Abstract
Ubiquitin and ubiquitin-like modifiers (UBLs) form covalent complexes with other proteins by isopeptide formation between their carboxyl (C)-termini and ɛ-amino groups of lysine residues of acceptor proteins. A hallmark of UBLs is a protruding C-terminal tail with a terminal glycine residue, which is required for ATP-dependent conjugation. Recently, the highly conserved protein HUB1 (homologous to ubiquitin 1) has been reported to function as a UBL following C-terminal processing. HUB1 exhibits sequence similarity with ubiquitin but lacks a C-terminal tail bearing a glycine residue. Here we show that HUB1 can form SDS-resistant complexes with cellular proteins, but provide evidence that these adducts are not formed through covalent C-terminal conjugation of HUB1 to substrates. The adducts are still formed when the C-terminus of HUB1 was altered by epitope tagging, amino-acid exchange or deletion, or when cells were depleted of ATP. We propose that HUB1 may act as a novel protein modulator through the formation of tight, possibly noncovalent interactions with target proteins.
Introduction
Modification of proteins by covalent attachment of ubiquitin occurs universally in eukaryotic cells. Proteins modified by ubiquitin tags are usually targeted for degradation by the proteasome. In other cases, this modification mediates protein sorting or regulates other functions (Hochstrasser, 1996). Eukaryotes also express ubiquitin-related proteins, which fall into two classes (Jentsch & Pyrowolakis, 2000). Proteins of the first category, termed ‘ubiquitin-like modifiers' or UBLs, function analogously to ubiquitin and become covalently conjugated to other proteins via their carboxyl (C)-termini. Proteins of the second class, called ‘ubiquitin-domain proteins' or UDPs, bear protein domains that are related to ubiquitin, but are not conjugated to other proteins.
UBLs (for example SUMO, RUB1/NEDD8, APG12) do not seem to promote proteasomal degradation, but regulate a variety of cellular functions. Like ubiquitin, some UBLs are expressed as inactive precursors; that is, as proteins with C-terminal extensions, which prevent conjugation. These tails, which can either be single amino acids or short peptides, are clipped off by the activity of specific proteases, thereby releasing the active UBL with a C-terminal glycine residue. Other UBLs are already expressed as mature proteins bearing an exposed C-terminal glycine residue (Furukawa et al., 2000). This glycine residue, often part of a di-glycine motif, appears to be crucial for conjugation and de-conjugation from substrates (Wilkinson & Audhya, 1981; Jentsch & Pyrowolakis, 2000). The conjugation pathways for UBLs resemble that of ubiquitin and utilize the same conserved mechanism. The first step involves an activating enzyme (E1), which hydrolyses ATP and forms an E1-UBL thiolester. UBLs are then transferred to conjugating enzymes (E2s), which results in a similar thiolester-linked complex. Finally, the UBLs are covalently attached to the substrates through the formation of an amide bond between their C-termini and a lysine side chain of the substrate protein. An exception is the UBL APG8, which is not attached to proteins but forms an amide bond with an amino group of a lipid (Ichimura et al., 2000).
Recently, a novel ubiquitin-like protein has been identified, which has been termed HUB1 (homologous to ubiquitin 1; also known as UBL5 or Beacon) (Jentsch & Pyrowolakis, 2000; Friedman et al., 2001; Dittmar et al., 2002; Kantham et al., 2003). HUB1 is a small, highly conserved protein, which shares about 22% sequence identity with ubiquitin. However, unlike ubiquitin and UBLs, HUB1 does not possess glycine residues at the C-terminal end but carries a conserved di-tyrosine motif, followed by a single, nonconserved C-terminal residue. Moreover, the HUB1 polypeptide chain is shorter than that of ubiquitin as it lacks four C-terminal residues compared to ubiquitin and UBLs (Fig. 1A). In fact, HUB1 completely lacks an exposed C-terminal tail, which is characteristic for ubiquitin and UBL proteins (Fig. 1B).
Figure 1.
