Abstract
RNA-mediated control can evolve far more rapidly than mechanisms that rely on proteins, creating selective advantages in adaptive gene regulation. Recently, evidence has emerged that messenger RNA is a source of cis-acting RNA elements that sense external signals and thereby regulate gene expression. With exquisite specificity, metabolite-sensing riboswitches control the formation or translation of prokaryotic mRNA. In eukaryotes, RNA sensors in human antiviral cytokine genes that encode tumour necrosis factor-α (TNF-α) and interferon-γ (IFN-γ) have been shown to activate strongly the RNA-dependent protein kinase PKR, a stress kinase that is also activated by double-stranded RNA—a hallmark of viral infection. These cis-acting RNA elements in the TNF-α and IFN-γ transcripts function as sensors of intracellular PKR levels and regulate gene expression at the level of mRNA splicing and translation, respectively. Although RNA sensors in bacteria may be remnants of an ancient RNA world, it is likely that they form an integral part of higher eukaryotic genomes as well.
Introduction
Noncoding RNAs can control gene expression by selectively targeting messenger RNA through RNA interference, a process that is based on pairing between specific, short RNA sequences (for a review, see Hannon, 2002). Such RNA-mediated control can evolve far more rapidly than mechanisms that rely on proteins, thus creating selective advantages in adaptive gene regulation. Recently, evidence has emerged that mRNA can harbour cis-acting RNA elements that are powerful regulators of expression. In both prokaryotic and eukaryotic genes, these elements act as sensors of external signals through mechanisms of molecular recognition that go beyond simple base pairing. These RNA sensors function within the cell to sense nutrients, temperature and stress.
Riboswitches
The plasticity of nucleic acids in molecular recognition is shown well by metabolite-sensing elements within RNA, known as riboswitches. Before the discovery of natural riboswitches, RNA oligonucleotides selected in vitro were found to fold into unique shapes that permit recognition of a target (for a review, see Wilson & Szostak, 1999), such as a particular coenzyme (Jadhav & Yarus, 2002). RNA aptamers were suggested to exist in vivo after genetic analysis identified conserved mRNA elements that were essential for feedback control, for example, in vitamin B12 and riboflavin synthesis (for a review, see Stormo & Ji, 2001). Indeed, aptamers selected in vitro for their ability to bind to aminoglycoside antibiotics or a dye can control translation in vivo when inserted into the 5′-untranslated region (UTR) of a reporter gene (Werstuck & Green, 1998). Phage phi29 encodes a 120-nucleotide (nt) packaging RNA (pRNA) that binds ATP tightly through a sequence that closely resembles an ATP aptamer selected in vitro (Shu & Guo, 2003).
During the past two years, natural riboswitches have been identified (for a review, see Lai, 2003). Recent studies have shown that riboswitches undergo a conformational change on the direct binding of a ligand and that these elements are used ubiquitously in prokaryotes to control the generation and translation of mRNA. For example, riboswitches that sense vitamin B12, thiamine pyrophosphate (TPP), flavin mononucleotide (FMN), S-adenosylmethionine (SAM), lysine, guanine or adenine are predicted to regulate 68 genes of Bacillus subtilis, and comprise nearly 2% of its genome (Mandal et al., 2003). Clearly, RNA has the ability to control fundamental biochemical pathways through its exquisite specificity and affinity for metabolites and coenzymes.
The simplest form of an RNA sensor could be the thermosensor that renders Listeria monocytogenes virulent at 37 °C. At 30 °C, the Shine–Dalgarno (SD) sequence in the mRNA that encodes a virulence-activating transcription factor is masked, because it pairs with an upstream sequence in the 5′-UTR; at 37 °C the helix opens up to permit translation (Johansson et al., 2002). A similar RNA-based thermosensor is predicted to function in Rhizobium (Nocker et al., 2001).
