
Yeast strains and media.
The wild-type, gcn4, gcn5 and snf2 strains used were those described in Syntichaki et al. (2000). MYC epitope tags were constructed following the method of Knop et al. (1999). High levels of Gcn4-dependent transcription were achieved by adding 10 mM 3-AT to minimal media. Low-phosphate media were prepared in accordance with the method of Almer et al. (1986).
DNA constructs.
The gene based on PHO5 was constructed by inserting a PCR fragment containing PHO5 sequences from nucleotides −392 to +3 (which includes both UAS1 and UAS2, the TATA element, the transcription start site and the initiator Met codon), as described in Syntichaki et al. (2000). A doublestranded oligonucleotide containing the GCRE (5'-CGATGACTCATATGCAT-3'; the GCRE is underlined) was inserted into the BstEII site in the linker region between nucleosomes −1 and −2. The TATA-box mutation (CGTA) was obtained by site-directed mutagenesis. The mononucleosomal construct was derived by transferring the DNA region encompassing nucleosome −2 and the GCRE to the polylinker of vector pRS316 (which carries ARS1, CEN6 and URA3).
Gene expression analysis.
Total yeast RNA was isolated as described previously (Larschan & Winston, 2001). Approximately 5 μg of RNA was loaded onto a 1.5% agarose–formaldehyde gel and hybridized following standard protocols.
Chromatin immunoprecipitation.
Occupancies of transcription factors and complexes were assayed at the following genomic regions: codon 816 of the POL2 ORF (total length, 2,223 codons), codon 192 of the FRE6 ORF (total length, 713 codons), codon 2 of the PHO8 ORF (total length, 513 codons) and at a position 2,773 bp from the end of chromosome VI (TEL6). Chromatin was immunoprecipitated as described in Kuo & Allis (1999) using antisera against the MYC epitope (Santa Cruz) or against diacetylated H3 (K9 and K14; Upstate Biotechnology, Inc.). The recovered DNA was subjected to quantitative real-time PCR analysis. The first positions of the 18-nucleotide primers used were: −146 and +143 for TRP3, +2,324 and +2,579 for POL2, +457 and +711 for FRE6, +1 and +247 for PHO8, and 267,261 and 267,514 for TEL6. Quantification reflected the enrichment of the tested DNA relative to a region of PHO5 ORF (primer starting positions +1,017 and +1,220) or a telomeric region referred as TEL (Vogelauer et al., 2000) after correction for the ratios of amplification achieved using input DNA. Each chromatin immunoprecipitation was repeated at least three times and the variation between experiments was ±8–10%.
Nucleosome remodelling.
Remodelling of nucleosome −2 was assayed by a restriction-enzyme accessibility assay (Almer et al., 1986). Accessibility was monitored after secondary digestions with EcoRI/HindIII or PvuII/HindIII (for the mononucleosomal construct) and probing with the EcoRI–HindIII PHO5 fragment. Mapping and remodelling of nucleosomes at PHO8 and POL2 were monitored by micrococcal nuclease digestions, followed by indirect end-labelling. For PHO8, secondary digestion was with NdeI at position +681 and probing was with a fragment corresponding to positions +649 to +370. For POL2, secondary digestion was with EcoRV at position +3,640 and probing was with a fragment corresponding to positions +3,640 to +3,062.
Acknowledgments
We thank I. Talianidis for critical reading of the manuscript and the Yeast group for helpful suggestions. This work was funded by a PENED 2001 GSRT grant.
References
- Almer A., Rudolph H., Hinnen A. & Horz W. ( 1986) Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J., 5, 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S.R. & Green M.R. ( 2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev., 15, 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E., Howe L., Sousa K., Alley S.C., Carrozza M.J., Tan S. & Workman J.L. ( 2001) Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science, 292, 2333–2337. [DOI] [PubMed] [Google Scholar]
- Bryant G.O. & Ptashne M. ( 2003) Independent recruitment in vivo by gal4 of two complexes required for transcription. Mol. Cell, 11, 1301–1309. [DOI] [PubMed] [Google Scholar]
- Carr A. & Biggin M.D. ( 1999) A comparison of in vivo and in vitro DNA-binding specificities suggests a new model for homeoprotein DNA binding in Drosophila embryos. EMBO J., 18, 1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez P.C., Frank S.R., Wang L., Schroeder M., Liu S., Greene J., Cocito A. & Amati B. ( 2003) Genomic targets of the human c-Myc protein. Genes Dev., 17, 1115–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.H., Neely K.E. & Workman J.L. ( 2001) Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell, 104, 817–827. [DOI] [PubMed] [Google Scholar]
- Hassan A.H., Prochasson P., Neely K.E., Galasinski S.C., Chandy M., Carrozza M.J. & Workman J.L. ( 2002) Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell, 111, 369–379. [DOI] [PubMed] [Google Scholar]
- Holstege F.C., Jennings E.G., Wyrick J.J., Lee T.I., Hengartner C.J., Green M.R., Golub T.R., Lander E.S. & Young R.A. ( 1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Iyer V.R., Horak C.E., Scafe C.S., Botstein D., Snyder M. & Brown P.O. ( 2001) Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature, 409, 533–538. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K. & Schiebel E. ( 1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Kuo M.H. & Allis C.D. ( 1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods, 19, 425–433. [DOI] [PubMed] [Google Scholar]
- Larschan E. & Winston F. ( 2001) The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev., 15, 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. & Johnston S.A. ( 2001) Are all DNA binding and transcription regulation by an activator physiologically relevant? Mol. Cell. Biol., 21, 2467–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb J.D., Liu X., Botstein D. & Brown P.O. ( 2001) Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nature Genet., 28, 327–334. [DOI] [PubMed] [Google Scholar]
- Mai X., Chou S. & Struhl K. ( 2000) Preferential accessibility of the yeast his3 promoter is determined by a general property of the DNA sequence, not by specific elements. Mol. Cell. Biol., 20, 6668–6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K., Jackson B.M., Zhou H., Winston F. & Hinnebusch A.G. ( 1999) Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell, 4, 657–664. [DOI] [PubMed] [Google Scholar]
- Neely K.E., Hassan A.H., Brown C.E., Howe L. & Workman J.L. ( 2002) Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol., 22, 1615–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P., Topalidou I. & Thireos G. ( 2000) The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature, 404, 414–417. [DOI] [PubMed] [Google Scholar]
- Vogelauer M., Wu J., Suka N. & Grunstein M. ( 2000) Global histone acetylation and deacetylation in yeast. Nature, 408, 495–498. [DOI] [PubMed] [Google Scholar]
- Yudkovsky N., Ranish J.A. & Hahn S. ( 2000) A transcription reinitiation intermediate that is stabilized by activator. Nature, 408, 225–229. [DOI] [PubMed] [Google Scholar]


