Abstract
In Saccharomyces cerevisiae, a large complex, known as the Ccr4–Not complex, containing two nucleases, is responsible for mRNA deadenylation. One of these nucleases is called Pop2 and has been identified by similarity with PARN, a human poly(A) nuclease. Here, we present the crystal structure of the nuclease domain of Pop2 at 2.3 Å resolution. The domain has the fold of the DnaQ family and represents the first structure of an RNase from the DEDD superfamily. Despite the presence of two non-canonical residues in the active site, the domain displays RNase activity on a broad range of RNA substrates. Site-directed mutagenesis of active-site residues demonstrates the intrinsic ability of the Pop2 RNase D domain to digest RNA. This first structure of a nuclease involved in the 3′–5′ deadenylation of mRNA in yeast provides information for the understanding of the mechanism by which the Ccr4–Not complex achieves its functions.
Introduction
mRNA degradation is an important step of gene expression, allowing to counter-balance or enhance the effect of transcription and thus contributing to overall gene regulation. In Saccharomyces cerevisiae, the deadenylation process (reviewed in Caponigro & Parker, 1996) is the first step of mRNA degradation. It is achieved by the Ccr4–Not complex (Daugeron et al., 2001; Tucker et al., 2001), a large multi-protein complex (Liu et al., 1997; Chen et al., 2001), built around a core of seven proteins: the carbon catabolite repressor 4 factor (Ccr4), first identified as a gene expression regulating protein (Denis, 1984), the Pop2 protein, also known as Caf1 (for Ccr4 associated factor 1; Draper et al., 1995) and five Not proteins (Not1–5) required for the proper function of the Ccr4–Not complex (Draper et al., 1994; Liu et al., 1998). Interactions between Ccr4, Pop2 and the Not proteins, together with other proteins present in the Ccr4–Not complex, have been characterized (Benson et al., 1998; Bai et al., 1999; Badarinarayana et al., 2000; Lemaire & Collart, 2000; Liu et al., 1997, 2001).
Ccr4 and Pop2 both contain a nuclease domain and are therefore potentially responsible for the deadenylation activity. The Ccr4 protein has been identified as a member of the Mg2+-dependent endonuclease-related protein family (Dlakic, 2000), and its activity has been characterized both in vivo (Daugeron et al., 2001; Chen et al., 2002; Tucker et al., 2001, 2002) and in vitro (Viswanathan et al., 2003). Disruption of the gene and various mutants of the protein active site show a strong decrease in mRNA deadenylation rates, indicating that the protein is an essential factor in this process (Daugeron et al., 2001; Tucker et al., 2001; Chen et al., 2002). The second protein, Pop2, is related to the ribonuclease (RNase) D family (Moser et al., 1997; Daugeron et al., 2001). While Pop2 has two non-conserved catalytic residues, the other Pop2/Caf1 family members display a conserved and thus potentially fully functional active site. In addition, Pop2 gene disruption affects the rate and the degree of deadenylation of reporter mRNAs (Daugeron et al., 2001; Tucker et al., 2001).
The RNase D family (Mian, 1997) belongs to the DEDD superfamily composed of RNases as well as deoxyribonucleases (DNases; Zuo & Deutscher, 2001). It is characterized by sequence motifs similar to the Exo I, II and III motifs of the 3′–5′ exodeoxyribonuclease (or proofreading) domains of DNA polymerases, which contain four conserved acidic residues, namely DEDD. These amino acids are responsible for binding the two metal ions involved in catalysis (reviewed in Joyce & Steitz, 1994). Three-dimensional (3D) structures have been determined for the DnaQ family (Ollis et al., 1985; Hamdan et al., 2002), which is a subgroup of the DEDD superfamily.
Here, we report the crystal structure of the RNase D domain of the yeast Pop2 protein. It represents the first structure of an RNase D protein as well as of an RNase from the DEDD superfamily. Despite the non-conservation of two catalytic residues from the DEDD signature sequence, we observe in vitro RNase activity towards poly(A), and also poly(C) and poly(U). Site-directed mutagenesis abolishes RNase activity, indicating that it is associated specifically with the Pop2 RNase D domain. Canonical active site residues are fully conserved within the Pop2/Caf1 family, strongly suggesting that RNase activity will be associated with all the family members.