Generally, riboswitches are bipartite in nature, containing an aptamer module that is a specific sensor and an expression module that transforms the change in RNA structure, as a result of ligand binding, into genetic control. This partition facilitates the evolution of aptamers independent of the expression module used to effect control. For example, when TPP binds to its 'thi-box' aptamer in the thiM 5′-UTR, this releases a 4-nt strand that can then anneal with the SD sequence and block translation (Winkler et al., 2002a). Similar results have been obtained with an Escherichia coli B12 riboswitch, in which B12 binding precludes ribosome binding (Nou & Kadner, 2000; Nahvi et al., 2002). The FMN aptamer in the riboflavin rib RNA 5′ leader forms an anti-terminator by annealing with a downstream sequence; when FMN binds, base pairing is precluded, and this results in premature termination (Winkler et al., 2002b; Mironov et al., 2002). Similarly, the SAM riboswitch abolishes an anti-terminator on binding of its ligand (McDaniel et al., 2003; Epshtein et al., 2003). Premature termination is also activated by the thi-box (Mironov et al., 2002).
Because of their more complex function in controlling gene expression, natural riboswitches are longer (up to 200 nt) than aptamers selected in vitro solely on the basis of their ligand binding affinity, which typically are <40 nt. However, the specificity of natural aptamers for their ligands is generally superior to that of aptamers selected in vitro. This allows riboswitches to discriminate between closely related metabolites. Thus, FMN and TPP riboswitches discriminate against the unphosphorylated precursors riboflavin and thiamine, respectively, by 1,000-fold (Mandal et al., 2003). The natural G (guanine) aptamer has an affinity of about 5 nM for G; changes to virtually any atom of this ligand lead to a drastic reduction in affinity, from tenfold for xanthine to >104-fold for 2-aminopurine. This implies that the aptamer must contact almost every atom in G. Notably, a single C to U change in the G-box core converts the guanine riboswitch into an adenine riboswitch (Mandal et al., 2003). This indicates that riboswitches can evolve rapidly to meet new needs.
RNA sensors in cytokine genes
The RNA-dependent protein kinase PKR is a major mediator of antiviral and inflammatory responses. This interferon (IFN)- and tumour necrosis factor (TNF)-inducible serine/threonine stress kinase is activated in the presence of double-stranded RNA (dsRNA), a hallmark of viral infection (Ben-Asouli et al., 2002). Once activated, PKR potently inhibits translation by phosphorylating the α-chain of eukaryotic initiation factor eIF2, which binds tRNAiMet in a GTP-dependent manner. Phospho-eIF2α sequesters eIF2B, the GDP/GTP exchange factor that is limiting in the cell but crucial for the recycling of eIF2 between successive rounds of initiation; thus, the phosphorylation of 10–20% of eIF2α is sufficient to block translation (Hinnebusch, 2000). This renders translation highly sensitive to inhibition by the activation of PKR. All cells express basal levels of PKR and use it to regulate their growth not only in response to stress but also under normal conditions.
PKR remains latent unless exposed to RNA, dsRNA being a powerful activator. Cells generally lack dsRNA but it is generated during the replication of RNA and DNA viruses. Activation of PKR by viral dsRNA leads to the shutdown of protein synthesis, resulting in a block of viral replication and selective apoptosis of the infected cell (Stark et al., 1998). The antiviral action of IFNs and TNF is based to a major extent on their ability to induce PKR to high levels, enabling a prompt shutoff of translation when dsRNA appears after infection.
PKR contains tandem dsRNA-binding motifs (dsRBMs). In the cell, most PKR is inactive because dsRBM2 interacts with the kinase domain and masks the active site. When a single molecule of dsRNA binds to both dsRBMs, the protein conformation relaxes, and the ATP binding and dimerization domains are revealed. RNA duplexes of at least 11–13 bp are needed for this to occur but their nucleotide sequence is not crucial (Bevilacqua & Cech, 1996). Once ATP is bound, the PKR dimer undergoes the trans-autophosphorylation necessary for its own activation.
Natural regulators of PKR have been discovered recently in the mRNAs that encode TNF-α and IFN-γ. These cis-acting RNA elements activate PKR even more potently than does dsRNA and function as sensors that respond to the level of PKR in the cell (Osman et al., 1999; Ben-Asouli et al., 2002). By locally activating PKR, these RNA sensors regulate TNF-α and IFN-γ gene expression at the level of mRNA splicing and translation, respectively. TNF-α pre-mRNA and IFN-γ mRNA mimic viral RNA as activators of PKR and may couple their regulated expression to the phosphorylation of eIF2α, a common outcome of different stress responses. Importantly, these novel RNA sensors respond to changes in intracellular levels of PKR that in turn fluctuate in response to the inflammatory microenvironment of the cell.
