Abstract
Menin is the product of the tumor suppressor gene Men1 that is mutated in the inherited tumor syndrome multiple endocrine neoplasia type 1 (MEN1). Menin has been shown to interact with SET-1 domain-containing histone 3 lysine 4 (H3K4) methyltransferases including mixed lineage leukemia proteins to regulate homeobox (Hox) gene expression in vitro. Using conditional Men1 knockout mice, we have investigated the requirement for menin in hematopoiesis and myeloid transformation. Men1 excision causes reduction of Hoxa9 expression, colony formation by hematopoietic progenitors, and the peripheral white blood cell count. Menin directly activates Hoxa9 expression, at least in part, by binding to the Hoxa9 locus, facilitating methylation of H3K4, and recruiting the methylated H3K4 binding protein chd1 to the locus. Consistent with signaling downstream of menin, ectopic expression of both Hoxa9 and Meis1 rescues colony formation defects in Men1-excised bone marrow. Moreover, Men1 excision also suppresses proliferation of leukemogenic mixed lineage leukemia-AF9 fusion-protein-transformed myeloid cells and Hoxa9 expression. These studies uncover an important role for menin in both normal hematopoiesis and myeloid transformation and provide a mechanistic understanding of menin's function in these processes that may be used for therapy.
Keywords: multiple endocrine neoplasia type 1, mixed lineage leukemia, Men1 gene, histone lysine methylation
Menin is a tumor suppressor encoded by Men1 that is mutated in the human inherited tumor syndrome multiple endocrine neoplasia type 1 (MEN1) (1, 2). This syndrome is characterized by tumor development in multiple endocrine organs (3, 4). Although the primary sequence of menin is highly conserved from fly to human, it does not bear obvious homology to known protein motifs, making it difficult to elucidate its biochemical function. Recent progress showed that menin regulates cell proliferation (5–7), apoptosis (8, 9), and genome stability (10–12). Many of these functions rely on the ability of menin to regulate transcription of various genes, including cyclin-dependent kinase inhibitors and caspase 8 (9, 13). Although menin directly binds DNA and associates with chromatin, no evidence shows that it binds to specific DNA sequences to regulate gene transcription (14).
Menin interacts with several transcription factors such as Jun D and NF-κB and a yeast SET1-like complex containing mixed-lineage leukemia (MLL) protein (15–19). MLL is a large protein with a highly conserved SET domain that specifically methylates histone 3 lysine 4 (H3K4) (20, 21). This protein is often fused with other proteins in MLL cells because of chromosomal translocations (22–24). These MLL fusion proteins play a crucial role in up-regulating the homeobox (Hox) genes, including Hoxa9, and the development of MLL. Wild-type MLL up-regulates transcription of Hox genes, presumably by binding to the promoters of these genes and methylating H3K4 (20, 21). Both MLL and its target Hoxa9, a Hox-containing transcription factor, are required for normal hematopoiesis (25, 26), and MLL fusion proteins may lead to dys-regulated expression of Hoxa9 and its cofactor Meis1, resulting in the development of MLL (27).
Although menin and its interaction with MLL are implicated in regulating several Hox genes (18, 19), it is unclear whether menin plays a role in either hematopoiesis or MLL oncoprotein-induced myeloid transformation. To examine the potential role of menin in hematopoiesis, we generated a system that can excise Men1 in a temporally controlled manner. Our studies revealed an important role for menin in bone marrow (BM) hematopoiesis and proliferation of MLL-AF9-transformed myeloid cells. These menin functions are executed, at least in part, by binding to the Hoxa9 locus and up-regulating Hoxa9 expression, via methylation of H3K4 at the Hoxa9 locus. These data provide a molecular model whereby menin-dependent modification of histones at certain Hox gene loci plays an important role in hematopoiesis and suggest that menin is required for proliferation of some MLL fusion protein-transformed cells.
Results
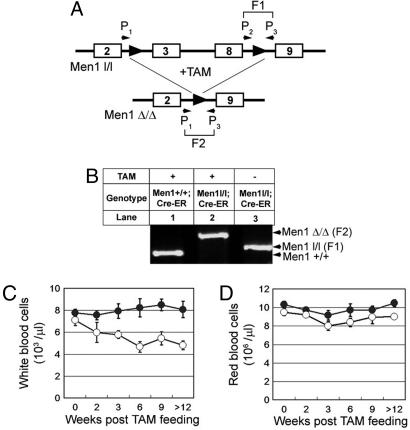
Men1 Excision from Men1l/l;Cre-ER Mice Reduces the Peripheral White Blood Cell (WBC) Count. Since homozygous Men1 deletion in mice leads to early embryonic lethality ∼day 11 because of unknown causes (28, 29), it is difficult to assess the role of menin in hematopoiesis. Although mice harboring conditional Men1 alleles (floxed Men1) and tissue-specific Cre were successfully used to determine the role of menin in tumor suppression in endocrine and liver cells (30–33), these models were unable to address menin function in hematopoiesis. Thus, we used a conditional system to delete Men1 in a temporally controlled manner via tamoxifen (TAM or 4-OHT) treatment. The floxed Men1 (Men1l/l) mice (30) were bred with mice expressing the Cre-ERT2 fusion protein that can be activated by TAM, and the transgene expressing Cre-ERT2 was driven by a ubiquitously active UBC9 promoter (34, 35) (E.J.B., unpublished data). To determine whether oral ingestion of TAM effectively excises the floxed Men1 from the Men1l/l;Cre-ER mice, BM genomic DNA was prepared from either untreated Men1l/l;Cre-ER mice or TAM-treated Men1+/+;Cre-ER and Men1l/l;Cre-ER mice for PCR genotyping (Fig. 1 A and B). Treating the Men1l/l;Cre-ER mice with TAM effectively excised the floxed Men1 from BM (Fig. 1B, lane 2), whereas the floxed Men1 was not excised in the untreated mice (Fig. 1B, lane 3), indicating a tight control of Men1 excision.