Results and Discussion
Structure determination and overall fold
The fragment of the yeast Pop2 protein corresponding to the RNase D domain was crystallized and its structure was determined using MAD data from a single Se-Met-substituted protein containing crystal. Native crystals belong to space group P212121 with two molecules in the asymmetric unit. The final model contains two calcium ions, two xenon atoms and 263 or 254 residues (for each respective molecule) representing all amino acids where the main chain was clearly defined (Fig. 1A). Three regions were poorly resolved and did not allow tracing (residues 1–3, 50–59 and 213–225). The two molecules constituting the asymmetric unit adopt very similar conformations with a root mean square deviation (r.m.s.d.) of 0.47 Å.
Figure 1.
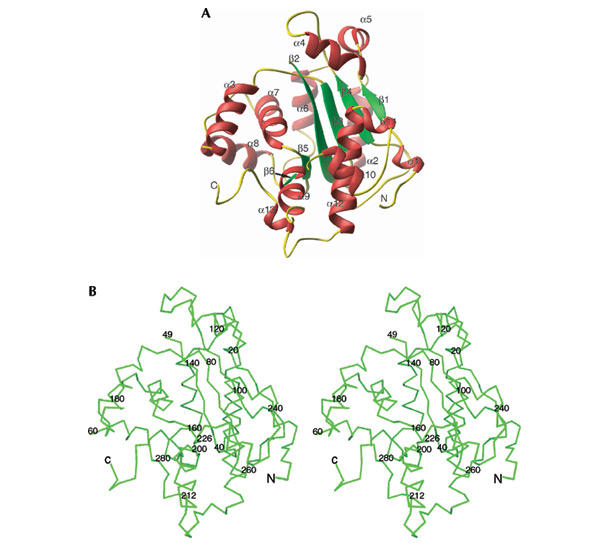
Structure of the nuclease domain of the Pop2 protein. (A) Ribbon plot representation with the secondary elements in the following colour code: α-helix, red; β-strands, green; and loops, yellow. (B) Crossed-eye stereo representation of the Cα trace is displayed with every 20th residue marked.
The overall structure contains 13 α-helices and six β-strands, forming a kidney-shaped structure (Fig. 1A). The β-strands form the central core flanked by the α-helices. Helices 2, 3, 6, 7, 8 and 13 form the near-side of the molecule, while the remaining helices build the two lobes creating the cavity of the kidney-shaped molecule (Fig. 1B).
Structural conservation within the DEDD nuclease family
During recent years, several structures of proofreading enzymes involved in DNA replication have been solved, revealing a common fold for the DnaQ subgroup of the DEDD superfamily (Zuo & Deutscher, 2001). While the sequence similarity between Pop2 and the ɛ-subunit of DNA polymerase III (PolIIIɛ) or the exonuclease domain of polymerase I (PolI) is only 15 or 20% (Fig. 2A), the overall superposition of the structures gives a r.m.s.d. of only 1.5 or 1.7 Å of the Cα trace over 134 and 128 residues, respectively (Fig. 2B; Ollis et al., 1985; Hamdan et al., 2002). The Pop2 structure provides the first crystallographic evidence that the DNase fold is also adopted by the RNase D, subgroup of the DEDD superfamily (Fig. 2B).
Figure 2.
