Abstract
Rad9 functions in the DNA-damage checkpoint pathway of Saccharomyces cerevisiae. In whole-cell extracts, Rad9 is found in large, soluble complexes, which have functions in amplifying the checkpoint signal. The two main soluble forms of Rad9 complexes that are found in cells exposed to DNA-damaging treatments were purified to homogeneity. Both of these Rad9 complexes contain the Ssa1 and/or Ssa2 chaperone proteins, suggesting a function for these proteins in checkpoint regula-tion. Consistent with this possibility, genetic experiments indicate redundant functions for SSA1 and SSA2 in survival, G2/M-checkpoint regulation, and phosphorylation of both Rad9 and Rad53 after irradiation with ultraviolet light. Ssa1 and Ssa2 can now be considered as novel checkpoint proteins that are likely to be required for remodelling Rad9 complexes during checkpoint-pathway activation.
Introduction
The DNA-damage checkpoint pathway somehow senses DNA lesions, inhibits the cell cycle and activates DNA-repair responses. The Saccharomyces cerevisiae RAD9 gene is the prototype DNA-damage checkpoint gene and is required for the DNA-checkpoint pathway throughout the cell cycle (for a review, see Lowndes & Murguia, 2000; Melo & Toczyski, 2002). Rad9 is phosphorylated during normal cell-cycle progression and is hyperphosphorylated after DNA damage, which is dependent on the protein kinases Mec1 and Tel1 (Vialard et al., 1998; Schwartz et al., 2002). Two distinct, large, soluble Rad9 complexes are found in yeast-cell extracts. The larger (≥850-kDa) complex, which is present in cells that have not been subjected to DNA damage, contains hypophosphorylated Rad9, whereas the smaller (560-kDa) complex, which is formed after DNA damage, contains hyperphosphorylated Rad9 and Rad53 (Gilbert et al., 2001). The smaller complex is capable of catalysing the phosphorylation and release of the active Rad53 kinase.
Here, we report the purification to homogeneity of the two main soluble Rad9 complexes. Mass spectrometric analysis shows that both Rad9 complexes contain the Ssa1 and/or Ssa2 chaperone proteins, whereas only the smaller complex contains Rad53. To test whether Ssa activity is required for Rad9 function, we constructed ssa1Δ, ssa2Δ and ssa1Δssa2Δ cells. Analysis of the mutant cells indicates that the chaperone activity of Ssa1 or Ssa2 is indeed required for the activation of Rad9 and Rad53.
Results and Discussion
Purification and analysis of Rad9 complexes
We estimated by quantitative western blotting that Rad9 is present at only 500–1,000 molecules per haploid cell (data not shown). A yeast strain that expresses normal levels of an epitope-tagged version of Rad9 was constructed (HH–Rad9; Fig. 1A; supplementary information online). Figure 1B shows that the level of expression of HH–Rad9, its cell-cycle phosphorylation and its DNA-damage-regulated hyperphosphorylation (Vialard et al., 1998) were essentially identical to those of untagged Rad9. Importantly, the tag did not significantly alter Rad9 function (Fig. 1C). We have shown previously that Rad9 exists in two soluble forms in whole-cell extracts: a large (≥850-kDa) complex that contains hypophosphorylated Rad9, and a smaller (560-kDa) complex that contains hyperphosphorylated Rad9 (Gilbert et al., 2001). Gel filtration (Fig. 1D) and glycerol-gradient sedimentation (Fig. 1E) analyses showed that there are no significant differences in the elution positions and sedimentation profiles of Rad9 or HH–Rad9, which indicates that the epitope tag does not alter Rad9 complex formation. Thus, HH–Rad9 is indistinguishable from Rad9 in terms of its abundance, regulation, function and presence in two large complexes.
Figure 1.