Fig. 1.
Excision of floxed Men1 in mice reduces the peripheral WBC counts. (A) A diagram for the floxed Men1 and primers for detecting excision of the floxed Men1 in genome. P1, P2, and P3 denote primers 1, 2, and 3, respectively. F1 and F2 are DNA fragments representing the unexcised and excised floxed Men1 loci, respectively. The numbered boxes denote exon numbers. (B) Tight control of TAM induced the floxed Men1 excision. Men1+/+;Cre-ER mice (lane 1) and Men1 l/l;Cre-ER mice (lane 2) were treated with TAM, whereas Men1l/l;Cre-ER mice (lane 3) were treated with corn oil. BM genomic DNA from each of the mice was used for genotyping. The results are representative of three mice for each condition. Men1Δ/Δ (F2), excised Men1 locus; Men1l/l (F1), unexcised floxed Men1.(C) Total peripheral WBC of control Men1+/+;Cre-ER mice (•) and Men1l/l;Cre-ER mice (○) are plotted, after treatment with TAM for various periods of time (weeks). (D) Time course of RBC counts in mice of both distinct genotypes. Five to 20 mice for each group were used for each time point over 12 weeks.
With this system, we first sought to determine whether Men1 excision caused abnormalities in the peripheral blood of these mice. Both Men1+/+;Cre-ER control mice and Men1l/l;Cre-ER mice were treated with TAM, and then monitored for changes in the numbers of peripheral WBC and red blood cells (RBC) for >12 weeks. The total WBC counts in the control and experimental mice were similar before TAM treatment, ∼7–8 × 103/μl (Fig. 1C). Excision of Men1 gradually decreased the total WBC, from 7.1 × 103/μl to 4.5 × 103/μl at the sixth week after TAM treatment, a 40% reduction (Fig. 1C, ○, P < 0.004), remaining low after >12 weeks, whereas the total WBC remained unchanged in control Men1+/+;Cre-ER mice after TAM treatment (Fig. 1C, •). However, Men1 excision did not significantly reduce RBC numbers (Fig. 1D) over 12 weeks. Collectively, these results indicate that menin is important for maintenance of steady-state peripheral WBC counts.
Men1 Excision in Vitro and in Vivo Reduces BM Colony Formation. We reasoned that the reduced peripheral WBC counts might be caused by dysfunction of BM progenitors, and thus sought to compare the function of hematopoietic progenitors from Men1-excised and Men1-intact mice by using colony formation assays, which are widely used to assess BM hematopoiesis (36, 37). BM was collected from Men1+/+;Cre-ER mice and Men1l/l;Cre-ER mice 7 days after TAM treatment and plated in methylcellulose medium for colony formation. BM cells from TAM-treated Men1+/+;Cre-ER mice formed 251 colonies, whereas Men1 excision in Men1l/l;Cre-ER mice markedly reduced colony formation to nine colonies, a reduction of 96% (Fig. 5 A–C, which is published as supporting information on the PNAS web site, P < 10–4). Notably, expression of Hoxa9, Hoxa7, Hoxc8, and Hoxc6, but not the control gene GAPDH, was also reduced, as shown by RT-PCR (Fig. 5D). As Hoxa9 is important for normal hematopoiesis (25), these data not only indicate that menin is important for BM colony formation, but also suggest that reduction of menin-dependent expression of Hox genes, including Hoxa9, may contribute to the defects in colony formation by the Men1-excised BM.
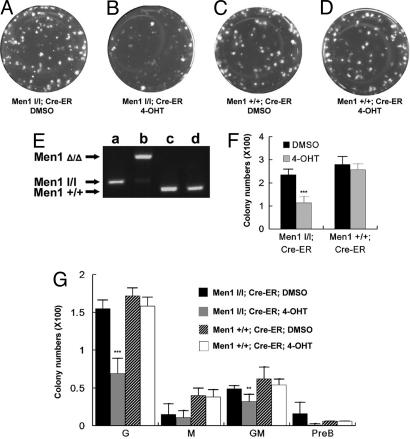
We sought to test whether the defective BM colony formation is caused by loss of an autonomous role of Men1 in BM cells. Thus, BM cells from both control Men1+/+;Cre-ER mice and Men1l/l;Cre-ER mice were plated in the methylcellulose-based medium with either DMSO or 4-OHT. After 12 days of culture, 4-OHT-mediated Men1 excision reduced colony formation of the Men1l/l;Cre-ER BM cells, from 235 colonies to 113 colonies, a 52% reduction (Fig. 2 A, B, and F, P < 0.001). It appeared that excision of menin preferentially reduced the number of big colonies. In contrast, the colony numbers from Men1+/+;Cre-ER BM treated with either DMSO or 4-OHT were 280 and 256, respectively, not significantly different (Fig. 2 C, D, and F, P > 0.2). The cells in those plates were harvested to determine the status of Men1 excision. Floxed Men1 was almost completely excised by 4-OHT treatment in Men1l/l;Cre-ER cells (Fig. 2E, lane b), but remained intact in the DMSO-treated cells (Fig. 2E, lane a). As expected, 4-OHT did not excise Men1 in Men1+/+;Cre-ER cells (Fig. 2E, lane d). Thus, we conclude that menin plays an autonomous role in maintaining the function of BM progenitors in colony formation. Men1 excision in vivo led to a 96% reduction in colony formation (Fig. 5C), but excision of the locus in vitro caused only 52% reduction (Fig. 2F). This difference is likely caused by the slow disappearance of the menin protein within the initial several days after the in vitro 4-OHT treatment, because low levels of menin are still detectable by Western blotting on day 4 after the 4-OHT treatment (data not shown). In addition, the total numbers of BM cells from the femur and tibias did not appear to be markedly different between the Men1-excised mice and Men1+/+ mice (data not shown, Y.-X.C., unpublished work), although the total progenitors from the BM were decreased when the floxed Men1 was excised, based on colony formation assays (Fig. 2 F and G).