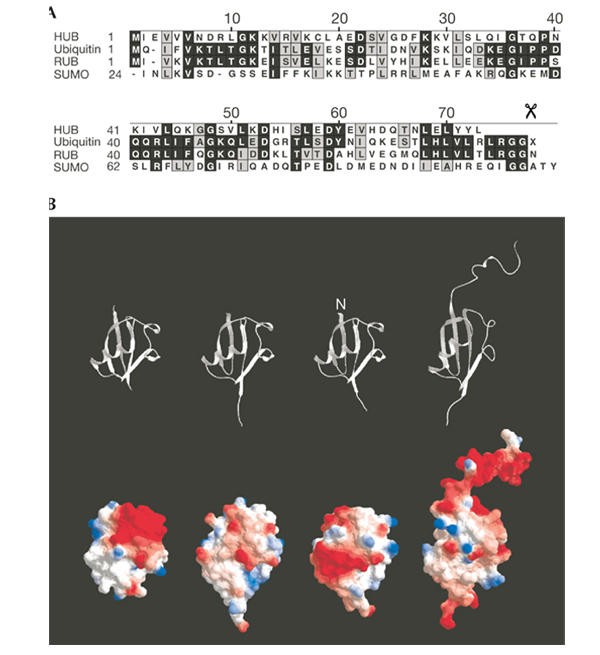
Protein sequences and three-dimensional structures of HUB1 (homologous to ubiquitin 1), ubiquitin and ubiquitin-like modifiers. (A) Sequence alignment of HUB1 with ubiquitin, RUB1 and SUMO from Saccharomyces cerevisiae. The scissors symbol indicates the processing site after the di-glycine motif in ubiquitin, RUB1 and SUMO. (B) Structural ribbon (top) and molecular surface representations (bottom) of HUB1 (S. cerevisiae; Ramelot et al., 2003), ubiquitin (Homo sapiens; Vijay-Kumar et al., 1987), RUB1/NEDD8 (H. sapiens; Whitby et al., 1998) and SUMO-1 (H. sapiens; Bayer et al., 1998).
Finley and co-workers reported recently that HUB1 acts as a covalent modifier in yeast (Dittmar et al., 2002). They also suggested that HUB1 is processed proteolytically, thereby exposing the second tyrosine residue of the conserved di-tyrosine motif at its C-terminus, which is then used for conjugation to substrate proteins. Here we present several lines of evidence, which are in disagreement with these conclusions. In contrast, our data indicate that HUB1 is able to form SDS-resistant protein adducts in the absence of ATP.
Results
Formation of SDS-resistant HUB1–protein adducts
When we expressed epitope-tagged HUB1 in yeast, we observed, in addition to the tagged protein of about 15 kDa, several slower migrating immunoreactive protein bands. These bands, which were absent in extracts from cells that do not express HUB1, persisted even after boiling in SDS-containing sample buffer or incubation with urea or under reducing conditions. Surprisingly, addition of the thiol-reactive agent N-ethylmaleimide (NEM) to the culture medium before harvesting the cells increased strongly the intensity of these slower migrating species in a time- and concentration-dependent manner (Fig. 2A,B). Because conjugation of ubiquitin and UBLs are NEM-sensitive reactions, this finding suggests that the HUB1-containing SDS-resistant adducts are generated by a mechanism dissimilar to ubiquitin/UBL conjugation.
Figure 2.