A positive feedback loop in mRNA splicing
A cis-acting element in the human TNF-α 3′-UTR renders the splicing of TNF-α mRNA not only dependent on the activation of PKR but also very efficient (Osman et al., 1999). Thus, TNF-α mRNA is spliced exceptionally well once PKR is activated, which fits with the key role of TNF-α in preventing autoimmunity (Via et al., 2001). When this cis-acting RNA element (2-aminopurine response element, 2-APRE) is present, mRNA splicing becomes sensitive to inhibition by 2-aminopurine, a PKR inhibitor, and to the expression of dominant-negative mutant PKR. Located upstream of the AU-rich instability element, the 104-nt 2-APRE contains a 5′-proximal stem–loop stabilized by 17 base pairs (Fig. 1). The apical region of the stem and the loop are phylogenetically conserved. 2-APRE RNA activates PKR more strongly than dsRNA, inducing eIF2α phosphorylation (Osman et al., 1999). Not only is the activation of PKR required for the splicing of precursor transcripts that contain the 2-APRE, but increased expression of PKR enhances their splicing by as much as 20-fold. By contrast, although it is closely related and homologous to the TNF-α gene, the TNF-β gene lacks this element. However, insertion of the 2-APRE into the TNF-β 3′-UTR leads to the requirement of PKR activation for TNF-β mRNA processing and concomitantly, to more efficient splicing (Osman et al., 1999).
Figure 1.
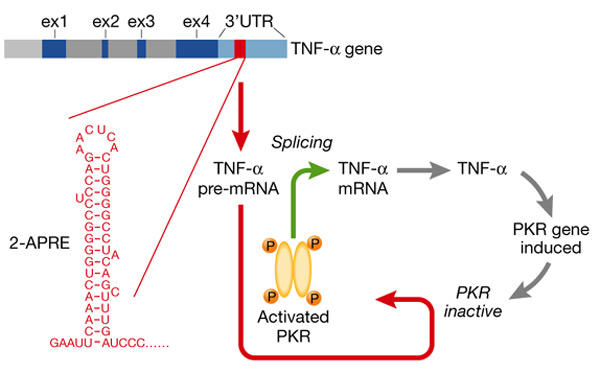
Human TNF-α pre-mRNA activates PKR through its 2-APRE sensor to enable splicing. The 2-aminopurine response element (2-APRE) in the tumour necrosis factor-α (TNF-α) 3′-untranslated region (UTR; of which the 5′ stem–loop is shown in red) acts as a strong local activator of RNA-dependent protein kinase PKR, allowing messenger RNA splicing to proceed and rendering it highly efficient. When the 2-APRE is prevented from activating PKR, splicing is blocked. Translation yields TNF-α, a secreted inflammatory mediator that can induce expression of more PKR in the cell, creating a positive feedback loop.
Because pre-mRNA that contains a 2-APRE is spliced even more efficiently when PKR levels rise, the induction of PKR by TNF-α or other signals during an inflammatory response creates a positive feedback loop (Fig. 1). The rate of TNF-α mRNA splicing is increased, mediated by the 2-APRE acting as a sensor for PKR. This could explain why TNF-α mRNA is expressed rapidly on immune stimulation (Jarrous & Kaempfer, 1994), whereas TNF-β mRNA is spliced sluggishly and expressed late (Osman et al., 1999). Although it promotes mRNA splicing by activating PKR, the 2-APRE does not reduce translation (Osman et al., 1999). In the cytoplasm, proteins may mask the 3′-UTR element, or induce refolding of its structure, to preclude PKR activation.