Fig. 2.
Men1 excision in vitro reduces colony formation of BM. (A–D) Representative plates with visible colonies from colony formation assays. BM pooled from Men1+/+;Cre-ER mice or Men1l/l;Cre-ER mice were plated (2.5 × 104 cells per plate) in complete medium supplemented with either DMSO or 4-OHT. The genotype of BM cells and treatment applied to the plate are indicated. (E) PCR genotyping with genomic DNA from the above plates as templates. (F) Summary of colony numbers from three independent experiments with duplicate plates. Asterisks indicate statistically significant differences between the adjacent groups (P < 3 × 10–4). (G) Average numbers of CFU-granulocytes (G), CFU-macrophages (M), CFU-granulocytes/macrophages (GM), and CFU-subset of B-lymphoid progenitors (PreB) were classified and recorded for each group from three independent experiments with duplicate plates as described in F. Identities of representative colonies were confirmed with Wright-Giemsa staining.
BM colony formation units (CFU) in the above plates were also quantified. The average numbers of CFU-granulocytes decreased from 155 to 69 (P < 0.0006) after Men1 excision. In addition, CFU-granulocytes/macrophages decreased from 15 to 11 (Fig. 2G, P < 0.01). Numbers of CFU-subset of B-lymphoid progenitors and CFU-macrophages were not markedly affected by Men1 excision (P > 0.075 and 0.65, respectively). However, we cannot rule out that the relatively low numbers of the two types of colonies masked a potential difference.
Ectopic Expression of Menin Rescues Defective Colony Formation of Men1-Excised BM Cells and Hoxa9 Expression. To further establish an essential and autonomous role for menin in maintaining normal BM precursors, we sought to “rescue” the colony formation defect in Men1-excised BM by ectopically expressing menin. BM cells from the Men1l/l;Cre-ER mice were transduced with either control vector retrovirus or retrovirus expressing menin. The transduced cells were assayed for colony formation in the presence of either DMSO or 4-OHT. Transduction by the control vector virus did not affect the reduction of colony formation caused by 4-OHT-mediated Men1 excision (Fig. 6 A, B, and E, which is published as supporting information on the PNAS web site, P < 0.001). However, the colony numbers from ectopic menin-expressing cells were quite similar between DMSO-treated and 4-OHT-treated Men1l/l;Cre-ER cells, i.e., 249 and 255, respectively (Fig. 6 C–E, P > 0.3). These studies indicate that exogenous menin genetically complements the functions of excised endogenous Men1 in colony formation of hematopoietic progenitors, whereas menin overexpression alone is insufficient to increase BM colony formation (Fig. 6E).
Real-time RT-PCR confirmed that 4-OHT treatment reduced expression of the endogenous Men1 but not ectopically transduced menin (Fig. 6F). Hoxa9 expression is reduced by 4-OHT treatment in vector-transduced BM cells by 85% (Fig. 6G Left), consistent with reduced Hoxa9 expression in primary BM cells with Men1-excised in vivo (Fig. 5D). Ectopic menin expression increased Hoxa9 mRNA levels by ∼70% and maintained this level even after the endogenous Men1 was excised (Fig. 6G Right).
Overexpression of Hoxa9 and Meis1 Rescues the Deficient Colony Formation of Men1-Excised BM. We explored the possibility that Hoxa9 is a downstream factor that mediates menin's function in hematopoiesis. Because Hoxa9 and its cofactor Meis1 together strongly stimulate proliferation of myeloid cells (27), we tested whether CFU activity of Men1-excised hematopoietic precursors can be maintained by overexpression of Hoxa9 and/or Meis1. BM cells from Men1l/l;Cre-ER mice were infected with vector retrovirus, virus expressing Hoxa9, virus expressing Meis1, or a combination of viruses expressing either Hoxa9 or Meis1. The transduced cells were selected and assayed for colony formation. Men1 excision by 4-OHT treatment reduced colony formation in vector-infected cells by 40% (Fig. 7 A, B, and I, which is published as supporting information on the PNAS web site, P < 0.001). Ectopic expression of Hoxa9 alone modestly increased colony formation from both Men1-intact cells and Men1-excised cells, by 24% and 41%, respectively (Fig. 7 C, D, and I, P < 0.07 and P < 0.02), whereas ectopic expression of Meis1 alone did not increase colony formation (Fig. 7 E, F, and I, P > 0.7 and 0.9, respectively). Importantly, coexpression of Hoxa9 and Meis1 not only markedly enhanced colony formation of the cells with or without menin expression, to 407 and 382 colonies, respectively, but also negated the dependence on menin in colony formation from 4-OHT-treated cells (Fig. 7 G–I, P > 0.30).
Real-time RT-PCR results confirmed that both Hoxa9 and Meis1 were overexpressed (Fig. 7 J and K). In addition, floxed Men1 excision in the 4-OHT-treated cells was confirmed by PCR genotyping and real-time RT-PCR (data not shown). Together, these results show that coexpression of both Hoxa9 and Meis1 prevents reduction of colony formation that results from loss of menin expression. Coupled with the finding that menin is important for Hoxa9 expression in BM (Fig. 6G), these results suggest that Hoxa9 and Meis1 are among the major effectors of menin in hematopoiesis. Because Men1 excision reduced expression of multiple Hox genes, including Hoxa9 (Fig. 5D) but not Meis1 (Fig. 7K), and Meis1 is a cofactor for Hoxa9, it is conceivable that coexpression of both Hoxa9 and Meis1 at an elevated level is necessary to compensate for the reduced colony formation in the absence of Men1.