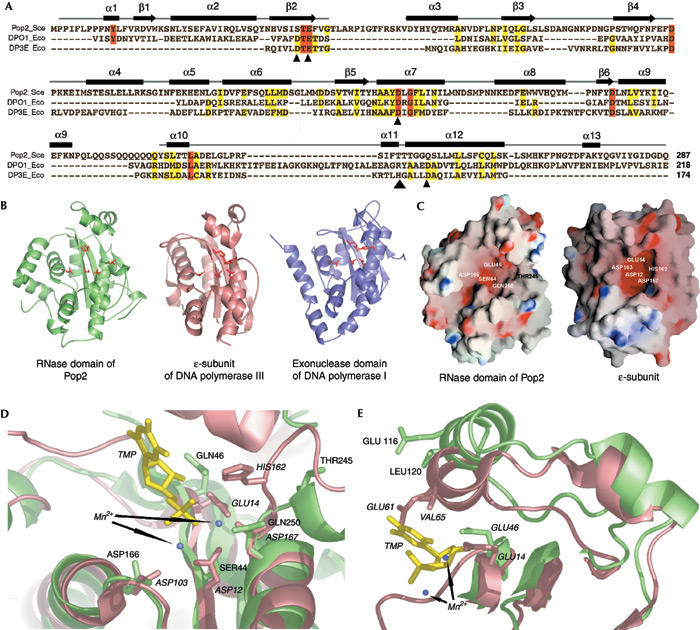
Structural homology of Pop2 with members of the DEDD nuclease superfamily. (A) Structure-based sequence alignment of Pop2, the exonuclease domain of PolI and the ɛ-subunit of PolIII. Sequence conservation is shown by colour coding: invariant residues are highlighted in red. Yellow highlights residues that have similar properties. Secondary structure elements of Pop2 are shown above the sequences. Small arrowheads indicate the conserved DEDD residues forming the catalytic site of the ɛ-subunit. (B) The three structures shown in the same relative orientation. (C) The electrostatic surface potentials of Pop2 and the ɛ-subunit indicate the location of the active site; catalytic residues are highlighted. (D) Close-up view of the active site of the ɛ-subunit (salmon colour; with bound TMP in yellow) superimposed with the Pop2 (light green) structure and (E) side view of the secondary structure elements interacting with the bound nucleotide. Bold and italic labels correspond to the amino acids from Pop2 or the ɛ-subunit, respectively.
A global view of the electrostatic surface potential of Pop2 indicates a highly negatively charged cavity (Fig. 2C). A similar feature can be observed in exonuclease I (Breyer & Matthews, 2000). These negatively charged regions in both proteins coincide with the active site in PolIII (Fig. 2C,D). A close look at the active site (Fig. 2D) shows that the Ser 44 and Gln 250 substitutions in the Pop2 amino-acid sequence could provide an oxygen atom able to interact with the active site magnesium. In line with this hypothesis, it is interesting to note that the two corresponding residues are phylogenetically conserved in yeast species that diverged 5–20 million years ago, thus supporting the importance of this combination for yeast Pop2 function. Given the number of negatively charged residues at the active site, it is also possible that Pop2 binds only one metal ion instead of two normally observed for the DNases, resulting in a different reaction mechanism.
Interestingly, the α-helices 4 and 5 (Fig. 2E) as well as residues from the loop between strands β2 and β3 contact the leaving nucleotide in the PolIII structure. These amino acids are identical, or at least similar, in the case of Pop2, suggesting an interaction with the 3′-terminal nucleotide during the nuclease reaction despite the somewhat different orientation observed in the crystal (Fig. 2E).
RNase activity of the Pop2 domain
The literature contains contradictory data about the RNase activity associated with the Pop2 protein from yeast. Under our in vitro conditions, Pop2 RNase activity is observed with poly(A), poly(U) and poly(C), but not with oligo(G) (Fig. 3A, lanes 2–3, 5–6, 8–9 and 11). However, more sensitive competition assays demonstrate a subtle preference for poly(A) (Daugeron et al., 2001). When the first two residues from the DEDD motif are mutated, that is, S44A and E46A, the activity is lost (Fig. 3B, compare lanes 13–14 and 16–17). The progressive appearance of shorter RNA fragments indicates the distributive activity of the Pop2 RNase D domain (Fig. 3C). The ability to degrade several types of RNA implies that the substrate RNA is not recognized in an absolute, specific manner but may involve a substantial contribution of van der Waals interactions, in agreement with the presence of a conserved hydrophobic patch following the β-strand 2 (Fig. 3E).
Figure 3.