The highly efficient splicing of TNF-α mRNA mediated by the 2-APRE is lost after the deletion of this element, whereupon splicing still occurs but at a much slower rate (Osman et al., 1999). Tight control by the 2-APRE implies that splicing only commences after this 3′-UTR element has been transcribed. Although splicing is mainly co-transcriptional (Neugebauer & Roth, 1997), the small size of the TNF-α gene (3.5 kb) favours the rapid completion of primary transcripts before splicing begins. The substrate of PKR in the splicing reaction remains to be identified. Both PKR (Jeffrey et al., 1995) and phospho-eIF2α (Kedersha et al., 2002) are abundant in the nucleus, but whether eIF2 has a role in splicing is not known. The activation of PKR controls splicing at all TNF-α introns (Jarrous et al., 1996) and may promote an early stage of spliceosome assembly.
A negative feedback loop in mRNA translation
IFN-γ is essential for protective immunity but when expressed in excess can induce autoimmune disease (Gerez et al., 1997) and even toxic shock (Arad et al., 2000). The human IFN-γ gene uses a novel strategy to attenuate its own expression. Through a pseudoknot in its 5′-UTR, IFN-γ mRNA activates PKR, which induces eIF2α phosphorylation and inhibits its translation. This is the first example of an mRNA that regulates its own translation by activating PKR and of PKR activation by an RNA pseudoknot (Ben-Asouli et al., 2002).
Translationally controlled tumour protein (P23/TCTP) mRNA may also reduce its translation by activating PKR (Bommer et al., 2002), but an RNA structure responsible for this activation has not been identified. RNA fragments from the 3′-UTR of cytoskeletal muscle mRNA (Davis & Watson, 1996; Nussbaum et al., 2002) and human immunodeficiency virus-1 trans-acting responsive (TAR) region (Edery et al., 1989; Maitra et al., 1994) induce autophosphorylation of PKR in vitro and can inhibit translation in reticulocyte lysate in trans. However, it remains to be shown whether PKR regulates translation of the corresponding mRNAs in vivo.
In IFN-γ mRNA, a type H pseudoknot, stabilized by a stem of at least 5 bp, is essential for PKR activation (Fig. 2A; Ben-Asouli et al., 2002). Mutations that impair pseudoknot formation abolish its ability to activate PKR and greatly increase the translation of IFN-γ mRNA, whereas compensatory mutations in the loop that restore pseudoknot structure activate PKR and translational repression. Correspondingly, a nonphosphorylatable mutant eIF2α, the deletion of the PKR gene and PKR inhibitors 2-aminopurine, the vaccinia E3L protein or a dominant-negative mutant PKR, enhance the translation of IFN-γ mRNA. Deletion or mutation of the first 14 nt of the IFN-γ 5′-UTR during evolution would have sufficed to eliminate the pseudoknot (Fig. 2A) and to enhance translation by as much as 30-fold. However, the pseudoknot is phylogenetically conserved, attesting to its crucial role in reducing IFN-γ mRNA translation to only a fraction of its full potential.
Figure 2.
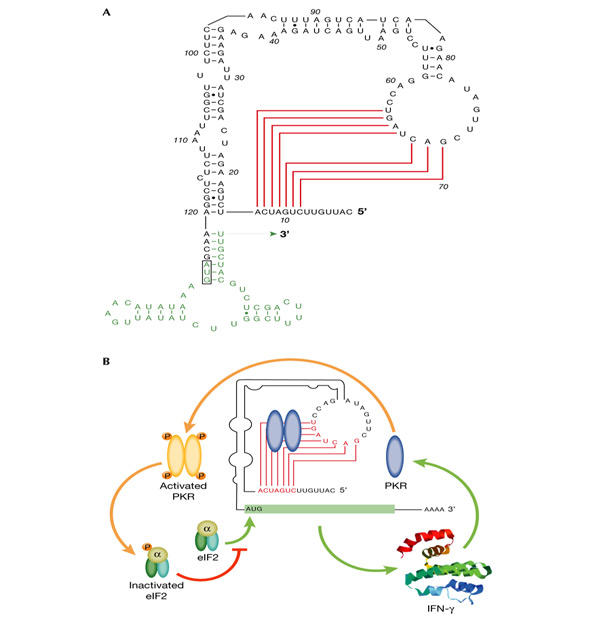
Human IFN-γ mRNA activates PKR through a pseudoknot sensor to inhibit its own translation. (A) Pseudoknot in the 5′-untranslated region of human interferon-γ (IFN-γ) messenger RNA. Base pairing in the pseudoknot stem is shown in red and the start of the open reading frame in green. (B) Autoregulation of IFN-γ synthesis. IFN-γ mRNA translation, which is dependent on eukaryotic initiation factor 2 (eIF2), yields IFN-γ that can induce more RNA-dependent protein kinase (PKR) expression in the cell, creating a negative feedback loop. Inactive PKR binds to the pseudoknot, leading to a local PKR activation. Resulting phosphorylation of eIF2α blocks further translation of IFN-γ mRNA.