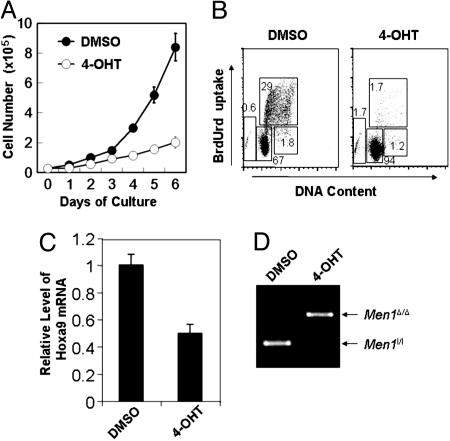
Menin Is Required for Optimal Proliferation of MLL-AF9-Transformed Myeloid Cells and Hoxa9 Expression. As our data indicate that menin is important for normal BM colony formation, we sought to determine whether menin is also important for growth of MLL oncoprotein-transformed myeloid cells. BM from Menl/l;Cre-ER mice was transformed by transduction with retroviruses expressing oncogenic MLL-AF9, an MLL fusion protein (23). Treatment of the cells with 4-OHT, but not DMSO, effectively excised floxed Men1 (Fig. 3D). Equal numbers of DMSO-treated or 4-OHT-treated cells were plated and cells were counted. By day 6 of culture, DMSO-treated cells reached a much higher number (8.7 × 105 cells) than the 4-OHT-treated cells (2.1 × 105 cells), a 4-fold difference (Fig. 3A). To determine whether loss of menin expression affected cell proliferation, apoptosis, or differentiation, the DMSO- or 4-OHT-treated cells were pulsed with BrdUrd, and BrdUrd-positive cells were determined by flow cytometry analysis. Men1-excised cells showed a marked decrease in cell proliferation (1.7% BrdUrd-positive cells) as compared with DMSO-treated cells (29% BrdUrd-positive cells) (Fig. 3B). Up to 10 days after 4-OHT treatment, the treated cells did not show either significant differentiation, by staining for the myeloid differentiation markers Mac-1 and Gr-1, or apoptosis, by staining for Annexin V (data not shown). Together, these results indicate that Men1 excision significantly inhibits proliferation but does not enhance apoptosis and differentiation of MLL-AF9-transformed cells. Moreover, Men1 excision in the cells also reduced expression of Hoxa9 mRNA (Fig. 3C).
Fig. 3.
Men1 excision inhibits proliferation of MLL-AF9-transformed myeloid cells and suppresses Hoxa9 expression. (A) BM from multiple Men1l/l;Cre-ER mice was transformed by transduction with retroviruses expressing MLL-AF9. The transformed BM cells from a representative mouse, AT-1, were plated in triplicate at 2.5 × 104 cells per well, treated with either DMSO or 4-OHT, and counted daily for 6 days. (B) The myeloid cells treated with either DMSO or 4-OHT for 4 days were pulsed with BrdUrd for 90 min, and then stained with an anti-BrdUrd antibody and 7-amino-actinomycin D, followed by flow cytometric analysis. (C) The transformed Men1l/l;Cre-ER cells were treated with either DMSO or TAM for 4 days, and then harvested for RNA isolation and real-time PCR to quantitate Hoxa9 mRNA. (D) Genomic DNA from the cells treated with either DMSO or 4-OHT was used for PCR to determine Men1 excision. Two independent experiments were performed for each of the above experiments.
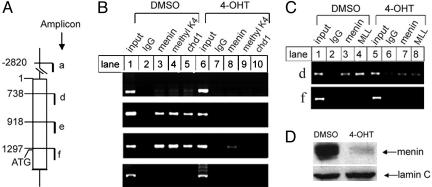
Menin Is Important for Methylation of H3K4 and Recruitment of Methylated H3K4-Binding Protein Chd1 to the Hoxa9 Locus. Regulation of Hoxa9 by menin is an attractive explanation for the effect of menin on normal hematopoiesis and myeloid transformation, because menin has been reported to bind MLL and been implicated in regulating Hoxa9 expression (19). Thus, we tested whether menin directly binds the Hoxa9 locus and is essential for methylation of H3K4 by using chromatin immunoprecipitation (ChIP) assays. Seven independent primer pairs for seven distinct amplicons (a–g) spanning over 5 kb in the Hoxa9 locus were synthesized to detect the potential location of menin in the locus in vivo (Fig. 8 Left, which is published as supporting information on the PNAS web site). ChIP assays with the transformed myeloid cells showed that menin primarily associated with the Hoxa9 locus from –1,221 to +1,109 bp (Fig. 8 Center, lane 3, amplicons a–e), but not with the coding sequence surveyed by the amplicons (Fig. 8 Center, lane 3, amplicons f and g). Almost superimposable on the pattern of menin association with the locus was the methylated H3K4, as detected by the antibody specific for the trimethylated lysine 4 of histone 3 (38, 39) (Fig. 8 Center, lane 4). Similarly, chd1, which specifically binds the methylated H3K4, remodels chromatin, and recruits histone acetylases (40), also overlaps with the binding patterns of menin and methylated H3K4 (Fig. 8 Center, lane 5). Because menin specifically binds serine 5-phosphorylated RNA polymerase II (18), we further tested whether RNA polymerase II associates with the Hoxa9 locus and its pattern of association with the locus. The ChIP assay showed that phosphorylated RNA polymerase II associated with the Hoxa9 locus in a pattern mimicking menin's distribution (Fig. 8 Right, lane 4). Because menin interacts with MLL and MLL2 that specifically methylates H3K4, our data suggest that menin and a H3K4 methyltransferase complex are recruited to the Hoxa9 locus at a 2-kb region that includes the proximal promoter and 5′ UTR region, but not the coding region, leading to H3K4 methylation.