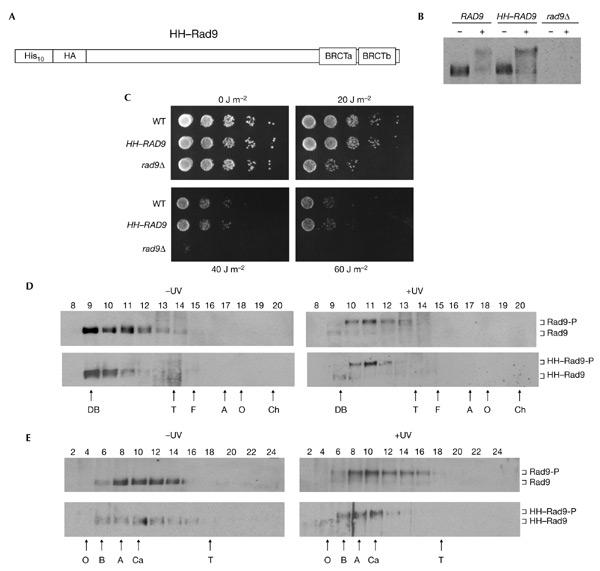
Characterization of epitope-tagged Rad9. (A) The relative position of the ten histidine residues, single haemagglutinin (HA) epitope and BRCA1 carboxy-terminal (BRCT) domains are indicated schematically. (B) Rad9 western blot of extracts from exponentially growing strains that were either mock treated (−) or irradiated with ultraviolet light (+). (C) Exponentially growing cells of the strains indicated were serially diluted onto YPD plates and irradiated with ultraviolet light at the doses indicated. (D) Rad9 western blot of fractions from a Superose 6 gel-filtration column loaded with crude extracts from wild-type cells. Blots from cells that were irradiated with ultraviolet light (+UV) or mock treated (−UV) are indicated. (E) Rad9 western blot of fractions from a 20–35% glycerol sedimentation gradient loaded with crude cell extracts from wild-type cells. Blots from cells that were irradiated with ultraviolet light (+UV) and mock treated (−UV) are indicated. Hyperphosphorylated and hypophosphorylated Rad9 and HH–Rad9 are indicated. The letters below the panels in (D) and (E) indicate the positions of standard proteins (masses, Stokes' radii and sedimentation coefficients are listed in the supplementary information online). A, aldolase; B, bovine serum albumin; Ch, chymotrypsinogen; DB, Dextran blue; F, ferritin; O, ovalbumin; T, thyroglobulin; WT, wild-type.
We purified HH–Rad9 in a complex that contained Ssa1, Ssa2 and Rad53 from whole-cell extracts (Fig. 2). Cell-cycle modified (hypophosphorylated) HH–Rad9 in extracts prepared from exponentially growing cells eluted from heparin–sepharose at ∼500 mM KCl (data not shown). However, HH–Rad9 from exponentially growing cells that had been irradiated with either ultraviolet light (data not shown) or γ-radiation (Fig. 2A) eluted in two distinct peaks, one eluting at ∼500 mM KCl (hypophosphorylated HH–Rad9) and one at ∼300 mM KCl (hyperphosphorylated HH–Rad9). These data further support the discrete nature of these two Rad9 complexes, which was previously suggested by hydrodynamic analysis (Gilbert et al., 2001; Fig. 1D,E). The pooled 300 mM and 500 mM peaks were independently subjected to 12CA5 immunoaffinity and Ni2+–agarose chromatography (see supplementary information online). Silver staining of the resulting purified proteins revealed the cell-cycle-phosphorylated and hyperphosphorylated forms of Rad9 and other bands of 175, 100, 97 and 70 kDa (Fig. 2B). Gradient gels did not show any polypeptides of lower molecular mass (data not shown). The p100 and p97 polypeptides were only detected as co-purifying bands with the hyperphosphorylated forms of Rad9 that were detected after irradiation with both ultraviolet light and γ-radiation. Peptide-mass fingerprinting using matrix-assisted laser desorption/ionization time-of-flight (MALDI–TOF) mass spectroscopy (Erdjument-Bromage et al., 1998) confirmed the identification of the hyperphosphorylated and cell-cycle-modified forms of Rad9 that were previously detected by western blotting. MALDI–TOF mass spectroscopy identified p175 as a protein that is encoded by yeast open reading frame YDL223c. However, p175 does not reproducibly co-purify with Rad9 (Fig. 2B,D), null alleles of YDL223c were not sensitive to DNA-damaging agents, and phosphorylation of Rad9 and Rad53 after DNA damage was normal (data not shown). Therefore, YDL223c was not analysed further. p100 and p97 were both identified as Rad53, which has been shown previously to interact with Rad9 (Vialard et al., 1998; Sun et al., 1998; Emili, 1998). Western blotting confirmed that p100 and p97 correspond to the phosphorylated and unphosphorylated forms, respectively, of Rad53 (data not shown). The p70 band was identified as Ssa1 and/or Ssa2. Given the high degree of conservation (97% identity) between Ssa1 and Ssa2, it was not possible to distinguish between them from the masses of the peptides obtained.