During the immune response, as IFN-γ and other inflammatory cytokines build up in the microenvironment of the cell, they induce higher levels of PKR and thus the pseudoknot activates PKR more extensively. With the resulting phosphorylation of eIF2α, a negative feedback loop is created and the production of IFN-γ is progressively attenuated (Fig. 2B). By contrast, overproduction of IFN-α or IFN-β is less directly associated with pathology and a pseudoknot motif that could mediate translational repression is not found in their 5′-UTRs (Ben-Asouli et al., 2002).
The IFN-γ pseudoknot activates PKR in the vicinity of IFN-γ mRNA, and gives rise to a localized inhibition of translation rather than a global effect. Translation of endogenous or foreign mRNA is insensitive to the nature of the IFN-γ mRNA co-expressed in the cell, whether it is wild type or mutated in the pseudoknot (Ben-Asouli et al., 2002). Hence, the pseudoknot in IFN-γ mRNA is a cis-acting element that regulates only its own translation through the local activation of PKR.
How can a pseudoknot be such a strong PKR activator? The kinase will bind to dsRNA at least 11–13 bp in length (Bevilacqua & Cech, 1996), and longer helices (85 bp) are required for maximal activation (Manche et al., 1992). However, the IFN-γ pseudoknot contains only a short helix motif in which single nucleotide changes can abolish its ability to activate PKR (Ben-Asouli et al., 2002). Folding of the pseudoknot may prevent helix relaxation that can occur near the ends of linear dsRNA and thus create a more stable helix for productive interaction with PKR. The pseudoknot stem is too short to provide the minimal 12 bp needed for binding to both dsRBMs in PKR. However, non-contiguous short RNA helices can cooperate in binding and activating PKR (Bevilacqua et al., 1998). Stacking of the pseudoknot stem with an adjacent helix (Fig. 2A) will extend the helical domain to a length that is sufficient to interact with both dsRBMs and to activate PKR.
IFN-γ mRNA is the first non-viral mammalian mRNA species in which a pseudoknot has been shown (http://wwwbio.leidenuniv.nl/~batenburg/PKBGet.html#s8). As the 5′-UTR of IFN-γ mRNA functions in translation by interacting with initiation factors and ribosomal subunits, dynamic and reversible changes in the pseudoknot structure will affect its ability to activate PKR, thus rendering it a sensor that responds to ribosomes as well as to stress conditions.
Perspectives
The low number of genes identified within mammalian genomes shifts the weight of controlling their expression towards the genes themselves. The discovery of RNA sensors within exonic portions of human inflammatory cytokine genes significantly expands the mechanisms within this regulatory repertoire. These findings show that RNA sensors are found not only in bacteria where conceivably they are remnants of an ancient RNA world, but also in higher eukaryotes where they probably form an integral part of their modern genomes.
Sequences homologous to the consensus bacterial TPP-dependent riboswitch occur in lower eukaryotes and plants, such as in the 3′-UTR of the ThiC gene in Arabidopsis and in a putative ThiC gene of bluegrass and rice, as well as in the first introns of the thiamine biosynthetic nmt-1 gene in Neurospora and of a putative ThiC gene in Fusarium (Sudarsan et al., 2003). Whether such sequences function as riboswitches needs to be shown but these findings suggest that introns could also serve as a source of RNA sensors.