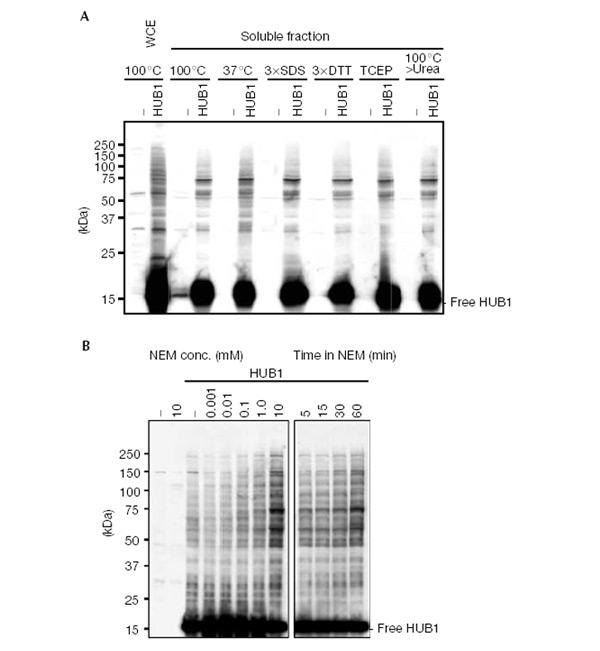
SDS-resistant HUB1–protein adducts. (A) Western blot analysis of hub1Δ cells overexpressing 3HAHUB1. Whole-cell extract (WCE) and a 100,000g supernatant (soluble fraction) were prepared from 3HAHUB1-expressing cells (HUB1) and control cells transformed with an empty plasmid (−). Proteins were denatured in standard sample buffer by boiling for 5 min (100°C) or for 30 min at 37°C. Other samples were denatured by boiling in sample buffers with the following modifications: a threefold higher SDS concentration (3 × SDS), a threefold higher DTT concentration (3 × DTT) and inclusion of tris(2-carboxyethyl)phosphine hydrochloride (TCEP) instead of DTT. One sample was first boiled in standard sample buffer, cooled to room temperature, and 8 M solid urea was added, dissolved and incubated for 30 min at 37°C. A monoclonal anti-HA antibody was used to detect 3HAHUB1. (B) Similar to the above, but WCEs were made after N-ethylmaleimide (NEM) was added directly to the culture medium at the indicated concentrations for 30 min (left panel) and at 10mM for the indicated times (right panel).
Adduct formation does not require HUB1's C-terminus
Dittmar et al. (2002) suggested that HUB1 is conjugated to proteins via the second tyrosine residue of the C-terminal di-tyrosine motif, after the last residue has been removed enzymatically. To investigate the role of HUB1's C-terminal residues, we probed HUB1 variants in a phenotypic complementation assay. HUB1 is not essential for viability (Dittmar et al., 2002). However, we noticed a growth defect of hub1 knockout cells in a specific strain background (Σ1285b; Liu et al., 1993), which could be easily monitored by comparing colony sizes (Fig. 3A). We constructed a truncated version in which the second tyrosine and the last residue were deleted (3HAHUB1ΔYL), and a derivative in which both tyrosine residues were replaced by alanines (3HAHUB1AAL). These constructs, bearing an amino (N)-terminal 3HA tag, were expressed from the genome driven by the HUB1 promoter. When we left the C-terminus intact (3HAHUB1), the protein was able to restore wild-type (WT) growth in hub1Δ cells, indicating that the N-terminal tag does not interfere with HUB1 function (Fig. 3A). Notably, both C-terminal HUB1 variants (3HAHUB1ΔYL and 3HAHUB1AAL) were also able, at least partially, to complement the growth phenotype of the deletion mutant and were able to form SDS-resistant protein adducts similar to WT HUB1 (Fig. 3B). Therefore, the C-terminal residues of HUB1 are not essential for adduct formation and may not even be involved. To address this issue directly, we constructed a HUB1 variant bearing a 3HA tag at its N-terminus and a protein-A tag at its C-terminus (3HAHUB1ProtA). Expression of this protein in yeast gave rise to the free form and additional, slower migrating HUB1-containing adducts (Fig. 4). Apparently as a consequence of the added protein-A tag, the pattern of bands appeared to be shifted to higher molecular mass compared to those generated by 3HAHUB1 (Fig. 4). Notably, we obtained a very similar immunoreactive protein pattern regardless of whether we probed the same sample for either the N-terminal or the C-terminal tag (or when we used a different C-terminal tag; Fig. 4). This indicates that HUB1 within these complexes carries not only the N-terminal but also the C-terminal tag. This finding disproves directly the assumption that the observed adducts are formed through covalent attachment of HUB1 to cellular proteins via HUB1's C-terminus (or one of its tyrosine residues). Moreover, it also indicates that C-terminal processing of HUB1 is not required for HUB1–protein complex formation. Given the known specificity of ubiquitin/UBL-activating enzymes, it seems extremely unlikely that a putative activating enzyme would tolerate a variety of different C-terminal residues or even unrelated sequences (for example, C-terminal protein-A sequences) as substrates.
Figure 3.