Figure 2.
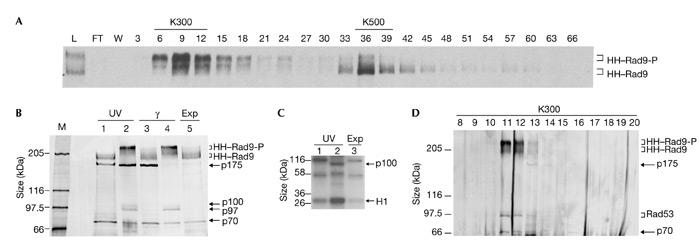
Purification of HH–Rad9 from untreated and DNA-damaged exponentially growing cells. (A) Rad9 western blot of fractions from a heparin–sepharose column loaded with clarified crude extracts from γ-irradiated cells (750 g). Every third fraction is shown. The 300 mM (K300) and 500 mM (K500) KCl pools and hyperphosphorylated (HH–Rad9-P) and hypophosphorylated (HH–Rad9) epitope-tagged Rad9 are indicated. (B) Silver-stained 6.5% SDS–polyacrylamide gel of the final Ni2+–NTA–agarose purification step. Rad9 and co-purifying polypeptides from cells irradiated with ultraviolet light (UV), γ-irradiated cells and exponentially growing (Exp) cells are indicated. The material loaded in lanes 1 and 3 was derived from K500 heparin–sepharose pools, which predominantly contained hypophosphoylated Rad9, and that in lanes 2 and 4 was derived from K300 heparin–sepharose pools, which predominantly contained hyperphosphoylated Rad9. The relative migration positions of co-purifying 175-, 100-, 97- and 70-kDa polypeptides are indicated. (C) Kinase assay using the purified material from cells irradiated with ultraviolet light and from exponentially growing cells. Lanes 1 and 2 contain material purified from the K500 and K300 heparin–sepharose pools, respectively. The migration position of exogenously added histone H1 and a 100-kDa band (p100) are indicated. (D) Silver staining of fractions obtained after Superose 6 (Pharmacia) gel-filtration of the purified Rad9 complex shown in lane 4 in (B). FT, flow-through; L, load; M, marker; W, wash with lysis buffer.
Gel filtration of the purified hyperphosphorylated Rad9 complex that was obtained from γ-irradiated cells (Fig. 2D) indicated that the Rad9 complex remained intact throughout the purification procedure. Furthermore, Ssa1/2 and the differentially modified forms of Rad53 co-purified with hyperphosphorylated Rad9, further supporting evidence for the integrity of this complex. Only a fraction of the total pool of the more abundant Ssa1/2 proteins associated with hyperphosphorylated Rad9 (data not shown). Furthermore, the distribution of Rad53 phosphoforms that co-purify with Rad9 was skewed towards hypophosphorylated Rad53 (Fig. 2B), whereas most of the Rad53 that was detected by western blotting in the input ultraviolet-light- and γ-irradiated extracts was fully phosphorylated (data not shown). Our data indicate that in exponentially growing cells hypophosphorylated Rad9 is a component of a large complex that contains Ssa1 and/or Ssa2 but lacks Rad53. After irradiation with ultraviolet light and γ-radiation, a smaller, hyperphosphorylated Rad9 complex forms, which contains Ssa1 and/or Ssa2, and hypophosphorylated forms of Rad53.
Consistent with the presence of the Rad53 protein kinase in the purified hyperphosphorylated Rad9 complex from cells irradiated with ultraviolet light (Fig. 2B), histone H1 kinase activity was elevated fourfold above background levels (Fig. 2C). A protein of ∼100 kDa, which was probably Rad53 itself, was also phosphorylated significantly in this kinase assay. The identities of the other phosphorylated species that were seen are unknown. We did not detect any ATPase activity in any of our purified Rad9 preparations. Nor was this activity detected in the presence of single- and double-stranded oligonucleotides or in structures corresponding to bubbles and 5' or 3' overhangs. Similarly, we did not detect binding of the Rad9 complex to any of these DNA structures in bandshift assays. In support of these DNA-binding studies, neither of the Rad9 complexes showed significant binding to single-stranded DNA–cellulose (data not shown).
