Abstract
The ubiquitous nuclear adaptor protein LIM-domain-binding protein 1 (Ldb1) was originally identified as a cofactor for LIM-homeodomain and LIM-only (LMO) proteins that have fundamental roles in development. In parallel, Ldb1 has been shown to have essential functions in diverse biological processes in different organisms. The recent targeting of this gene in mice has revealed roles for Ldb1 in neural patterning and development that have been conserved throughout evolution. Furthermore, the elucidation of the three-dimensional structures of LIM–Ldb1 complexes has provided insight into the molecular basis for the ability of Ldb1 to contact diverse LIM-domain proteins. It has become evident that Ldb1 is a multi-adaptor protein that mediates interactions between different classes of transcription factors and their co-regulators and that the nature of these complexes determines cell fate and differentiation.
Introduction
Gene regulation is in part determined by the interactions of distinct cofactor complexes with transcription factors. It is becoming increasingly clear that the same transcription factor/cofactor can interact with different higher-order protein complexes, allowing it to regulate a range of processes in distinct cell types and organs. One important cofactor that is involved in the assembly of multi-protein complexes is LIM-domain-binding protein 1 (Ldb1, also known as CLIM2 and NLI). LIM domains are double zinc-binding motifs that are found in diverse proteins (Dawid et al., 1998), and Ldb1 was first discovered because of its ability to bind LIM-homeodomain (LIM-HD) and LIM-only (LMO) proteins (Agulnick et al., 1996; Bach et al., 1997; Jurata et al., 1996; Visvader et al., 1997). Ldb1 has no known enzymatic or nucleic-acid-binding function. Rather, it seems to act as a specific protein-binding interface.
Ldb proteins are found in species ranging from Caenorhabditis elegans to man, and are widely expressed throughout embryonic and adult tissues. Nematodes, Drosophila and Xenopus each bear a single ldb gene, mammals carry two Ldb genes, and zebrafish have four ldb genes, consistent with genome duplication events. On the basis of sequence homology, the Ldb family can be divided into two main groups, Ldb1 and Ldb2, with Ldbs from Drosophila (also known as Chip) and nematodes being the most divergent (Fig. 1A). Alternative splice variants have been identified for the ldb genes. All known Ldb proteins carry a conserved nuclear localization sequence and are found predominantly in the nucleus (Jurata et al., 1996). Sequence comparisons between species indicate the presence of two other well-conserved domains: an amino-terminal homodimerization domain and a carboxy-terminal LIM interaction domain (LID). The LID interacts with LIM-HD and LMO proteins, which have important roles in cell-fate determination, tissue development and cytoskeleletal organization. The physiological relevance of LIM–Ldb interactions has now been established in several developmental systems. This review summarizes recent data indicating that the many cellular functions of Ldb1 reflect its remarkable capacity to participate in different protein complexes.
Figure 1.

Species relationships and domain organization of LIM-domain-binding proteins. (A) Phylogenetic tree constructed from the aligned protein sequences. Nematodes and Drosophila carry one, vertebrates two, and zebrafish four LIM-domain-binding protein (Ldb) genes. (B) Schematic diagram showing domain organization. The dimerization domain (DD), nuclear localization sequence (NLS) and LIM interaction domain (LID) are shown for mouse Ldb1 and Ldb2, Drosophila Ldb/Chip and Caenorhabditis elegans LDB (using the numbering of the Ldb1A splice variant). The other interaction domain (OID, boxed) comprises residues 438–456 in Drosophila Ldb/Chip (Torigoi et al., 2000) and the Ldb1/Chip conserved domain (LLCD, dashed box) comprises residues 201–249 in Ldb1 and 387–426 in Chip (Van Meyel et al., 2003).