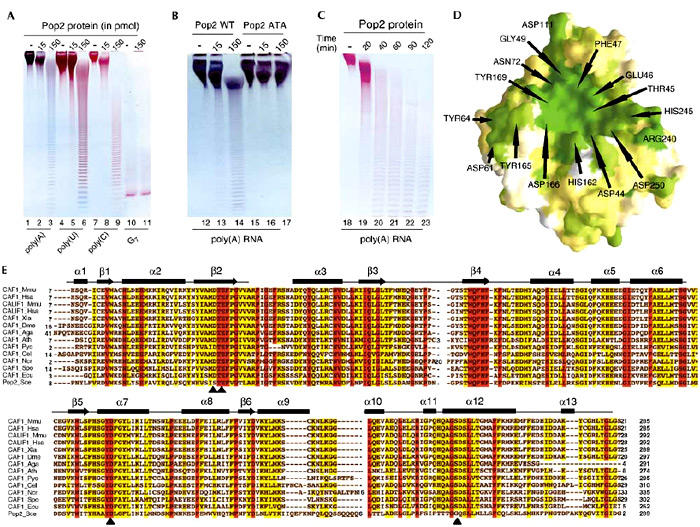
RNase activity of Pop2 and conservation within the Pop2/Caf1 family. (A) Activity tests showing the RNase activity of Pop2 WT with poly(A), poly(U), poly(C) and G7 RNA substrates. (B) The double-mutant S44A/E46A (Pop ATA) does not degrade poly(A) RNA. (C) The time course of the RNase reaction demonstrates that the Pop2 nuclease is distributive. (D) Surface representation of the sequence conservation within the Pop2/Caf1 family is shown and is based on the sequence alignment shown in (E). Green colour indicates conserved residues, and white for non-conserved. Numbering is according to the Pop2 sequence and the residue name is according to the conserved amino acid in the Pop2/Caf1 family. (E) Sequence alignment of the eukaryotic Pop2/Caf1 family with Mus musculus CAF1 (accession number NP_035265) and CALIF1 (NP_081225), Homo sapiens CAF1 (L46722) and CALIF1 (Q9UFF9), Xenopus laevis CAF1 (AAH41239), Drosophila melanogaster CAF1 (AE003543), Anopheles gambiae CAF1 (EAA12934), Arabidopsis thaliana CAF1 (AY070420), Plasmodium yoelii yoelii CAF1 (EAA20457), Caenorhabditis elegans CAF1 (U21854), Neurospora crassa CAF1 (EAA29793), Schizosaccharomyces pombe CAF1 (NP_588385), Encephalitozoon cuniculi CAF1 (NP_597215) and the RNase D domain of Saccharomyces cerevisiae Pop2 (P39008), colour coded as in Fig. 2A. DEDD catalytic site residues are indicated with arrowheads.
The structure of the Pop2 nuclease domain represents the first 3D model of a catalytic component of the yeast deadenylase complex.
Pop2/Caf1 family
Since Pop2 is the least conserved protein within the Pop2/Caf1 family (Fig. 3E; Daugeron et al., 2001), the fold conservation between the DNases and Pop2 suggests that the nuclease domain of the Pop2/Caf1 family members will have the same fold as the Pop2 protein. According to the sequence alignment of the Pop2/Caf1 family, the other members do have a canonical active site, that is, the DEDD signature sequence (Fig. 3E, arrowheads). Therefore, these proteins are expected to display at least the same or even higher RNase activity than Pop2. This has been verified in the case of human CAF1 (F. Mauxion and B. Séraphin, unpublished data). In addition, the conserved amino acids within the Pop2/Caf1 family line the entire cavity, indicating a functionally important surface likely to correspond to the catalytic and the RNA interacting sites (Fig. 3D).
Deadenylation and mRNA degradation
The role of Pop2 in mRNA deadenylation has remained unclear, while it is essential for deadenylation in vivo (Daugeron et al., 2001; Tucker et al., 2001) and was reported to have RNase activity in vitro (Daugeron et al., 2001), analysis of point mutants failed to reveal the requirement for a specific step in mRNA decay (Chen et al., 2002; Tucker et al., 2002). The crystal structure of the nuclease domain of Pop2 in conjunction with mutagenesis and in vitro assays demonstrate definitively that yeast Pop2 contains a nuclease domain displaying RNase activity of broad specificity.
The requirement for two nucleases in the Ccr4–Not complex to mediate mRNA deadenylation remains puzzling. Published data suggest that Ccr4 provides most of the nuclease activity in yeast (Chen et al., 2002; Tucker et al., 2002). However, as yeast Pop2 has an RNase activity in vitro and a non-canonical active site, this situation may differ in other organisms. Indeed, the presence of two variations in the active site of Pop2 may strongly influence its efficiency compared to human CAF1 for example. It would thus be of interest to evaluate the relative contribution of human CCR4 and human CAF1 to exonucleolytic poly(A) degradation. Similarly, it would be of interest to test whether the N-terminal domain of full-length Pop2 inhibits RNase activity in vivo and whether this is affected by the phosphorylation event known to occur in this region (Moriya et al., 2001).