Rad9 function requires chaperone activity
The copurification of Ssa1 and/or Ssa2 with both forms of the Rad9 complex suggests that Ssa activity is required for Rad9 function. We tested this hypothesis by constructing ssa1Δ, ssa2Δ and ssa1Δssa2Δ cells. The single null alleles were not sensitive to irradiation with ultraviolet light. However, ssa1Δssa2Δ double-mutant cells were more sensitive to irradiation with ultraviolet light than were isogenic wild-type cells, although they were not as sensitive as rad9Δ cells (Fig. 3A; and data not shown). Furthermore, triple-mutant ssaΔssa2Δrad9Δ cells were more sensitive than either rad9Δ single mutants or ssa1Δssa2Δ double mutants. Thus, SSA1 and SSA2 must have other functions in cell survival after irradiation with ultraviolet light other than their proposed role in Rad9 function. Also consistent with such a function, ssa1Δssa2Δ cells, like rad9Δ cells, are not particularly sensitive to hydroxyurea, but are sensitive to methyl methanesulphate (data not shown). To determine whether ssa1Δssa2Δ cells are defective in RAD9-specific functions, we examined the G2/M checkpoint in these cells relative to wild-type, rad9Δ and ssa1Δssa2Δrad9Δ cells (Fig. 3B). Exponentially growing cells were irradiated with ultraviolet light, and large-budded cells with a single nucleus in the neck of the cell were scored using fluorescence microscopy. As reported previously (Aboussekhra et al., 1996; de la Torre-Ruiz et al., 1998), wild-type cells were clearly proficient for G2/M arrest, reaching a peak of G2/M cells after 3 h, whereas rad9Δ cells were partially defective for this checkpoint. In ssa1Δssa2Δ cells, the G2/M checkpoint was also partially defective, with 27% G2/M cells at 3 h, compared with 53% and 32% for wild-type and rad9Δ cells, respectively. The ssa1Δssa2Δrad9Δ triple mutant had a more defective checkpoint response than either the ssa1Δssa2Δ or rad9Δ mutants, giving only 12% G2/M cells after 3 h. We also confirmed the G2/M checkpoint defect of ssa1Δssa2Δ cells by blocking the cells in G2 using nocadazole, irradiating the G2-blocked cells with ultraviolet light and releasing them into the cell cycle (see supplementary information online). Thus, SSA1 and SSA2 are both required for a normal G2/M-checkpoint response. Furthermore, as RAD9 and SSA1 plus SSA2 might not be entirely epistatic, these data suggest that SSA1 and SSA2 might have functions in the checkpoint response that are distinct from their role in the Rad9 complex.
Figure 3.
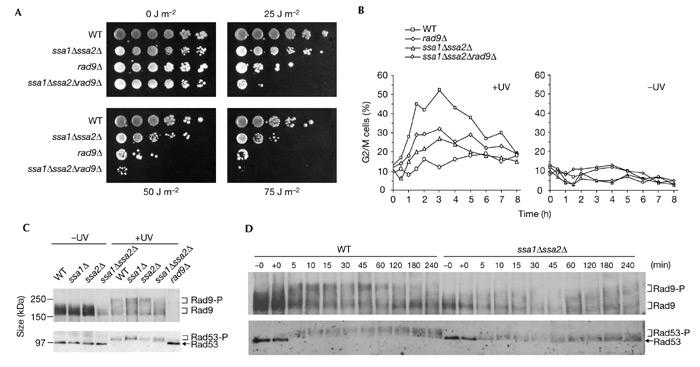
The SSA1 and SSA2 genes are redundantly required for normal survival and G2/M-checkpoint regulation after DNA damage. (A) Drop-test analysis of the strains indicated irradiated with ultraviolet light at the doses indicated. (B) G2/M-checkpoint analysis of the strains indicated after irradiation with ultraviolet light (50 J m−2) of exponentially growing cells. For clarity, the non-irradiated controls (−UV) are shown in a separate panel. (C) Western blot analysis of Rad9 and Rad53 at 60 min after irradiation with ultraviolet light (60 J m−2) in the strains indicated. (D) Western blot analysis of the kinetics of Rad9 hyperphosphorylation and Rad53 phosphorylation after irradiation with ultraviolet light (50 J m−2) in wild-type (WT) and ssa1Δ/ssa2Δ cells. Samples taken prior to irradiation and immediately after irradiation are indicated as −0 and +0. Note that serum specific to unphosphorylated Rad9 and Rad53 was used. The extent of mobility shift, as well as the reduction in signal, is indicative of the extent of modification.