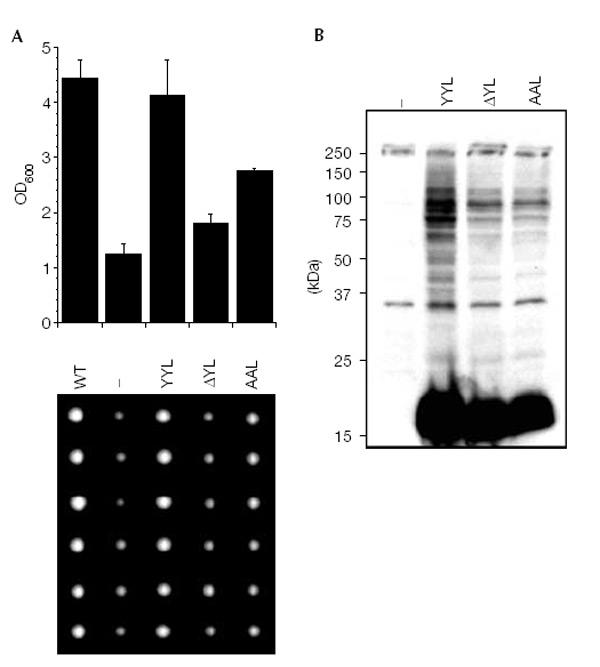
C-terminal HUB1 mutants. (A) The growth of WT Σ1285b cells was compared to hub1Δ (Σ1285b) cells transformed with an empty plasmid (−) or plasmids coding for 3HAHUB1 (YYL), or mutants 3HAHUB1ΔYL (ΔYL) and 3HAHUB1AAL (AAL) under control of the HUB1 promoter. Cells were grown to saturation and then diluted in fresh YPD medium. After 24 h of growth at 30°C, the optical densities were determined at 600 nm (upper panel). Alternatively, single cells from saturated cultures were placed on an agar plate using a micromanipulator and grown at 30 °C until colonies became visible (lower panel). (B) Formation of HUB1–protein adducts with the HUB1 variants described in (A).
Figure 4.
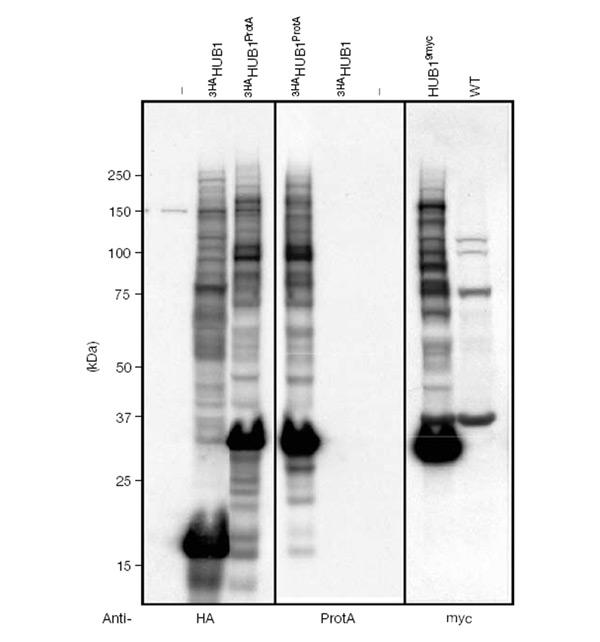
C-terminal HUB1 extensions. Western blot (left and centre) of hub1Δ cells containing empty vector (−) or overexpressing N-terminally HA-tagged HUB1 (3HAHUB1) and N- and C-terminally tagged HUB1 (3HAHUB1ProtA). The same blot was probed with monoclonal anti-HA (left) or anti-ProtA antibodies (centre). A mirror image of the blot is shown in the central panel for easy comparison. The Western blot on the right is of hub1Δ cells expressing C-terminally 9myc-tagged HUB1 from its own promoter and probed with a monoclonal anti-myc antibody. The monoclonal anti-HA antibody used does not recognize the ProtA tag.