To determine whether menin is required for H3K4 methylation at the Hoxa9 locus and binding of the methylated H3K4 binding protein chd1 to the locus, menin expression was markedly reduced in MLL-AF9-transformed Men1l/l;Cre-ER myeloid cells with 4-OHT treatment, as shown by Western blotting (Fig. 4D). ChIP assays show that menin, methylated H3K4, and chd1 bind the Hoxa9 locus in menin-expressing myeloid cells, in a region covered by amplicons d and e (Fig. 4B, lanes 3–5); Men1 excision led to the marked reduction of menin binding to the Hoxa9 locus (Fig. 4B, lane 8). Additionally, association of methylated H3K4 and chd1 to the Hoxa9 locus was markedly reduced after Men1 excision (Fig. 4B, lanes 9 and 10). ChIP assays also showed that both menin and MLL bound to the Hoxa9 locus (Fig. 4C, lanes 3 and 4), but Men1 excision markedly reduced the binding of both menin and MLL to the locus (Fig. 4C, lanes 7 and 8). Together, these data indicate that menin is important for binding of MLL, its substrate H3K4, and chd1 to the Hoxa9 locus and suggests a molecular link between these proteins in the Hoxa9 locus.
Fig. 4.
Menin is important for methylation of H3K4 in the Hoxa9 locus and for binding of MLL and cdh1 to the locus. (A) A diagram of the murine Hoxa9 locus. (B and C) Myeloid cells harboring Men1l/l;Cre-ER were treated with DMSO or 4-OHT, and then processed for ChIP assays by using the indicated antibodies. Results are representative of two independent experiments. (D) Men1 excision induced by 4-OHT was confirmed by Western blotting using an antimenin antibody. Equal loading of the samples was revealed by blotting for Lamin C.
Discussion
We identified an important and autonomous role for menin in BM hematopoiesis, at least in part, through regulating Hox genes, including Hoxa9. We also found that menin is important for the proliferation of MLL oncoprotein-transformed myeloid cells, pointing to a paradoxically oncogenic role for the tumor suppressor menin in proliferation of transformed myeloid cells. This finding is consistent with a recent report showing that menin is required for maintenance of myeloid transformation by the oncogenic MLL-ENL (41). Moreover, our studies uncovered a molecular link between menin, methylation of H3K4, and the recruitment of chd1 to the Hoxa9 locus, suggesting a role of this cascade in hematopoiesis and myeloid transformation.
Because modification or a combination of modifications of histones may serve as a “code” to regulate transcription from the genome (42), menin-dependent modification of H3K4 may serve as a platform for maintenance of Hoxa9 transcription. Thus, MLL may require menin for binding to the Hoxa9 locus and to methylate the local H3K4. The methylated H3K4 may further recruit chd1, a protein that specifically binds the methylated H3K4 via its second chromodomain and is also a component of large chromatin remodeling and acetylation complexes such as SAGA and SLIK (40). Our data indicate that chd1 associates with the Hoxa9 locus, overlapping with the region bound by methylated H3K4 (Fig. 8), and chd1 binding to the locus also depends on the presence of menin (Fig. 4B). This finding suggests an important role for menin in lysine 4 methylation by MLL and/or other H3K4 methyltransferases and subsequent binding of the methylated H3K4 by chd1. A recent report shows that WDR5, a component of the MLL complex, binds the methylated H3K4 and activates Hoxa9 transcription (43). Our finding, together with that report, highlights the importance of the methylated H3K4 in regulating Hox gene expression and hematopoiesis. The relative contribution of either chd1 or WDR5 to hematopoiesis is unclear. Because menin does not bind to specific DNA sequence but is recruited specifically to the Hoxa9 locus (Fig. 4), it is likely that menin is recruited either directly or indirectly by a DNA sequence-specific factor to the Hoxa9 locus to promote Hoxa9 transcription. Further work is required to decipher the detailed mechanisms governing the specific recruitment of menin, MLL complex, and the methylated H3K4 proteins to the Hoxa9 locus and their role in hematopoiesis and myeloid transformation.
Although we observed an important role for menin in hematopoietic CFU activity and peripheral WBC, hematological abnormalities in either MEN1 patients or Men1+/– mice have not yet been reported to our knowledge. It is possible that MEN1 patients or Men1+/– mice have mild blood abnormalities or alternatively are more vulnerable to BM stress. Mutations of Men1, a bona fide tumor suppressor, result in development of tumors in multiple endocrine organs (4). Yet Men1 promotes proliferation of MLL-AF9-transformed cells (Fig. 3). Thus, the role of menin in tumorigenesis is context-dependent; it serves as a tumor suppressor in endocrine organs but a tumor promoter in myeloid cells. It remains to be delineated what determines the tissue-specific activities of menin and whether the menin/MLL/Hox gene pathway plays a role in suppressing tumorigenesis in endocrine organs.