The 70-kDa Ssa3 and Ssa4 proteins, which are usually stress-inducible, are constitutively expressed in ssa1Δssa2Δ cells (Boorstein & Craig, 1990a,b). Therefore, it is possible that ssa1Δssa2Δ cells are not as sensitive to ultraviolet radiation as are rad9Δ cells because the highly conserved Ssa3 and Ssa4 proteins might partially rescue some Ssa1/2 functions that are required for survival after irradiation with ultraviolet light. However, the induction of Ssa3 and Ssa4 in ssa1Δssa2Δ cells is not sufficient to rescue the G2/M checkpoint. In fact, ssa1Δssa2Δ cells seem to be more checkpoint defective than rad9Δ cells (Fig. 3B). Therefore, the functions of SSA1 and SSA2 in G2/M checkpoint regulation cannot be rescued effectively by the elevated levels of Ssa3/4 that are seen in ssa1Δssa2Δ cells. We have purified the Rad9 complex from exponentially growing, non-irradiated ssa1Δssa2Δ cells. A 70-kDa polypeptide that is present at similar levels to the Ssa1/2 band detected in wild-type cells was revealed by silver staining. Western blotting with serum that is specific for Ssa3 and Ssa4 indicated that the 70-kDa band corresponded to Ssa3/4 (see supplementary information online). Thus, although Ssa3 and Ssa4 can be recruited to the Rad9 complex in ssa1Δssa2Δ cells, they cannot fully substitute for Ssa1/2 in this complex. Furthermore, less Rad9 is present in ssa1Δssa2Δ cells (Fig. 3C,D).
The checkpoint defect in ssa1Δssa2Δ cells was confirmed by examining the phosphorylation of both Rad9 and Rad53 after DNA damage. Sixty minutes after irradiation, hyperphosphorylation of Rad9 and phosphorylation of Rad53 were partially defective in the ssa1Δssa2Δ double mutant relative to wild-type cells, whereas these modifications were essentially normal in the ssa1Δ and ssa2Δ single mutants (Fig. 3C). To further characterize Rad9 and Rad53 modification after DNA damage, we examined the kinetics of Rad9 and Rad53 phosphorylation after irradiation with ultraviolet light of exponentially growing wild-type and ssa1Δssa2Δ cells (Fig. 3D). As previously observed (Vialard et al., 1998), Rad9 was rapidly phosphorylated in wild-type cells, reaching maximal levels of the most slowly migrating hyperphosphorylated form after only 5 min. By contrast, hyperphoshorylated Rad9 was not detected at early time points and was only weakly detected towards the end of the time course in ssa1Δssa2Δ cells. Although some Rad53 phosphorylation was clearly evident in ssa1Δssa2Δ cells at early time points, this modification was much less extensive than that seen in similarly treated wild-type cells. The presence of some Rad53 phosphorylation might be due to the separate, additive Rad24 pathway that also activates Rad53 (de la Torre-Ruiz et al., 1998). Single mutants for either RAD9 or RAD24 are not completely defective for checkpoint activity, whereas in double-mutant cells the checkpoint is almost abolished. Alternatively, the residual Rad53 phosphorylation in ssa1Δssa2Δ cells could be because of some compensatory activity of the Ssa3 and Ssa4 proteins, as discussed above, or a combination of both these possibilities. Our data indicate that SSA1 or SSA2 is required for normal Rad9 and Rad53 phosphorylation after irradiation with ultraviolet light. This requirement supports the previous survival and G2/M-checkpoint experiments, which also indicated that either SSA1 or SSA2 function is required for normal survival and G2/M-checkpoint regulation after irradiation with ultraviolet light. Moreover, biochemical analysis of the function of hyperphosphorylated Rad9 in catalysing the activation of Rad53 indicated a preference for potassium ions (Gilbert et al., 2001), which is consistent with the known potassium-dependent ATPase activation of Ssa1 and Ssa2 (Fung et al., 1996).