In eukaryotes, the concept that cellular genes encode RNA elements that regulate mRNA splicing and translation by activating PKR is new. This opens a line of investigation into the nature of RNA structures that regulate this kinase and their role as sensors of cellular stress and of the inflammatory microenvironment of the cell. An intriguing question is why the genes for TNF-α and IFN-γ, a synergistic pair of immunomodulators crucial for cellular immunity and antiviral responses, activate PKR for their control, a strategy usually associated with viral replication. Rather than these cytokine genes mimicking viruses, it is tempting to speculate that viruses may have borrowed the ability to activate PKR from cellular genes. Beyond inflammatory cytokine genes, natural RNA regulators of PKR may be found more widely in those transcripts whose highly efficient splicing or tightly controlled translation pose selective advantages.
Use of RNA sensors as ligand-dependent switches in prokaryotes, and more broadly as regulators of eukaryotic mRNA formation and function, provides an enormous potential for controlling gene expression, given the exquisite sensitivity and specificity of RNA in molecular sensing. Such RNA elements could also provide a potent new means for controlling gene expression by design and they may constitute a new class of drug targets, for example, for antimicrobial or anti-inflammatory agents.

References
- Arad G., Levy R., Hillman D. & Kaempfer R. ( 2000) Superantigen antagonist protects against lethal shock and defines a new domain for T-cell activation. Nature Med., 6, 414–421. [DOI] [PubMed] [Google Scholar]
- Ben-Asouli Y., Banai Y., Pel-Or Y., Shir A. & Kaempfer R. ( 2002) Human interferon-γ mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell, 108, 221–232. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P.C. & Cech T.R. ( 1996) Minor-groove recognition of double stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry, 35, 9983–9994. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P.C., George C.X., Samuel C.E. & Cech T.R. ( 1998) Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry, 37, 6303–6316. [DOI] [PubMed] [Google Scholar]
- Bommer U.A., Borovjagin A.V., Greagg M.A., Jeffrey I.W., Russell P., Laing K.G., Lee M. & Clemens M.J. ( 2002) The mRNA of the translationally controlled tumor protein P23/TCTP is a highly structured RNA, which activates the dsRNA-dependent protein kinase PKR. RNA, 4, 478–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S. & Watson J.C. ( 1996) In vitro activation of the interferon-induced, double-stranded RNA-dependent protein kinase PKR by RNA from the 3′ untranslated regions of human α-tropomyosin. Proc. Natl Acad. Sci. USA, 93, 508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I., Petryshyn R. & Sonenberg N. ( 1989) Activation of double-stranded RNA-dependent kinase (dsI) by the TAR region of HIV-1 mRNA: a novel translational control mechanism. Cell, 56, 303–312. [DOI] [PubMed] [Google Scholar]
- Epshtein V., Mironov A.S. & Nudler E. ( 2003) The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl Acad. Sci. USA, 100, 5052–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerez L., Shkolnik T., Hirschmann O., Lorber M., Arad G. & Kaempfer R. ( 1997) Hyperinducible expression of the interferon-γ (IFN-γ) gene and its suppression in systemic lupus erythematosus (SLE). Clin. Exp. Immunol., 109, 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G.J. ( 2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. ( 2000) in Translational Control of Gene Expression (eds Sonenberg, N., Hershey, J.W.B. & Mathews, M.B.) 185–243. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA. [Google Scholar]
- Jadhav V.R. & Yarus M. ( 2002) Coenzymes as coribozymes. Biochimie, 84, 877–888. [DOI] [PubMed] [Google Scholar]
- Jarrous N. & Kaempfer R. ( 1994) Induction of human interleukin-1 gene expression by retinoic acid and its regulation at processing of precursor transcripts. J. Biol. Chem., 269, 23141–23149. [PubMed] [Google Scholar]
- Jarrous N., Osman F. & Kaempfer R. ( 1996) 2-Aminopurine selectively inhibits splicing of tumor necrosis factor-α mRNA. Mol. Cell. Biol., 16, 2814–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey I.W., Kadereit S., Meurs E.F., Metzger T., Bachmann M., Schwemmle M., Hovanessian A.G. & Clemens M.J. ( 1995) Nuclear localization of the interferon-inducible protein kinase PKR in human cells and transfected mouse cells. Exp. Cell. Res., 218, 17–27. [DOI] [PubMed] [Google Scholar]