Interaction of Ccr4 and Pop2 with the Not factors may also play an important role in regulating Pop2 activity and subsequently the deadenylation process. In the case of the human poly(A) nuclease PARN, reported to be similar to Pop2 as well as a member of the DEDD family, the protein contains a large domain belonging to the R3H family (Daugeron et al., 2001), which is able to bind single-stranded RNA on its own. Since the Pop2/Caf1 family does not contain any additional RNA-binding domain, it is tempting to speculate that Pop2 interactions with the Ccr4–Not complex may target it to the poly(A) tail.
Further structural studies on the various complexes formed between Ccr4, the Not proteins, mRNA and Pop2 are likely to give important information for the understanding of the deadenylase complex in yeast and other eukaryotic species. This information could be used in order to better understand how the deadenylation process is coupled to translation as well as to decapping processes before the degradation occurs.
Methods
Expression and crystallization
The yeast Pop2 RNase D domain (amino acids (aa) 147–433) was expressed from a pET24d derivative (Novagen) with an amino (N)-terminal 6xHis tag-TEV protease site extension. After overexpression in Escherichia coli BL21-CodonPlus (DE3)-RIL, cells were lysed with a French press in buffer A (50 mM NaH2PO4 (pH 7.5), 200 mM NaCl, 10 mM β-mercaptoethanol) in the presence of protease inhibitor cocktail (Promega) and 10 μg ml−1 of RNase A (Sigma). After binding to Ni-NTA beads (Quiagen), the protein was eluted as indicaded. Pop2-containing fractions were passed onto a Superdex 75 column (Amersham-Pharmacia) in buffer A. The tag was then removed by TEV protease cleavage at 16°C (enzyme:substrate ratio 1:100 w/w) followed by chromatography on Ni-NTA beads. A second gel filtration step in 20 mM Tris/Cl (pH 7.5) and 200 mM NaCl led to a >95% pure sample, which was concentrated to 9 mg ml−1. A Se-Met derivative was prepared similarly using the B834 E. coli host strain grown in minimal medium containing L-seleno-methionine (50 μg ml−1).
Crystals of the Pop2 nuclease domain were grown by vapour diffusion (hanging drops) at 4 °C by mixing an equal volume of protein and reservoir solution containing 2.5% PEG 3350 MW, 100 mM Hepes (pH 7.0), 80 mM calcium acetate and 16.5% glycerol. Drops were immediately micro-seeded and, after 7–8 days, transferred over a well containing a similar solution and 27.5% glycerol. Crystals were then directly flash-frozen in liquid nitrogen. Crystals from the Se-Met derivative were grown using the following conditions: 2% PEG 3350 MW, 120 mM calcium acetate and protein at 5 mg ml−1. Xenon derivatization was carried out on native crystals using a pressurized cell at 10 bar for 10 min (Djinovic-Carugo et al., 1998).
A complete 2.3 Å data set was collected under cryogenic conditions using the ESRF ID29 beamline. A three-wavelength MAD experiment was performed on a single Se-Met-substituted protein crystal at the PX06 beamline at SLS. Data were processed using the HKL package (Otwinowski & Minor, 1997). The native and the Se-Met-substituted protein crystallized in space groups P212121 (a=78.5 Å, b=79.4 Å, c=101.3 Å, α=90°, β=90°, γ=90°) with two molecules per asymmetric unit, and P41212 (a=78.2 Å, b=78.2 Å, c=102.6 Å, α=90°, β=90°, γ=90°) with one molecule per asymmetric unit, respectively.
Structure determination and refinement Phases were obtained at 3.3 Å with SOLVE (Terwilliger & Berendzen, 1999) and the electron density map was improved by solvent flattening using the program RESOLVE (Terwilliger, 2000). Only 40% of the protein could be traced with program O, version 7 (Jones et al., 1991). DMMULTI from the CCP4 suite (CCP4, 1994) was used to extend the phases to 2.3 Å in P212121. Then, the density modification procedure implemented in CNS (Brunger et al., 1998) and based on the phases calculated from the partial model provided additional electron density. After three cycles of model building, annealing, individual B-factor refinement and density modification, 263 and 254 residues for each molecule in the asymmetric unit could be built (residues 4–49, 60–213 and 226–289 for molecule 1 and residues 4–49, 62–207 and 228–289 for molecule 2). The final Rfree is 26.3%, and the model shows good stereochemistry as indicated by PROCHECK (Laskowski et al., 1993). Table 1 shows data collection and refinement statistics.