Ssa1 and Ssa2 belong to a family of four (Ssa1–Ssa4) highly related molecular chaperones of the heat-shock protein 70 (Hsp70) family that are located in both the cytoplasm and the nucleus (Bukau & Horwich, 1998). Ssa1 and Ssa2 are 97% identical and are constitutively expressed to high levels (∼60,000 and ∼66,000 molecules per cell, respectively; Norbeck & Blomberg, 1997). Ssa3 and Ssa4 have ∼80% identity to Ssa1/2 and are not detectable in exponentially growing cells, but are induced to high levels after heat shock. Mutant yeast strains that lack both SSA1 and SSA2 are characterized by slow growth, temperature sensitivity and constitutive expression of SSA3 and SSA4, which suggests that under these conditions SSA3 and SSA4 only partially substitute for SSA1 and SSA2 (Craig & Jacobsen, 1984; Baxter & Craig, 1998). Mutation of all four SSA genes is lethal. The Hsp70 molecular chaperones function in various cellular compartments to guide the folding of proteins and aid in translocation across membranes (Bukau & Horwich, 1998). Moreover, these proteins have also been found to have functions in signal transduction and cell-cycle progression. Ssa1 positively controls the cyclic AMP pathway through its interactions with cell-division cycle 25 (Cdc25; Geymonat et al., 1998), and both Ssa1 and Ssa2 co-purify with the G1 cyclin, Cln3 (Yaglom et al., 1996). From the point of view of this study, it is interesting that this latter interaction is thought to facilitate phosphorylation of Cln3 by Cdc28, leading to Cln3 degradation. In general, chaperone proteins have been proposed to function in signal transduction by regulating transitions between active and inactive states (for a review, see Rutherford & Zucker, 1994), for example, by modulating the state of oligomerization or folding of mature signalling molecules, thereby allowing regulated progression to the next step of the pathway. Hydrodynamic data clearly indicate that after irradiation with ultraviolet light, the Rad9 complex loses mass and changes conformation, becoming less elongated, which is reflected by its smaller Stokes' radius and reduced frictional coefficient (Gilbert et al., 2001). It is tempting to speculate that this substantial change in the Rad9 complex could be mediated by, and dependent on, Ssa1/2-dependent chaperone activity.
Methods
Strains and plasmids.
All strains used in this study were constructed in the W303-1a background (MATa; ade2-1; trp1-1; leu2-3,112; his3-11, 15; ura3). The rad9Δ strain has been described previously (de la Torre-Ruiz et al., 1998). The ssa1Δssa2Δ and rad9Δ ssa1Δssa2Δ strains were obtained as meiotic segregants from a cross between MATa; ssa1::kanr and MATa; rad9::HIS3; ssa2::kanr. The HH–RAD9 strain was generated by site-specific integration of plasmid pHH–RAD9 (linearized with PshA1) into the RAD9 locus. This plasmid was derived from pRS303– 10HisRad9 (Vialard et al., 1998) by inserting oligonucleotides corresponding to a single haemagglutinin (HA) epitope into the unique BamH1 site of pRS303–10HisRad9. These oligonucleotides (5'-CGCGGATCCTACCCATACGACGTCCCAGACTACGCTGGATCCCGC-3' and 5'-GCGGGATCCAGCGTAGTCTGGGACGTCGTATGGGTAGGATCCGCG-3') were annealed by heating them at 95 °C for 5 min and allowing them to cool slowly to 21 °C before ligation. The resulting HH–Rad9 protein, which contains ten histidine residues and a single HA epitope at its amino terminal, was expressed from the RAD9 promoter at the RAD9 locus.
Survival after ultraviolet irradiation, checkpoint analysis, western blotting and kinase assays.
To assess cell survival after irradiation with ultraviolet light, serial dilutions were made from mid-log cultures and plated on to YPD plates using a replica plater (Sigma; catalogue number R2383). The YPD plates were irradiated with the indicated doses of ultraviolet light and incubated at 30 °C until colonies appeared. Analysis of the G2/M checkpoint, preparation of clarified, crude cell extracts, Rad53 kinase assays and western blotting of Rad9 with serum NLO5 and Rad53 with serum NLO16 have been described previously (Vialard et al., 1998; de la Torre-Ruiz et al., 1998). In Fig. 2, the kinase assays used one-fifth of the material shown in the silver-stained gels.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/4-embor935-s1.pdf).
Supplementary Material
Supplementary information
Acknowledgments
We thank our colleagues for their critical reading of the manuscript and E. Craig for serum specific to Ssa3 and Ssa4.