Similar to the observations of Dittmar et al. (2002), we also observed that C-terminally truncated HUB1 variants lacking the tyrosine residues are less functional than WT HUB1 (Fig. 3). However, we do not interpret this ineffectiveness as evidence for the importance of tyrosine residues for conjugation. Rather, we suggest that the truncations might have a profound influence on the folding and half-life of the protein. Indeed, deletion of the last three residues of HUB1 results in an unstable protein, which cannot be detected in Western blots (data not shown). Furthermore, the recently solved NMR structure of HUB1 suggests that the C-terminal residues of HUB1 contribute to the globular ubiquitin-like fold of HUB1 (McNally et al., 2003; Ramelot et al., 2003).
ATP-independent formation of HUB1 adducts
UBL conjugation is an ATP-dependent reaction (Fig. 5A, upper right panel). In contrast, ATP depletion had no effect on the formation of HUB1 adducts (Fig. 5A, upper left panel). Moreover, NEM-induced elevation of steady-state levels of HUB1 adducts was not diminished by ATP depletion (Fig. 5A, lower left panel).
Figure 5.
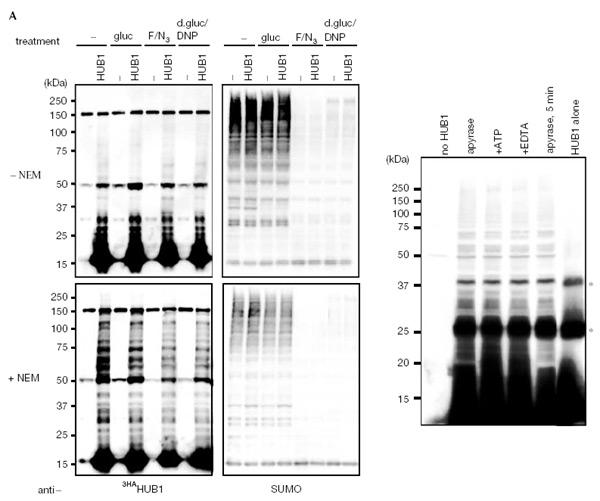
ATP-independent HUB1–protein adduct formation. (A) Western blots of extracts from cells expressing HA-tagged HUB1. Cells were grown in YP medium lacking (−) or containing glucose (gluc). For ATP depletion, cells were grown either in NaF and NaN3 (F/N3) or deoxyglucose and dinitrophenol (d-gluc/DNP) containing YP medium. Samples for the bottom panels were treated additionally with N-ethylmaleimide (NEM) for 30 min. The blots were probed either for HUB1 with anti-HA antibodies (left panels) or for SUMO/SMT3 using affinity-purified SUMO antibodies. Note that ATP depletion leads to a loss of SUMO conjugates. (B) In vitro adduct formation of purified recombinant HUB1 with proteins from yeast extracts (see Methods). The extracts were treated with the indicated reagents (25 U ml−1 apyrase, 5 mM EDTA, 10 mM NEM) and incubated for 30 min at 30°C or for 5 min on ice. The bands labelled with asterisks are HUB1 dimers and trimers formed in vitro even without added extract (right lane). Free HUB1 runs at the bottom of the gel.
A potential explanation of the previous results is that the observed HUB1–protein adducts are formed nonenzymatically. To address this possibility, we investigated HUB1 adduct formation in vitro by adding purified recombinant HisHAHUB1 to a yeast extract. We either depleted ATP from the extract by treatment with apyrase, or added EDTA, or we replenished the extract with ATP prior to HisHAHUB1 addition. As shown in Fig. 5B, HUB1 adduct formation was detectable in vitro as well. In addition to a pattern of HUB1–protein adducts, HUB1 pronouncedly formed two major adducts in vitro. These bands represent SDS-resistant HUB1 dimers and trimers as they are also formed with purified, recombinant HUB1 in vitro (Fig. 5B, right lane). HUB1 adduct formation in vitro was apparently ATP-independent and insensitive to treatment with EDTA. Notably, adduct formation was evidently independent of the incubation time and also occurred on ice (Fig. 5B). These results argue strongly against a ubiquitin-like conjugation mechanism for HUB1, but rather suggest that they are formed by a different, possibly nonenzymatic mechanism.