Men1 excision potently inhibits proliferation of MLL-AF9-transformed cells and Hoxa9 expression (Fig. 3). Hoxa9 is often up-regulated in MLL cells or by retroviral insertion-induced leukemias (44), and it is also required for MLL-ENL-mediated myeloid transformation (45). Menin may promote proliferation of MLL-AF9-transformed cells by up-regulating Hoxa9 and other genes through recruiting MLL, because loss of menin markedly reduces binding of MLL to the Hoxa9 locus (Fig. 4C). This result is also consistent with a recent report showing that MLL-AF6 is also recruited to the Hoxa9 locus (41). Hence, menin may cooperate with MLL to control expression of a group of genes, including Hoxa9. Thus menin may act as a convergent point in regulating a group of Hox genes to control normal hematopoiesis and myeloid transformation, which raises the interesting possibility that blocking menin's function in MLL cells may lead to a novel therapy for this leukemia.
Materials and Methods
Mice, Genotyping, and Treatment with TAM. All laboratory mice were maintained on a 12-h light-dark cycle in the animal facility at the University of Pennsylvania in compliance with related guidelines. Men1l/l;Cre-ER mice, in a mixed genetic background, were generated by crossing floxed Men1 mice (Sv129) and Cre-ER transgenic mice (C57BL/6). Floxed Men1 mice were kindly provided by Francis Collins (National Institute for Human Genome Research, Bethesda) (30), in which exons 3–8 of Men1 were flanked by two loxP sites. Mice expressing a transgene, Cre-ERT2 driven by the pan-active UBC9 promoter, were generated by lentitransgenesis, as described (34, 35) (E.J.B., unpublished data). The mice were genotyped by PCR using the tail genomic DNA; the primer sequences will be provided on request. Genotyping and excision of the floxed Men1 are described in Supporting Text, which is published as supporting information on the PNAS web site.
Analysis of Peripheral Blood Cells and Statistic Analysis. Blood samples collected from the retroorbital sinus of mice were stored in Microtainer tubes with EDTA (BD Biosciences, Franklin Lakes, NJ). The hematology profile of each mouse was analyzed immediately on a Hemavet blood cell counter (CDC Technologies, Oxford, CT). Student's t tests were used to analyze the differences in blood cell counts and BM colony formation.
Generation of Recombinant Retroviruses, Infection of Primary BM Cells, and Transformation of Myeloid Cells. Plasmids for generating retroviruses, pMX-Vector-puro, pMX-menin, pMSCVpgk-Hoxa9-GFP, pMSCV-MLL-AF9, and pMSCVpacMeis1A were generated as described (11, 20, 27). The viruses were packaged in 293T cells as described (9). Men1l/l;Cre-ER and Men1+/+;Cre-ER mice were injected with 5-fluorouracil (American Pharmaceutical Partners, Schaumburg, IL) i.p. at a dose of 50 mg/kg body weight. Four days later, BM cells were collected from femurs and tibias of the mice and prestimulated with 10 ng/ml IL-3, 10 ng/ml IL-6, and 100 ng/ml stem cell factor (PeproTech, Rocky Hill, NJ) overnight, as described (46). In vitro infection was performed on 2 consecutive days by incubating BM cells with retroviral supernatant with 4 μg/ml polybrene and spinoculating at 1,300 × g for 90 min at 25°C. The cells were seeded at 1 × 105 cells per 35-mm dish in methyl-cellulose-based medium supplemented with IL-3, IL-6, and stem cell factor (MethoCult GF M3534; StemCell Technologies, Vancouver). The cells infected with MSCV-MLL-AF9 viruses were replated weekly in the same medium with 1 mg/ml G418. After the third plating, the transformed myeloid cells were transferred to liquid medium (Iscove's modified Dulbecco's medium supplemented with 15% fetal calf medium, 1% Pen/Strep, 1% l-glutamine, and 10 ng/ml IL-3) as described (46).
Colony Formation Assay and Analysis of Cell Growth. BM cells used for colony formation assays were harvested from BM as described above or retrovirally infected myeloid cells expressing ectopic genes. Cells (2.5 × 104) were seeded in a 35-mm Petri dish in 1.1 ml of MethoCult GF M3534 methylcellulose medium (StemCell Technologies). 4-OH (Sigma) dissolved in DMSO was added to the medium at 1:500 dilution to reach 200 nM to excise floxed Men1, and an equal volume of DMSO was added to control dishes. After 12-day incubation at 37°C, the dishes were photographed, and the total colony numbers and types of colonies in plates were evaluated under a microscope, and colonies with >50 cells were scored. Criteria for classifying various CFU were as described and confirmed by Wright-Giemsa staining (47). For evaluating cell growth, MLL-AF9-transformed Men1l/l;Cre-ER cells were maintained in Iscove's modified Dulbecco's medium with 15% FCS and 10 ng/ml IL-3 and treated with either 200 nM 4-OHT or DMSO for 4 days. PCR genotyping was performed to confirm Men1 excision. Men1-intact and Men1-excised cells were seeded in 6-well plates in the same medium at a cell density of 2.5 × 104 cells per well in triplicate and counted in a hemacytometer daily.
PCR, RT-PCR, Taqman Real-Time RT-PCR, Western Blotting, and Flow Cytometry Analysis. Total RNA from tissues or cells was prepared by using an RNeasy extraction kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. For classical RT-PCR, 1 mg of RNA from each sample was amplified by using the Titan One-Tube RT-PCR system (Roche Applied Science). For real-time RT-PCR, 5 μg of total RNA from each sample was used to synthesize cDNA by using SuperScript II RNase H reverse transcriptase (Invitrogen) with oligo(dT) primers. Real-time PCR was performed in triplicate tubes on the AB 7500 real-time PCR system (Applied Biosystems) by using the indicated FAM primer pairs (Applied Biosystems). The average Δ-Δ cycle threshold of different samples was analyzed to calculate the relative quantities of mRNA levels, using either actin or GAPDH mRNAs as control. Western blot and flow cytometry analysis were performed as described (14). All of the PCR primers were synthesized by Integrated DNA Technologies (Coralville, IA).