Table 1.
Data collection, phasing and refinement statistics
|
Se-Met data sets |
||||
|---|---|---|---|---|
| Native | Peak | Inflection | Remote | |
| Values for the outermost resolution shell are shown in parentheses. FOM, figure of merit; Rfree: R factor for 5% of the reflections excluded from the refinement. | ||||
| Rsym, ∑hkl∣I−(I)∣/∑hklI. | ||||
| Data collection and phasing statistics | ||||
| λ (Å) | 1.8461 | 0.9795 | 0.9797 | 0.9689 |
| Space group | P212121 | P41212 | P41212 | P41212 |
| Resolution (Å) | 2.3 | 3.15 | 3.20 | 3.25 |
| (2.38–2.3) | (3.26–3.15) | (3.31–3.2) | (3.37–3.25) | |
| Reflection (unique) | 52,320 | 10,414 | 10,007 | 9,508 |
| Completeness (%) | 95.9 (76.6) | 99.2 (100) | 99.5 (100) | 99.6 (100) |
| Redundancy | 6.7 | 4 | 4 | 4 |
| Average I/σ(I) | 11.9 (2.9) | 11.9 (4.6) | 11.7 (4.4) | 11.4 (3.9) |
| Rsym | 4.0 (30.9) | 7.1 (30.5) | 7.6 (32.8) | 7.6 (34.9) |
| FOMMAD | — | — | — | 0.49 |
| FOM after solvent flattening | — | — | — | 0.65 |
| Refinement statistics | ||||
| No. of reflections used | 51,486 | — | — | — |
| Rvalue (%) | 23.8 | — | — | — |
| Rfree (%) | 26.0 | — | — | — |
| Residues (out of 289) | 263 (mol. 1)/254 (mol. 2) | — | — | — |
| Number of protein/solvent/ion atoms | 4,214/46/4 | — | — | — |
| Average B factor (Å2) | 51 | — | — | — |
| r.m.s.d. Δbond lengths (Å) | 0.008 | — | — | — |
| r.m.s.d. Δbond angles (degree) | 1.4 | — | — | — |
The coordinates have been deposited in the Protein Data Bank under accession code 1UOC.
RNaseassays Polynucleotides (Amersham-Pharmacia) were stored as recommended. In vitro RNase assays were performed in 20 mM Tris/Cl (pH 7.0), 150 mM NaCl, 2 mM MgCl2, 5 U RNasin (Promega), 3 mM RNA substrate and indicated amounts of purified Pop2. A volume of 10 μl of the reaction mixture was incubated at 25 °C for 1 h unless otherwise stated. Reactions that were stopped by the addition of formamide/EDTA buffer were loaded onto 8 M urea/18% acrylamide (19:1) gels that were stained with toluidine blue.
Acknowledgments
We thank the staff at the European Synchrotron Radiation facility (ESRF), Swiss Light Synchrotron (SLS) and Deutsches Elekronen Synchrotron (DESY) for assistance during data collection. The MAD data sets were collected at SLS, Paul Schanen Institute, Switzerland. We also thank C. Schulze-Briex for support and acknowledge J. Basquin and J. Féthière for data collection of the MAD data sets as well as A. Popov for support at the X13 beamline at DESY/EMBL-Hamburg. We also thank E. Ennifar for useful crystallographic discussions and C. Temme for help with mutagenesis. Work in B.S.'s lab is supported by La Ligue contre le Cancer, CNRS and the Ministry for Research (PRFMMIP).