Developmental roles of Ldb1 in different organisms
Ldb1 has fundamental roles in development, many of which have been conserved across species. In Xenopus, Ldb1 was first shown to be necessary for the function of the LIM-HD protein X-Lim1 (Agulnick et al., 1996). Co-injection of X-Lim1 and Ldb1 into Xenopus embryos induced secondary axis formation, whereas neither protein alone showed this activity. In Drosophila, Chip/Ldb was initially described as a facilitator of enhancer–promoter interactions (Morcillo et al., 1997) and has been shown to be essential for normal segmentation, neuronal axon guidance and wing morphology. Chip functionally interacts with the LIM-HD protein Apterous (Ap; see below) in a dosage-dependent manner to regulate neuronal specification and wing disc development. Single-stranded DNA-binding proteins (Ssdp) have also been recently shown to interact with Chip and to regulate the activity of Chip–Ap protein complexes. Moreover, hypomorphic alleles of the ssdp gene lead to wing defects that mirror those seen in Chip and Ap mutants (Chen et al., 2002; van Meyel et al., 2003).
In C. elegans, LIM-HD proteins have been implicated in the development and function of neuronal, gonadal and vulval cells (Cassata et al., 2000; Gupta et al., 2003). LDB-1 modulates some but not all of these functions. This adaptor protein is essential for specific neuronal functions including motility and touch (Cassata et al., 2000) and seems to have a role in determining post-mitotic neuronal function rather than early specification. A requirement for LDB-1 in vulval morphogenesis has been shown, and this function is mediated through its direct binding to LIN-11 (Gupta et al., 2003). Ldbs have also been ascribed several roles in zebrafish development, many of which are linked to LIM-HD function (Becker et al., 2002). Forced expression of a dominant-negative Ldb1, which prevents all Ldb–LIM interactions, leads to the inhibition of zebrafish eye and midbrain–hindbrain boundary development. A crucial role for Ldb1 has also been established in axonal outgrowth of peripheral, trigeminal, sensory and motor neurons, which parallels the phenotypes observed in LIM-HD knockout mice.
Finally, ablation of the Ldb1 gene in mice has firmly established a key role for Lbd1 in diverse developmental processes (Mukhopadhyay et al., 2003). Ldb1-null mice display severe anterior–posterior patterning defects, leading to the truncation of head structures, posterior axis duplication and a lack of heart anlage. Although there is overlap between these phenotypes and those seen in LIM-HD knockout mice, distinct defects are also evident in the Ldb1 mutants. For example, these mice display defects in the generation of early heart precursor cells, and this might reflect interactions between Ldb1 and proteins such as the homeodomain protein Nkx2.5. Furthermore, defects in patterning along the anterior–posterior axis may be attributable to decreased expression of inhibitors of Wnt signalling. The plethora of phenotypes observed in Ldb1-deficient embryos indicates that Ldb1 is a cofactor that is required for the normal function of many different transcription factors. In contrast with findings from Drosophila, in which embryos heterozygous for Chip die without wing-segment formation or at the larval stage (Morcillo et al., 1996), mice heterozygous for Ldb1 appear normal. This observation in mice suggests that a level of redundancy has evolved within the Ldb family to protect mammals from gene dosage effects during normal development.
The stoichiometry of Ldb–LIM protein complexes
The absolute levels of Ldb family members and their associated proteins seem to be important for proper cellular function. This has been elegantly demonstrated using wing development in Drosophila as the model system. Either a deficiency or an excess of Chip in Drosophila mimics the Ap lack-of-function phenotype (Fernandez-Funez et al., 1998; Rincon-Limas et al., 2000). The level of Ap in these Chip mutants remains normal, indicating that Ap function is dependent on the correct amount of cofactor expression. Importantly, phenotypes that arise from excess Chip can be rescued by mutations that promote Ap gain-of-function. However, defects elicited by Chip mutants that lack the ability to bind LIM domains, and therefore cannot bind Ap, cannot be rescued. The functional complex in these cells is thought to be a Chip–Ap tetramer, comprised of two molecules of each protein (Fig. 2A; Milan & Cohen, 1999). This stoichiometry is also important for determining axon guidance in Drosophila (van Meyel et al., 2000) in which excess Chip leads to the formation of inactive complexes (Fig. 2B). Ap activity is also governed by the expression of Drosophila LMO, which competes with Ap for binding to Chip (Fig. 2C; Milan et al., 1998; Shoresh et al., 1998; Zeng et al., 1998).