ChIP. ChIP assays were performed as described with certain modifications (48). The detailed procedures are described in Supporting Text.
Supplementary Material
Acknowledgments
We thank Dr. Francis Collins for generously providing the mice with the floxed Men1 locus; Drs. Gary Koretzky, Celeste Simon, and Craig Thompson for critically reading the manuscript; Bob Schnepp for stimulating discussions; and Sirin Suchintabandid for technical assistance. This work was supported by National Institutes of Health Grant CA100912 (to X.H., W.S.P., and J.L.H.), American Cancer Society Grant RSG-03-055-01-LIB (to X.H.), a Rita Allen Foundation scholar award (to X.H.), a Leukemia and Lymphoma Society SCOR grant (to X.H., W.S.P., and J.L.H.), and a fellowship from the Leukemia and Lymphoma Society (to K.K.).
Conflict of interest statement: No conflicts declared.
Abbreviations: MEN1, multiple endocrine neoplasia type 1; MLL, mixed lineage leukemia; H3K4, histone 3 lysine 4; Hox, homeobox; BM, bone marrow; TAM, tamoxifen; CFU, colony formation units; ChIP, chromatin immunoprecipitation.
References
- 1.Lemmens, I., Van de Ven, W. J., Kas, K., Zhang, C. X., Giraud, S., Wautot, V., Buisson, N., De Witte, K., Salandre, J., Lenoir, G., et al. (1997) Hum. Mol. Genet. 6, 1177–1183. [DOI] [PubMed] [Google Scholar]
- 2.Chandrasekharappa, S. C., Guru, S. C., Manickam, P., Olufemi, S. E., Collins, F. S., Emmert-Buck, M. R., Debelenko, L. V., Zhuang, Z., Lubensky, I. A., Liotta, L. A., et al. (1997) Science 276, 404–407. [DOI] [PubMed] [Google Scholar]
- 3.Marx, S. J. & Stratakis, C. A. (2005) J. Intern. Med. 257, 2–5. [DOI] [PubMed] [Google Scholar]
- 4.Pannett, A. A. & Thakker, R. V. (1999) Endocr. Relat. Cancer 6, 449–473. [DOI] [PubMed] [Google Scholar]
- 5.Poisson, A., Zablewska, B. & Gaudray, P. (2003) Cancer Lett. 189, 1–10. [DOI] [PubMed] [Google Scholar]
- 6.Chandrasekharappa, S. C. & Teh, B. T. (2003) J. Intern. Med. 253, 606–615. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal, S. K., Kennedy, P. A., Scacheri, P. C., Novotny, E. A., Hickman, A. B., Cerrato, A., Rice, T. S., Moore, J. B., Rao, S., Ji, Y., et al. (2005) Horm. Metab. Res. 37, 369–374. [DOI] [PubMed] [Google Scholar]
- 8.Sayo, Y., Murao, K., Imachi, H., Cao, W. M., Sato, M., Dobashi, H., Wong, N. C. & Ishida, T. (2002) Endocrinology 143, 2437–2440. [DOI] [PubMed] [Google Scholar]
- 9.Schnepp, R. W., Mao, H., Sykes, S. M., Zong, W. X., Silva, A., La, P. & Hua, X. (2004) J. Biol. Chem. 279, 10685–10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busygina, V., Suphapeetiporn, K., Marek, L. R., Stowers, R. S., Xu, T. & Bale, A. E. (2004) Hum. Mol. Genet. 13, 2399–2408. [DOI] [PubMed] [Google Scholar]
- 11.Jin, S., Mao, H., Schnepp, R. W., Sykes, S. M., Silva, A. C., D'Andrea, A. D. & Hua, X. (2003) Cancer Res. 63, 4204–4210. [PubMed] [Google Scholar]
- 12.Scappaticci, S., Maraschio, P., del Ciotto, N., Fossati, G. S., Zonta, A. & Fraccaro, M. (1991) Cancer Genet. Cytogenet. 52, 85–92. [DOI] [PubMed] [Google Scholar]
- 13.Milne, T. A., Hughes, C. M., Lloyd, R., Yang, Z., Rozenblatt-Rosen, O., Dou, Y., Schnepp, R. W., Krankel, C., Livolsi, V. A., Gibbs, D., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.La, P., Silva, A. C., Hou, Z., Wang, H., Schnepp, R. W., Yan, N., Shi, Y. & Hua, X. (2004) J. Biol. Chem. 279, 49045–49054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agarwal, S. K., Guru, S. C., Heppner, C., Erdos, M. R., Collins, R. M., Park, S. Y., Saggar, S., Chandrasekharappa, S. C., Collins, F. S., Spiegel, A. M., et al. (1999) Cell 96, 143–152. [DOI] [PubMed] [Google Scholar]
- 16.Heppner, C., Bilimoria, K. Y., Agarwal, S. K., Kester, M., Whitty, L. J., Guru, S. C., Chandrasekharappa, S. C., Collins, F. S., Spiegel, A. M., Marx, S. J. & Burns, A. L. (2001) Oncogene 20, 4917–4925. [DOI] [PubMed] [Google Scholar]
- 17.Lin, S. & Elledge, S. J. (2003) Cell 113, 881–889. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, C. M., Rozenblatt-Rosen, O., Milne, T. A., Copeland, T. D., Levine, S. S., Lee, J. C., Hayes, D. N., Shanmugam, K. S., Bhattacharjee, A., Biondi, C. A., et al. (2004) Mol. Cell 13, 587–597. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama, A., Wang, Z., Wysocka, J., Sanyal, M., Aufiero, D. J., Kitabayashi, I., Herr, W. & Cleary, M. L. (2004) Mol. Cell. Biol. 24, 5639–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milne, T. A., Briggs, S. D., Brock, H. W., Martin, M. E., Gibbs, D., Allis, C. D. & Hess, J. L. (2002) Mol. Cell 10, 1107–1117. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura, T., Mori, T., Tada, S., Krajewski, W., Rozovskaia, T., Wassell, R., Dubois, G., Mazo, A., Croce, C. M. & Canaani, E. (2002) Mol. Cell 10, 1119–1128. [DOI] [PubMed] [Google Scholar]