Discussion
HUB1 has been reported to function as a UBL (Dittmar et al., 2002). Although we cannot rule out this possibility formally, our results are in clear conflict with this conclusion. Dittmar et al. (2002) identified two proteins, SPH1 and HBT1, as substrates of HUB1 modification. However, the assays used by these authors could not distinguish between covalent and noncovalent HUB1 interactions. Moreover, we could not confirm conjugation of these proteins to HUB1 by western blotting or immunoprecipitations using the reagents and protocols of Dittmar et al. (data not shown), but we cannot exclude the possibility that a very small fraction of these proteins binds HUB1. We also explored the possibility that HUB1 adducts are formed through disulphide linkages as HUB1 contains a cysteine residue. However, HUB1–protein adducts were still detectable after boiling in DTT-containing buffers (Fig. 2A). Moreover, a HUB1 mutant protein, in which the cysteine residue was replaced by alanine (HUB1C−A), formed HUB1–protein adducts in vivo similar to WT HUB1 (supplementary figure 1 online). Being able to detect HUB1 adducts after their separation from free HUB1 by gel filtration (supplementary figure 2 online) demonstrates that the detection of these adducts does not depend on the presence of a large excess of HUB1 in the sample.
Our data indicate that HUB1 is a UDP and that the HUB1–protein adducts arise through probably noncovalent protein–protein interactions. Several other proteins can form SDS-resistant protein complexes, for example, SNARE proteins, phage tail spike proteins, nucleoporines, prion-like proteins and protein adducts formed under stress conditions (Goldenberg et al., 1982; Fasshauer et al., 1998; Kopito, 2000; Favreau et al., 2001; Speransky et al., 2001; Waelter et al., 2001). An attractive speculation is that HUB1 may form tight associations with proteins or protein complexes in order to regulate their function or stability. Indeed, gel filtration experiments indicate that HUB1–protein adducts may be part of larger protein complexes (supplementary figure 2 online). In such complexes, HUB1 may attract specific enzymes, for example chaperones, for complex disassembly. Our observation that the steady-state level of HUB1 adducts in living cells is induced strongly by NEM may in fact point to a role for NEM-sensitive enzymes, for example AAA-type ATPases, in this process. Interesting candidates are ubiquitin-selective chaperones like CDC48/p97 (Rape et al., 2001) or the 19S cap of the proteasome. Possibly because of the large number of different adducts and their low abundance, we were so far unsuccessful in purifying HUB1-containing complexes from cells for further analysis. It would be interesting to see whether some of the binding partners of HUB1 are indeed components of the ubiquitin system.
Methods
Cloning and yeast techniques
Yeast techniques have been described previously (Guthrie & Fink, 1991). Strains are derivatives of DF5 (Finley et al., 1987) or haploid cells of Σ1285b (Liu et al., 1993). A DF5 hub1Δ mutant was created by gene replacement using LEU2, and the Σ1285b hub1Δ deletion by PCR-based replacement (Knop et al., 1999) using a kanMX6 resistance gene. 3HA-tagged HUB1 (3HAHUB1) were constructed in YIplac211 and placed under the control of the GAL1/10 promoter, the ADH1 promoter or its own promoter by inserting a 170 bp fragment of the 5′ untranslated region of the HUB1 gene. C-terminal variants of HUB1 were created by introducing a stop codon after the codons for the first Y of the YY motif (HUB1ΔYL) or by changing the YY codons into AA codons (HUB1AAL). A strain expressing HUB19myc was obtained by chromosomal tagging of the HUB1 gene in DF5. Cells overexpressing HUB1 with both N-terminal and C-terminal tags (3HAHUB1ProtA) under control of the GAL1-10 promoter were obtained by chromosomal tagging (Knop et al., 1999) of 3HAHUB1 encoded by an integrated plasmid in a DF5 hubΔ deletion strain. For expression of HisHA-tagged HUB1 in Escherichia coli, a DNA fragment coding for HAHUB1 was inserted into the pQE30 vector (Qiagen).
Protein techniques and antibodies Antibodies used for western analysis are monoclonal mouse anti-HA (12CA5), monoclonal mouse anti-myc (9E10), polyclonal peroxidase-coupled rabbit anti-protein-A antibodies and affinity-purified polyclonal rabbit anti-SUMO antibodies (Hoege et al., 2002). Peroxidase-coupled goat anti-rabbit IgG and goat anti-mouse IgG antibodies were used as secondary antibodies (Jackson Immunoresearch).