Figure 2.
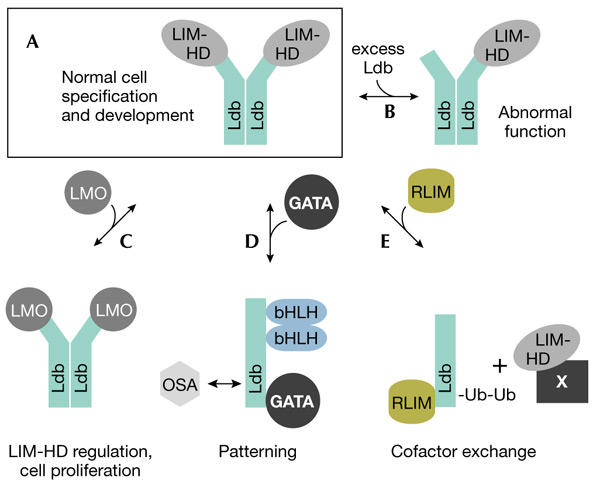
The stoichiometry and composition of Ldb1 complexes. (A) Ldb1–LIM-HD tetramers have major roles in cellular development. (B) Incorrect stoichiometry, for example, by overexpression of Ldb1, leads to abnormal development. (C) LMO proteins regulate LIM-HD activity by effectively competing with LIM-HDs for binding to the LID. Overexpression of LMOs can lead to uncontrolled cell proliferation. (D) Other active complexes involve monomeric Ldb1. (E) Cofactor exchange by RLIM can regulate LIM-HD activity. bHLH, basic helix–loop–helix; Ldb1, LIM-domain-binding protein 1; LIM-HD, LIM-homeodomain; LMO, LIM-only protein; Ub, ubiquitin.
The composition and stoichiometry of Ldb1-containing complexes in mammalian systems are not as well defined. Despite no apparent defects in Ldb1 heterozygous mice, the absolute levels of Ldb and LIM-domain proteins are important for the normal function of specific cell types. The overexpression of Ldb1 in either immature erythroid cells or mammary epithelial cells blocks their terminal differentiation (Visvader et al., 1997, 2001). Similarly, forced expression of LMO2 in erythroid or LMO4 in mammary epithelial cells markedly curtails their differentiation. These findings indicate that one function of LMO–Ldb1 complexes is to maintain the proliferative rather than differentiative state. Changes in the stoichiometry of these complexes may perturb the normal pathways acting within these cells and promote the immature phenotype. For example, overexpression of LMO1 or LMO2 is associated with acute T-cell leukaemia (Rabbitts, 1998). Whether Ldb1 is also involved in haemopoietic malignancies is not known. Interestingly, however, overexpression of Ldb1 has been reported in LMO1-induced T-cell leukaemia (Valge-Archer et al., 1998) and squamous-cell carcinomas of the oral cavity (Mizunuma et al., 2003).
Additional functions of LIM-domain-binding proteins
Drosophila embryos that lack Chip activity are devoid of segments (Morcillo et al., 1997). However, there are no known LIM-domain proteins that control segmentation, suggesting that Chip interacts with other factors during early embryogenesis to alter chromatin structure and regulate enhancer activity. The observation that Chip colocalizes with polytene chromosomes supports this idea and indeed, Chip has been shown to interact functionally with a number of regulatory proteins in Drosophila. These include the homeodomain proteins Bicoid (Bcd) and Fushi tarazu (Ftz), and proteins involved in enhancer–promoter interactions, Suppressor of Hairy wing (SuHw) and Modifier of Mdg4 (Mod(Mdg4)) proteins (Torigoi et al., 2000). Chip also forms a complex with the GATA factor Pannier and the Achaete–Scute complex–Daughterless (ASC–Da) bHLH (basic helix–loop–helix) heterodimer (Ramain et al., 2000). This complex increases enhancer–promoter interactions, leading to the activation of genes required for proneural development and thorax patterning. Ap is thought to antagonize these functions by competing with Pannier for Chip, and the precise stoichiometry between these three proteins determines normal patterning. Moreover, recruitment of the negative regulator Osa to this complex (Fig. 2D) results in the repression of proneural development (Heitzler et al., 2003).