- 22.Pui, C. H., Relling, M. V. & Downing, J. R. (2004) N. Engl. J. Med. 350, 1535–1548. [DOI] [PubMed] [Google Scholar]
- 23.Rowley, J. D. (1999) Semin. Hematol. 36, 59–72. [PubMed] [Google Scholar]
- 24.Popovic, R. & Zeleznik-Le, N. J. (2005) J. Cell. Biochem. 95, 234–242. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence, H. J., Helgason, C. D., Sauvageau, G., Fong, S., Izon, D. J., Humphries, R. K. & Largman, C. (1997) Blood 89, 1922–1930. [PubMed] [Google Scholar]
- 26.Ernst, P., Fisher, J. K., Avery, W., Wade, S., Foy, D. & Korsmeyer, S. J. (2004) Dev. Cell 6, 437–443. [DOI] [PubMed] [Google Scholar]
- 27.Zeisig, B. B., Milne, T., Garcia-Cuellar, M. P., Schreiner, S., Martin, M. E., Fuchs, U., Borkhardt, A., Chanda, S. K., Walker, J., Soden, R., et al. (2004) Mol. Cell. Biol. 24, 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertolino, P., Radovanovic, I., Casse, H., Aguzzi, A., Wang, Z. Q. & Zhang, C. X. (2003) Mech. Dev. 120, 549–560. [DOI] [PubMed] [Google Scholar]
- 29.Crabtree, J. S., Scacheri, P. C., Ward, J. M., Garrett-Beal, L., Emmert-Buck, M. R., Edgemon, K. A., Lorang, D., Libutti, S. K., Chandrasekharappa, S. C., Marx, S. J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crabtree, J. S., Scacheri, P. C., Ward, J. M., McNally, S. R., Swain, G. P., Montagna, C., Hager, J. H., Hanahan, D., Edlund, H., Magnuson, M. A., et al. (2003) Mol. Cell. Biol. 23, 6075–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biondi, C. A., Gartside, M. G., Waring, P., Loffler, K. A., Stark, M. S., Magnuson, M. A., Kay, G. F. & Hayward, N. K. (2004) Mol. Cell. Biol. 24, 3125–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertolino, P., Tong, W. M., Herrera, P. L., Casse, H., Zhang, C. X. & Wang, Z. Q. (2003) Cancer Res. 63, 4836–4841. [PubMed] [Google Scholar]
- 33.Scacheri, P. C., Rozenblatt-Rosen, O., Caplen, N. J., Wolfsberg, T. G., Umayam, L., Lee, J. C., Hughes, C. M., Shanmugam, K. S., Bhattacharjee, A., Meyerson, M. & Collins, F. S. (2004) Proc. Natl. Acad. Sci. USA 101, 1892–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feil, R., Wagner, J., Metzger, D. & Chambon, P. (1997) Biochem. Biophys. Res. Commun. 237, 752–757. [DOI] [PubMed] [Google Scholar]
- 35.Lois, C., Hong, E. J., Pease, S., Brown, E. J. & Baltimore, D. (2002) Science 295, 868–872. [DOI] [PubMed] [Google Scholar]
- 36.Orkin, S. (1995) Curr. Opin. Cell Biol. 7, 870–877. [DOI] [PubMed] [Google Scholar]
- 37.Lensch, M. & Daley, G. (2004) Curr. Top. Dev. Biol. 60, 127–196. [DOI] [PubMed] [Google Scholar]
- 38.Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C., Schreiber, S. L., Mellor, J. & Kouzarides, T. (2002) Nature 419, 407–411. [DOI] [PubMed] [Google Scholar]
- 39.Schneider, R., Bannister, A. J., Myers, F. A., Thorne, A. W., Crane-Robinson, C. & Kouzarides, T. (2004) Nat. Cell Biol. 6, 73–77. [DOI] [PubMed] [Google Scholar]
- 40.Pray-Grant, M. G., Daniel, J. A., Schieltz, D., Yates, J. R., III, & Grant, P. A. (2005) Nature 433, 434–438. [DOI] [PubMed] [Google Scholar]
- 41.Yokoyama, A., Somervaille, T. C., Smith, K. S., Rozenblatt-Rosen, O., Meyerson, M. & Cleary, M. L. (2005) Cell 123, 207–218. [DOI] [PubMed] [Google Scholar]
- 42.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 43.Wysocka, J., Swigut, T., Milne, T. A., Dou, Y., Zhang, X., Burlingame, A. L., Roeder, R. G., Brivanlou, A. H. & Allis, C. D. (2005) Cell 121, 859–872. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura, T., Jenkins, N. A. & Copeland, N. G. (1996) Oncogene 13, 2235–2242. [PubMed] [Google Scholar]
- 45.Ayton, P. M. & Cleary, M. L. (2003) Genes Dev. 17, 2298–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lavau, C., Szilvassy, S. J., Slany, R. & Cleary, M. L. (1997) EMBO J. 16, 4226–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hess, J. L., Yu, B. D., Li, B., Hanson, R. & Korsmeyer, S. J. (1997) Blood 90, 1799–1806. [PubMed] [Google Scholar]
- 48.Kurdistani, S. K. & Grunstein, M. (2003) Methods 31, 90–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.