Whole-cell extracts (WCE) for western blots were prepared from yeast grown in rich media containing 2% glucose or 2% galactose to induce expression from the GAL promoter. To induce HUB1–protein adducts, NEM was added directly to the culture medium prior to harvesting the cells at an OD600 of 1–2. Cell pellets were then processed by NaOH/β-mercaptoethanol treatment, TCA precipitation and solubilization of the precipitated proteins in high-urea (HU) sample buffer as described previously (Knop et al., 1999). Alternatively, to test for the stability of HUB1–protein adducts, cells were harvested, treated with zymolyase and lysed in a hypotonic buffer (20 mM HEPES, pH 7.2, 150 mM potassium acetate, 2 mM MgCl2) containing complete protease inhibitors (Roche). The lysate was centrifuged at 100,000g at 4 °C for 1 h. The supernatant was mixed with (Laemmli) sample buffer (final concentrations: 60 mM Tris–HCl, pH 6.8, 2% SDS, 100 mM DTT, 10% glycerol, 0.01% bromophenol blue) or variations thereof, followed by incubation at 100 or 37 °C for 5 or 30 min, respectively. Samples were analysed by SDS–PAGE and western blotting using standard procedures.
For the in vitro formation of HUB1–protein adducts, HisHAHUB1 was expressed in E. coli M15 cells (Qiagen) and recombinant protein was purified by chromatography on NiNTA-agarose (Qiagen). After elution of HUB1 from the beads, the preparation was dialysed against 20 mM HEPES (pH 7.2) and 150 mM NaCl, and concentrated by ultrafiltration. Cell extracts were prepared from DF5 hub1Δ cells by lysis of spheroplasts in lysis buffer (see above) and centrifugation at 20,000g for 30 min. ATP in the supernatant was depleted and/or inactivated by treatment that routinely prevents ubiquitin or SUMO conjugation, that is, incubating the sample at 30 °C for 15 min with 25 U ml−1 apyrase (Sigma) or by adding 5 mM EDTA. For other experiments, 2 mM fresh ATP was added. Then, purified HisHAHUB1 was added and the mixture containing 5 μg HisHAHUB1 and about 100 μg cellular protein from the extract was incubated for 30 min at 30 °C. Samples were boiled after the addition of sample buffer and analysed by SDS–PAGE and western blotting.
ATP depletion ATP depletion in yeast cells was achieved by two different treatments. Logarithmically growing cells were pelleted, washed twice in YP medium lacking glucose, and then resuspended in YP medium containing either 20 mM NaF and 20 mM NaN3, or alternatively in 20 mM dinitrophenol and 0.2 mM deoxyglucose. Cells were incubated on a shaker for 4 h at 30 °C. Cell extracts were prepared by the TCA lysis method (Knop et al., 1999; see above) either directly or after adding 10 mM NEM to the culture for 30 min. Samples were analysed by SDS–PAGE and western blotting.
Monitoring the growth of yeast strains Σ1285b WT cells and Σ1285b hub1Δ cells expressing 3HA-tagged HUB1 or C-terminal mutants from an integrated plasmid were grown to saturation in YPD. Fresh cultures were inoculated and grown overnight in YPD at 30 °C. The next morning, OD600 was determined and monitored hourly when the cells had reached an OD600 of approximately 0.1.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/4-7400025-s1.pdf).
Supplementary Material
Supplementary Figures
Acknowledgments
The HUB1 gene was discovered in the lab of S.J. by D. Liakopoulos. We thank him for discussions and his contribution to this study. We thank D. Kempe for technical assistance, G. Fink for the gift of the Σ1285 strain, and D. Finley and G. Dittmar for HUB1 materials. S.J. was supported by Fonds der Chemischen Industrie.