Ldb1 as a transcriptional modulator
There is increasing evidence that LIM-domain proteins and Ldb family members can exert both positive and negative regulatory influences on transcription. The LIM domains in LIM-HD transcriptional regulators can inhibit the DNA-binding activity of the HD (Sanchez-Garcia et al., 1993), but can also increase the transcriptional activity of LIM-HDs in synergy with other classes of transcription factors (Bach et al., 1997; German et al., 1992). Likewise, Ldb2/CLIM1 and Xenopus Ldb1 mediate the synergistic activation of specific promoters by LIM-HD and Otx-HD transcription factors (Bach et al., 1997; Mochizuki et al., 2000). Conversely, Ldb1 has been shown to repress the synergistic activation of the insulin enhancer by Lmx1 and E47 (Jurata & Gill, 1997). Although the mechanisms underlying these observations are not known, the functional effects of the Ldb proteins seem to be dependent on the partners with which they liase.
Ldb1 is itself subject to regulation. RLIM, a protein that contains a RING domain with E3 ubiquitin ligase activity, is able to interact with and ubiquitinate Ldb cofactors bound to LIM-HD proteins, and thus target Ldb1 for degradation by the 26S proteasome (Ostendorff et al., 2002). Moreover, Ldb1 and LIM domains compete for RLIM binding, providing a mechanism for cofactor exchange on LIM-HD proteins (Fig. 2E).
The dimerization domain of Ldb1
Most binding functions of Ldbs can be ascribed to specific structural domains (Table 1). Their homodimerization through an N-terminal domain (Fig. 1B) triggers the activation of LIM-HDs. The effective dimerization of LIM-HDs (Fig. 3A) might facilitate binding to multiple homeoboxes, which could either enhance DNA binding or bring distal sites into close proximity. HDs from LIM-HD proteins tend to be biologically inactive in the absence of either their LIM domains or Ldbs, but can be rescued in vivo by chimaeras in which the HD is fused to the dimerization domain of Ldb1 (Thaler et al., 2002; van Meyel et al., 2000). Thus, an Ldb dimer has the potential to bind different LIM-containing proteins (Jurata et al., 1998). For example, this dimer is thought to form the nucleus of an Ldb–LIM-HD hexamer, in which Isl1 binds Ldb1 directly, and the Lhx3 LIM domains bind the Isl-HD (Fig. 3B; Thaler et al., 2002). The ability of Ldb1 to act in a combinatorial manner with LIM-HDs is likely to contribute to the specification and development of a large number of cell types, particularly in the central nervous system (Thaler et al., 2002). By contrast, the activation of Pannier by ASC and the formation of the Pannier–Chip–ASC–Da complex is thought to require a Chip monomer (Fig. 2D; Ramain et al., 2000).
Table 1.
Binding partners of LIM-domain-binding protein 1
| Protein | Function | Reference |
|---|---|---|
| Binds to the Ldb1 dimerization domain (DD) | ||
| Pannier | Proneural development and thorax patterning | Ramain et al., 2000 |
| Osa | Repression of proneural development | Heitzler et al., 2003 |
| RLIM | Cofactor exchange, neuronal development | Ostendorff et al., 2002 |
| Binds to the Ldb1 LIM interaction domain (LID) | ||
| LIM-HD (for example, Isl, Lhx, Lmx) | Cell specification and development | Reviewed in Bach, 2000; Hobert & Westphal, 2000 |
| LMO | Cell proliferation | Reviewed in Bach, 2000 |
| ASC–Da heterodimer | Proneural development, thorax patterning | Ramain et al., 2000 |
| Binds to the other interaction domain (OID) | ||
| SuHw, Modifier of Mdg4 | Enhancer–promoter communication | Gause et al., 2001; Torigoi et al., 2000 |
| Bcd, Ftz | Segmentation in Drosophila | Torigoi et al., 2000 |
| Chip | Homodimerization | Torigoi et al., 2000 |
| Binds to Ldb1/Chip conserved domain (LLCD) | ||
| Ssdp | Axis induction, wing development | Chen et al., 2002; Van Meyel et al., 2003 |
| Undefined binding site on Ldb1 | ||
| LIN-11 | Vulval morphogenesis | Gupta et al., 2003 |
ASC, Achaete–Scute complex; Bcd, Bicoid; Da, Daughterless; Ftz, Fushi tarazu; HD, homeodomain; LMO, LIM-only protein; Ssdb, single-stranded DNA-binding proteins; SuHw, Suppressor of Hairy wing.