- Johansson J., Mandin P., Renzoni A., Chiaruttini C., Springer M. & Cossart P. ( 2002) An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell, 110, 551–561. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J. & Anderson P. ( 2002) Evidence that ternary complex (eIF2–GTP–tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell, 13, 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.C. ( 2003) RNA sensors and riboswitches: self-regulating messages. Curr. Biol., 13, R285–291. [DOI] [PubMed] [Google Scholar]
- Maitra R.K., McMillan N.A., Desai S., McSwiggen J., Hovanessian A.G., Sen G., Williams B.R. & Silverman R.H. ( 1994) HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology, 204, 823–827. [DOI] [PubMed] [Google Scholar]
- Manche L., Green S.R., Schmedt C. & Mathews M.B. ( 1992) Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol., 12, 5238–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., Boese B., Barrick J.E., Winkler W.C. & Breaker R.R. ( 2003) Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell, 113, 577–586. [DOI] [PubMed] [Google Scholar]
- McDaniel B.A.M., Grundy F.J., Artsimovitch I. & Henkin T.M. ( 2003) Transcription termination control of the S-box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl Acad. Sci. USA, 100, 3083–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov A.S., Gusarov I., Rafikov R., Lopez L.E., Shatalin K., Kreneva R.A., Perumov D.A. & Nudler E. ( 2002) Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell, 111, 747–756. [DOI] [PubMed] [Google Scholar]
- Nahvi A., Sudarsan N., Ebert M.S., Zou X., Brown K.L. & Breaker R.R. ( 2002) Genetic control by a metabolite binding RNA. Chem. Biol., 9, 1043–1049. [DOI] [PubMed] [Google Scholar]
- Neugebauer K.M. & Roth M.B. ( 1997) Transcription units as RNA processing units. Genes Dev., 11, 3279–3285. [DOI] [PubMed] [Google Scholar]
- Nocker A., Hausherr T., Balsiger S., Krstulovic N.P., Hennecke H. & Narberhaus F. ( 2001) A mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res., 29, 4800–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nou X. & Kadner R.J. ( 2000) Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc. Natl Acad. Sci. USA, 97, 7190–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum J.M., Gunnery S. & Mathews M.B. ( 2002) The 3′-untranslated regions of cytoskeletal muscle mRNAs inhibit translation by activating the double-stranded RNA-dependent protein kinase PKR. Nucleic Acids Res., 30, 1205–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F., Jarrous N., Ben-Asouli Y. & Kaempfer R. ( 1999) A cis-acting element in the 3′-untranslated region of human TNF-α mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev., 13, 3280–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D. & Guo P. ( 2003) A viral RNA that binds ATP and contains a motif similar to an ATP-binding aptamer from SELEX. J. Biol. Chem., 278, 7119–7125. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Kerr I.M., Williams B.R., Silverman R.H. & Schreiber R.D. ( 1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Stormo G.D. & Ji Y. ( 2001) Do mRNAs act as direct sensors of small molecules to control their expression? Proc. Natl Acad. Sci. USA, 98, 9465–9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N., Barrick J.E. & Breaker R.R. ( 2003) Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA, 9, 644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via C.S., Shustov A., Rus V., Lang T., Nguyen P. & Finkelman F.D. ( 2001) In vivo neutralization of TNF-α promotes humoral autoimmunity by preventing the induction of CTL. J. Immunol., 167, 6821–6826. [DOI] [PubMed] [Google Scholar]
- Werstuck G. & Green M.R. ( 1998) Controlling gene expression in living cells through small molecule-RNA interactions. Science, 282, 296–298. [DOI] [PubMed] [Google Scholar]
- Wilson D.S. & Szostak J.W. ( 1999) In vitro selection of functional nucleic acids. Annu. Rev. Biochem., 68, 611–647. [DOI] [PubMed] [Google Scholar]
- Winkler W.C., Nahvi A. & Breaker R.R. ( 2002a) Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature, 419, 952–956. [DOI] [PubMed] [Google Scholar]
- Winkler W.C., Cohen-Chalamish S. & Breaker R.R. ( 2002b) An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. USA, 99, 15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]