References
- Aboussekhra A., Vialard J.E., Morrison D.E., de la Torre-Ruiz M.A., Cernakova L., Fabre F. & Lowndes N.F. ( 1996) A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J., 15, 3912–3922. [PMC free article] [PubMed] [Google Scholar]
- Baxter B.K. & Craig E.A. ( 1998) Suppression of an Hsp70 mutant phenotype in Saccharomyces cerevisiae through loss of function of the chromatin component Sin1p/Spt2p. J. Bacteriol., 180, 6484–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein W.R. & Craig E.A. ( 1990a) Structure and regulation of the SSA4 HSP70 gene of Saccharomyces cerevisiae. J. Biol. Chem., 265, 18912–18921. [PubMed] [Google Scholar]
- Boorstein W.R. & Craig E.A. ( 1990b) Transcriptional regulation of SSA3, an HSP70 gene from Saccharomyces cerevisiae. Mol. Cell. Biol., 10, 3262–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B. & Horwich A.L. ( 1998) The Hsp70 and Hsp60 chaperone machines. Cell, 92, 351–366. [DOI] [PubMed] [Google Scholar]
- Craig E.A. & Jacobsen K. ( 1984) Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell, 38, 841–849. [DOI] [PubMed] [Google Scholar]
- de la Torre-Ruiz M.A., Green C.M. & Lowndes N.F. ( 1998) RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J., 17, 2687–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A. ( 1998) MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell, 2, 183–189. [DOI] [PubMed] [Google Scholar]
- Erdjument-Bromage H., Lui M., Lacomis L., Grewal A., Annan R.S., McNulty D.E., Carr S.A. & Tempst P. ( 1998) Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A, 826, 167–181. [DOI] [PubMed] [Google Scholar]
- Fung K.L., Hilgenberg L., Wang N.M. & Chirico W.J. ( 1996) Conformations of the nucleotide and polypeptide binding domains of a cytosolic Hsp70 molecular chaperone are coupled. J. Biol. Chem., 271, 21559–21565. [DOI] [PubMed] [Google Scholar]
- Geymonat M., Wang L., Garreau H. & Jacquet M. ( 1998) Ssa1p chaperone interacts with the guanine nucleotide exchange factor of ras Cdc25p and controls the cAMP pathway in Saccharomyces cerevisiae. Mol. Microbiol., 30, 855–864. [DOI] [PubMed] [Google Scholar]
- Gilbert C.S., Green C.M. & Lowndes N.F. ( 2001) Budding yeast RAD9 is an ATP-dependent activating machine. Mol. Cell, 8, 129–136. [DOI] [PubMed] [Google Scholar]
- Lowndes N.F. & Murguia J.R. ( 2000) Sensing and responding to DNA damage. Curr. Opin. Genet. Dev., 10, 17–25. [DOI] [PubMed] [Google Scholar]
- Melo J. & Toczyski D. ( 2002) A unified view of the DNA-damage checkpoint. Curr. Opin. Cell. Biol., 14, 237–245. [DOI] [PubMed] [Google Scholar]
- Norbeck J. & Blomberg A. ( 1997) Two-dimensional electrophoretic separation of yeast proteins using a non-linear wide range (pH 3–10) immobilized pH gradient in the first dimension; reproducibility and evidence for isoelectric focusing of alkaline (pI > 7) proteins. Yeast, 13, 1519–1534. [DOI] [PubMed] [Google Scholar]
- Rutherford S.L. & Zuker C.S. ( 1994) Protein folding and the regulation of signaling pathways. Cell, 79, 1129–1132. [DOI] [PubMed] [Google Scholar]
- Schwartz M.F., Duong J.K., Sun Z., Morrow J.S., Pradhan D. & Stern D.F. ( 2002) Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell., 9, 1055–1065. [DOI] [PubMed] [Google Scholar]
- Sun Z., Hsiao J., Fay D.S. & Stern D.F. ( 1998) Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science, 281, 272–274. [DOI] [PubMed] [Google Scholar]
- Vialard J.E., Gilbert C.S., Green C.M. & Lowndes N.F. ( 1998) The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J., 17, 5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaglom J.A., Goldberg A.L., Finley D. & Sherman M.Y. ( 1996) The molecular chaperone Ydj1 is required for the p34CDC28-dependent phosphorylation of the cyclin Cln3 that signals its degradation. Mol. Cell. Biol., 16, 3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information