Figure 3.
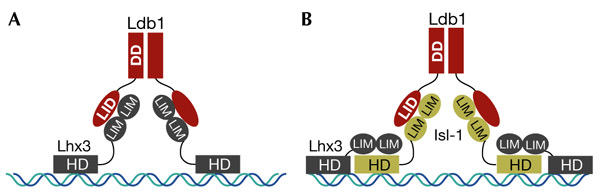
Stoichiometry and binding domains of Ldb1–LIM-HD complexes. (A) Ldb1–Lhx3 tetrameric complex in V2 interneuron development. (B) Ldb1–Isl-1–Lhx3 hexameric complexes in motor-neuron development. DD, dimerization domain; HD, homeodomain; Lbd1, LIM domain binding protein 1; LID, LIM interaction domain.
The LIM interaction domain: LID
The shortest region of Ldb1 that has been shown to be sufficient for binding LIM domains comprises residues 300–338 of mouse Ldb1 (Jurata & Gill, 1997). The recently solved structures of the LIM domains from LMO2 and LMO4 in complex with the LID (Deane et al., 2003) show that residues 333–340 of Ldb1 do not form part of the interaction interface. Protein sequence alignments of Ldbs show high homology for residues 300–332 but not for 333–340, suggesting that the LID may be more accurately defined by this shorter region (Cassata et al., 2000; Jurata & Gill, 1997).
LIM domains are protein-binding motifs that are found in proteins with diverse functions, ranging from transcription factors to regulators of the actin cytoskeleton. This motif binds a number of different protein types, including bHLH and zinc-finger transcription factors, protein kinases and other LIM-containing proteins, in addition to the Ldbs (reviewed by Bach, 2000). LIM proteins have been classified into three groups according to their sequence similarity and function (Dawid et al., 1998). The Ldb-LID interacts only with LMO and LIM-HD proteins from group 1 LIM proteins, but does not bind LIM domains from other groups. Nor does it bind the group 1 protein LIM-kinase. The selective binding of Ldbs to LMOs and LIM-HD proteins reflects the importance of these cofactors in regulating transcriptional processes mediated by the nuclear LIM proteins. However, the sequences of LIM domains from LMO and LIM-HD proteins vary. How can a relatively short peptide sequence, which shows both high affinity and specificity, recognize a subset of proteins that have a poorly defined consensus sequence? Structural studies have provided important insights into this question.
The LID is largely unstructured in solution (Deane et al., 2003). On binding to the N-terminal LIM domains (LIM1) of LMO2 and LMO4, the LID takes up an extended conformation (Fig. 4A), stretching along the face of the LIM domain and adding an additional β-strand to a β-hairpin found in LIM1 (Deane et al., 2003). Thus, the interaction is mediated by main-chain hydrogen bonds, which suggests a mechanism for the interaction of LID with a range of LIM domains. Specificity and stability in the LMO–Ldb1 complex come from both electrostatic interactions distal to, and hydrophobic side-chain–side-chain interactions near to, the β-sheet in LID. A few of these bulky side chains are lacking in LIM-kinase (Fig. 4B), and mutation of those LMO4 residues to the corresponding LIM-kinase residues abrogates Ldb binding. Thus, Ldb–LIM1 binding is mediated by a combination of main-chain hydrogen bonding, hydrophobic and electrostatic interactions. It seems likely that similar modes of binding will be seen for the binding of Ldb-LIDs to other LIM domains.
Figure 4.