References
- Badarinarayana V., Chiang Y.C. & Denis C.L. ( 2000) Functional interaction of CCR4–NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast. Genetics, 155, 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Salvadore C., Chiang Y.C., Collart M.A., Liu H.Y. & Denis C.L. ( 1999) The CCR4 and CAF1 proteins of the CCR4–NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol., 19, 6642–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson J.D., Benson M., Howley P.M. & Struhl K. ( 1998) Association of distinct yeast Not2 functional domains with components of Gcn5 histone acetylase and Ccr4 transcriptional regulatory complexes. EMBO J., 17, 6714–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyer W.A. & Matthews B.W. ( 2000) Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nature Struct. Biol., 7, 1125–1128. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. ( 1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Caponigro G. & Parker R. ( 1996) Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev., 60, 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4, C.C.P. ( 1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chen J., Rappsilber J., Chiang Y.C., Russell P., Mann M. & Denis C.L. ( 2001) Purification and characterization of the 1.0 MDa CCR4–NOT complex identifies two novel components of the complex. J. Mol. Biol., 314, 683–694. [DOI] [PubMed] [Google Scholar]
- Chen J., Chiang Y.C. & Denis C.L. ( 2002) CCR4, a 3′–5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J., 21, 1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron M.C., Mauxion F. & Seraphin B. ( 2001) The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res., 29, 2448–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C.L. ( 1984) Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics, 108, 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djinovic-Carugo K., Everitt P. & Tucker P.A. ( 1998) A cell for producing xenon-derivative crystals for cryocrystallographic analysis. J. Appl. Crystallogr., 31, 812–814. [Google Scholar]
- Dlakic M. ( 2000) Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem. Sci., 25, 272–273. [DOI] [PubMed] [Google Scholar]
- Draper M.P., Liu H.Y., Nelsbach A.H., Mosley S.P. & Denis C.L. ( 1994) CCR4 is a glucose-regulated transcription factor whose leucine-rich repeat binds several proteins important for placing CCR4 in its proper promoter context. Mol. Cell. Biol., 14, 4522–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper M.P., Salvadore C. & Denis C.L. ( 1995) Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol. Cell. Biol., 15, 3487–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan S., Carr P.D., Brown S.E., Ollis D.L. & Dixon N.E. ( 2002) Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure (Camb.), 10, 535–546. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W. & Kjeldgaard, M. ( 1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Joyce C.M. & Steitz T.A. ( 1994) Function and structure relationships in DNA polymerases. Annu. Rev. Biochem., 63, 777–822. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S. & Thornton J.M. ( 1993) PROCHECK: a program to check the stereochemical quality of protein structure. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lemaire M. & Collart M.A. ( 2000) The TATA-binding protein-associated factor yTafII19p functionally interacts with components of the global transcriptional regulator Ccr4–Not complex and physically interacts with the Not5 subunit. J. Biol. Chem., 275, 26925–26934. [DOI] [PubMed] [Google Scholar]
- Liu H.Y., Toyn J.H., Chiang Y.C., Draper M.P., Johnston L.H. & Denis C.L. ( 1997) DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J., 16, 5289–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.Y., Badarinarayana V., Audino D.C., Rappsilber J., Mann M. & Denis C.L ( 1998) The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J., 17, 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.Y., Badarinarayana V., Audino D.C., Rappsilber J., Mann M. & Denis C. ( 2001) Characterization of CAF4 and CAF16 reveals a functional connection between the CCR4–NOT complex and a subset of SRB proteins of the RNA polymerase II holoenzyme. J. Biol. Chem., 276, 7541–7548. [DOI] [PubMed] [Google Scholar]
- Mian I.S. ( 1997) Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res., 25, 3187–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H., Shimizu-Yoshida Y., Omori A., Iwashita S., Katoh M. & Sakai A. ( 2001) Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev., 15, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M.J., Holley W.R., Chatterjee A. & Mian I.S. ( 1997) The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res., 25, 5110–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D.L., Brick P., Hamlin R., Xuong N.G. & Steitz T.A. ( 1985) Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature, 313, 762–766. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. & Minor W. ( 1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. ( 2000) Maximum-likelihood density modification. Acta Crystallogr. D, 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. & Berendzen J. ( 1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez M.A., Staples R.R., Chen J., Denis C.L. & Parker R. ( 2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell, 104, 377–386. [DOI] [PubMed] [Google Scholar]
- Tucker M., Staples R.R., Valencia-Sanchez M.A., Muhlrad D. & Parker R. ( 2002) Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J., 21, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan P., Chen J., Chiang Y.C. & Denis C.L. ( 2003) Identification of multiple RNA features that influence CCR4 deadenylation activity. J. Biol. Chem., 278, 14949–14955. [DOI] [PubMed] [Google Scholar]
- Zuo Y. & Deutscher M.P. ( 2001) Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res., 29, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]