References
- Bayer P., Arndt A., Metzger S., Mahajan R., Melchior F., Jaenicke R. & Becker J. ( 1998) Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol., 280, 275–286. [DOI] [PubMed] [Google Scholar]
- Dittmar G.A., Wilkinson C.R., Jedrzejewski P.T. & Finley D. ( 2002) Role of a ubiquitin-like modification in polarized morphogenesis. Science, 295, 2442–2446. [DOI] [PubMed] [Google Scholar]
- Fasshauer D., Eliason W.K., Brunger A.T. & Jahn R. ( 1998) Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry, 37, 10354–10362. [DOI] [PubMed] [Google Scholar]
- Favreau C., Bastos R., Cartaud J., Courvalin J.C. & Mustonen P. ( 2001) Biochemical characterization of nuclear pore complex protein gp210 oligomers. Eur. J. Biochem., 268, 3883–3889. [DOI] [PubMed] [Google Scholar]
- Finley D., Özkaynak E. & Varshavsky A. ( 1987) The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell, 48, 1035–1046. [DOI] [PubMed] [Google Scholar]
- Friedman J.S., Koop B.F., Raymond V. & Walter M.A. ( 2001) Isolation of a ubiquitin-like (UBL5) gene from a screen identifying highly expressed and conserved iris genes. Genomics, 71, 252–255. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Mizushima N., Noda T. & Ohsumi Y. ( 2000) A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem., 275, 7462–7465. [DOI] [PubMed] [Google Scholar]
- Goldenberg D.P., Berget P.B. & King J. ( 1982) Maturation of the tail spike endorhamnosidase of Salmonella phage P22. J. Biol. Chem., 257, 7864–7871. [PubMed] [Google Scholar]
- Guthrie C. & Fink G.R. (eds) ( 1991) Guide to Yeast Genetics and Molecular biology. Methods in Enzymology, Vol 194. Academic, San Diego, California, USA. [PubMed] [Google Scholar]
- Hochstrasser M. ( 1996) Ubiquitin-dependent protein degradation. Annu. Rev. Genet., 30, 405–439. [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.-L., Pyrowolakis G. & Jentsch S. ( 2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature, 419, 135–141. [DOI] [PubMed] [Google Scholar]
- Ichimura Y. et al. ( 2000) A ubiquitin-like system mediates protein lipidation. Nature, 408, 488–492. [DOI] [PubMed] [Google Scholar]
- Jentsch S. & Pyrowolakis G. ( 2000) Ubiquitin and its kin: how close are the family ties? Trends Cell Biol., 10, 335–342. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K. & Schiebel E. ( 1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Kopito R.R. ( 2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol., 10, 524–530. [DOI] [PubMed] [Google Scholar]
- Liu H., Styles C.A. & Fink G.R. ( 1993) Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science, 262, 1741–1744. [DOI] [PubMed] [Google Scholar]
- McNally T., Huang Q., Janis R.S., Liu Z., Olejniczak E.T. & Reilly R.M. ( 2003) Structural analysis of UBL5, a novel ubiquitin-like modifier. Protein Sci., 7, 1562–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramelot T.A., Cort R.J., Yee A.A., Semesi A., Edwards A.M., Arrowsmith C.H. & Kennedy M.A. ( 2003) Solution structure of the yeast ubiquitin-like modifier protein Hub1. J. Struct. Funct. Genom., 4, 25–30. [DOI] [PubMed] [Google Scholar]
- Rape M., Hoppe T., Gorr I., Kalocay M., Richly H. & Jentsch S. ( 2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48UFD1/NPL4, a ubiquitin-selective chaperone. Cell, 107, 667–677. [DOI] [PubMed] [Google Scholar]
- Speransky V.V., Taylor K.L., Edskes H.K., Wickner R.B. & Steven A.C. ( 2001) Prion filament networks in [URE3] cells of Saccharomyces cerevisiae. J. Cell Biol., 153, 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg C.E. & Cook W.J. ( 1987) Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol., 194, 531–544. [DOI] [PubMed] [Google Scholar]
- Waelter S., Boeddrich A., Lurz R., Scherzinger E., Lueder G., Lehrach H. & Wanker E.E. ( 2001) Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell, 12, 1393–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby F.G., Xia G., Pickart C.M. & Hill C.P. ( 1998) Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem., 273, 34983–34991. [DOI] [PubMed] [Google Scholar]
- Wilkinson K.D. & Audhya T.K. ( 1981) Stimulation of ATP-dependent proteolysis requires ubiquitin with the COOH-terminal sequence Arg-Gly-Gly. J. Biol. Chem., 256, 9235–9241. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures