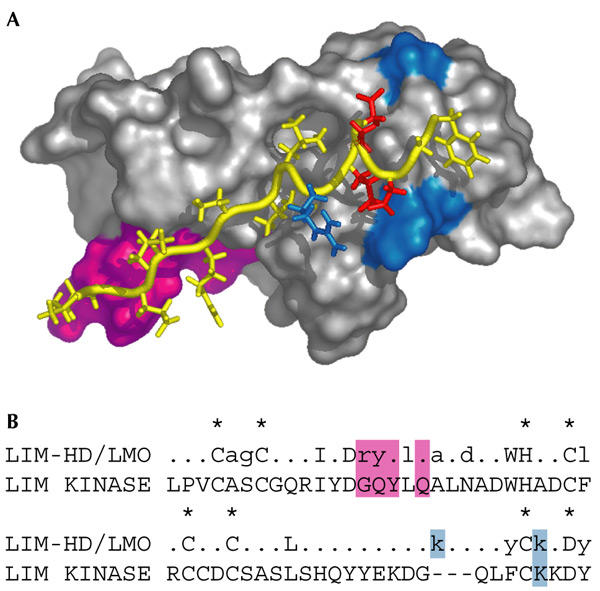
Recognition of LIM domains by Ldb–LID. (A) Structure of a complex formed by LMO4-LIM1 (shown in surface representation) and the Ldb1–LID (in yellow). Side chains involved in electrostatic interactions are red (negative) and blue (positive). Residues in group 1 LIM domains that specify binding to Ldb are magenta. (B) Consensus sequence of the LIM1 domains from group 1 proteins (fully conserved residues are upper case, highly conserved residues are lower case) and the sequence of LIM-kinase are shown. Zinc-binding residues are marked by an asterisk. Boxed residues are coloured as in A. Ldb1, LIM-domain-binding protein 1; LID, LIM interaction domain; LIM-HD, LIM-homeodomain; LMO, LIM-only protein.
Other interaction domains: OID and LLCD
Additional interaction domains have been described for Ldb1 and Chip. The other interaction domain, OID, overlaps with the nuclear localization sequence and has been shown to bind the homeodomain proteins, Bcd and Ftz, as well as SuHw and Mod(Mdg4) proteins (Torigoi et al., 2000). The OID may also contribute to homodimerization of Chip. Whether the OID is also present in other Ldbs has yet to be established. A separate binding site for Ssdp proteins has been identified in a highly conserved region of mouse Ldb1 and Chip (van Meyel et al., 2003). This Ldb1/Chip conserved domain (LLCD) lies close to, but is distinct from, the OID (Fig. 1B).
Conclusions
Ldb1 and LIM domain proteins have emerged as key molecular adaptors with a remarkable potential to mediate protein–protein interactions. The ability of Ldb1 to contact different protein partners and its apparent role in cofactor exchange provide a molecular basis for its pleiotropic functions in several different organisms. Ldb1 is central to the activity of many transcription factors, forming higher-order complexes whose stability is modulated by other cofactors. The composition of Ldb1 complexes almost certainly varies with cell type and is likely to dictate whether Ldb1 acts as a positive or negative modulator of transcription. Although many of the functions that Ldb1 exerts will reflect those of its partners, elucidation of the precise mechanisms by which Ldb1 influences cell fate and differentiation remains a challenge for the future.


Acknowledgments
The authors acknowledge the work of many other researchers that could not be cited due to space constraints. We thank J. Mackay for critical comments on this manuscript. J.M.M. is an Australian Research Council Research Fellow. J.E.V. is supported by the Victorian Breast Cancer Research Consortium.
References
- Agulnick A.D., Taira M., Breen J.J., Tanaka T., Dawid I.B. & Westphal H. ( 1996) Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature, 384, 270–272. [DOI] [PubMed] [Google Scholar]
- Bach I. ( 2000) The LIM domain: regulation by association. Mech. Dev., 91, 5–17. [DOI] [PubMed] [Google Scholar]
- Bach I., Carriere C., Ostendorff H.P., Andersen B. & Rosenfeld M.G. ( 1997) A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and OTX homeodomain proteins. Genes Dev., 11, 1370–1380. [DOI] [PubMed] [Google Scholar]
- Becker T., Ostendorff H.P., Bossenz M., Schluter A., Becker C.G., Peirano R.I. & Bach I. ( 2002) Multiple functions of LIM domain-binding CLIM/NLI/Ldb cofactors during zebrafish development. Mech. Dev., 117, 75–85. [DOI] [PubMed] [Google Scholar]
- Cassata G., Rohrig S., Kuhn F., Hauri H.P., Baumeister R. & Burglin T.R. ( 2000) The Caenorhabditis elegans Ldb/NLI/CLIM orthologue Ldb-1 is required for neuronal function. Dev. Biol., 226, 45–56. [DOI] [PubMed] [Google Scholar]
- Chen L., Segal D., Hukriede N.A., Podtelejnikov A.V., Bayarsaihan D., Kennison J.A., Ogryzko V.V., Dawid I.B. & Westphal H. ( 2002) Ssdp proteins interact with the LIM-domain-binding protein Ldb1 to regulate development. Proc. Natl Acad. Sci. USA, 99, 14320–14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I.B., Breen J.J. & Toyama R. ( 1998) Lim domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet., 14, 156–162. [DOI] [PubMed] [Google Scholar]
- Deane J.E., Mackay J.P., Kwan A.H., Sum E.Y., Visvader J.E. & Matthews J.M. ( 2003) Structural basis for the recognition of Ldb1 by the N-terminal LIM domains of LMO2 and LMO4. EMBO J., 22, 2224–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Funez P., Lu C.H., Rincon-Limas D.E., Garcia-Bellido A. & Botas J. ( 1998) The relative expression amounts of apterous and its co-factor dLdb/Chip are critical for dorso-ventral compartmentalization in the Drosophila wing. EMBO J., 17, 6846–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M., Morcillo P. & Dorsett D. ( 2001) Insulation of enhancer–promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol. Cell. Biol., 21, 4807–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German M.S., Wang J., Chadwick R.B. & Rutter W.J. ( 1992) Synergistic activation of the insulin gene by a LIM-homeodomain protein and a basic helix–loop–helix protein: building a functional insulin minienhancer complex. Genes Dev., 6, 2165–2176. [DOI] [PubMed] [Google Scholar]
- Gupta B.P., Wang M. & Sternberg P.W. ( 2003) The C. elegans LIM homeobox gene lin-11 specifies multiple cell fates during vulval development. Development, 130, 2589–2601. [DOI] [PubMed] [Google Scholar]
- Heitzler P., Vanolst L., Biryukova I. & Ramain P. ( 2003) Enhancer–promoter communication mediated by Chip during Pannier-driven proneural patterning is regulated by OSA. Genes Dev., 17, 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. & Westphal H. ( 2000) Functions of LIM-homeobox genes. Trends Genet., 16, 75–83. [DOI] [PubMed] [Google Scholar]
- Jurata L.W. & Gill G.N. ( 1997) Functional analysis of the nuclear LIM domain interactor NLI. Mol. Cell. Biol., 17, 5688–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata L.W., Kenny D.A. & Gill G.N. ( 1996) Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc. Natl Acad. Sci. USA, 93, 11693–11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata L.W., Pfaff S.L. & Gill G.N. ( 1998) The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of lim domain transcription factors. J. Biol. Chem., 273, 3152–3157. [DOI] [PubMed] [Google Scholar]
- Milan M. & Cohen S.M. ( 1999) Regulation of LIM homeodomain activity in vivo: a tetramer of dLdb and apterous confers activity and capacity for regulation by dLMO. Mol. Cell, 4, 267–273. [DOI] [PubMed] [Google Scholar]
- Milan M., Diaz-Benjumea F.J. & Cohen S.M. ( 1998) Beadex encodes an LMO protein that regulates apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev., 12, 2912–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunuma H., Miyazawa J., Sanada K. & Imai K. ( 2003) The LIM-only protein, LMO4, and the LIM domain-binding protein, Ldb1, expression in squamous cell carcinomas of the oral cavity. Br. J. Cancer, 88, 1543–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki T. et al. ( 2000) Xlim-1 and LIM domain binding protein 1 cooperate with various transcription factors in the regulation of the goosecoid promoter. Dev. Biol., 224, 470–485. [DOI] [PubMed] [Google Scholar]
- Morcillo P., Rosen C. & Dorsett D. ( 1996) Genes regulating the remote wing margin enhancer in the Drosophila cut locus. Genetics, 144, 1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo P., Rosen C., Baylies M.K. & Dorsett D. ( 1997) Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev., 11, 2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay M. et al. ( 2003) Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development, 130, 495–505. [DOI] [PubMed] [Google Scholar]
- Ostendorff H.P., Peirano R.I., Peters M.A., Schluter A., Bossenz M., Scheffner M. & Bach I. ( 2002) Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature, 416, 99–103. [DOI] [PubMed] [Google Scholar]
- Rabbitts T.H. ( 1998) LMO T-cell translocation oncogenes typify genes activated by chromosomal translocations that alter transcription and developmental processes. Genes Dev., 12, 2651–2657. [DOI] [PubMed] [Google Scholar]
- Ramain P., Khechumian R., Khechumian K., Arbogast N., Ackermann C. & Heitzler P. ( 2000) Interactions between Chip and the Achaete/Scute-Daughterless heterodimers are required for Pannier-driven proneural patterning. Mol. Cell, 6, 781–790. [DOI] [PubMed] [Google Scholar]
- Rincon-Limas D.E., Lu C.H., Canal I. & Botas J. ( 2000) The level of dLdb/Chip controls the activity of the LIM homeodomain protein Apterous: evidence for a functional tetramer complex in vivo. EMBO J., 19, 2602–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia I., Osada H., Forster A. & Rabbitts T.H. ( 1993) The cysteine-rich LIM domains inhibit DNA binding by the associated homeodomain in Isl-1. EMBO J., 12, 4243–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoresh M., Orgad S., Shmueli O., Werczberger R., Gelbaum D., Abiri S. & Segal D. ( 1998) Overexpression Beadex mutations and loss-of-function heldup-a mutations in Drosophila affect the 3′ regulatory and coding components, respectively, of the dLMO gene. Genetics, 150, 283–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler J.P. et al. ( 2002) LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein–protein interactions. Cell, 110, 237–249. [DOI] [PubMed] [Google Scholar]
- Torigoi E., Bennani-Baiti I.M., Rosen C., Gonzalez K., Morcillo P., Ptashne M. & Dorsett D. ( 2000) Chip interacts with diverse homeodomain proteins and potentiates bicoid activity in vivo. Proc. Natl Acad. Sci. USA, 97, 2686–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valge-Archer V., Forster A. & Rabbitts T.H. ( 1998) The LMO1 and Ldb1 proteins interact in human T cell acute leukaemia with the chromosomal translocation t(11;14)(p15;q11). Oncogene, 17, 3199–3202. [DOI] [PubMed] [Google Scholar]
- van Meyel D.J., O'Keefe D.D., Thor S., Jurata L.W., Gill G.N. & Thomas J.B. ( 2000) Chip is an essential cofactor for apterous in the regulation of axon guidance in Drosophila. Development, 127, 1823–1831. [DOI] [PubMed] [Google Scholar]
- van Meyel D.J., Thomas J.B. & Agulnick A.D. ( 2003) Ssdp proteins bind to LIM-interacting co-factors and regulate the activity of LIM-homeodomain protein complexes in vivo. Development, 130, 1915–1925. [DOI] [PubMed] [Google Scholar]
- Visvader J.E., Mao X., Fujiwara Y., Hahm K. & Orkin S.H. ( 1997) The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc. Natl Acad. Sci. USA, 94, 13707–13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader J.E. et al. ( 2001) The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc. Natl Acad. Sci. USA, 98, 14452–14457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Justice N.J., Abdelilah S., Chan Y.M., Jan L.Y. & Jan Y.N. ( 1998) The Drosophila LIM-only gene, dLMO, is mutated in Beadex alleles and might represent an evolutionarily conserved function in appendage development. Proc. Natl Acad. Sci. USA, 95, 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]


