Abstract
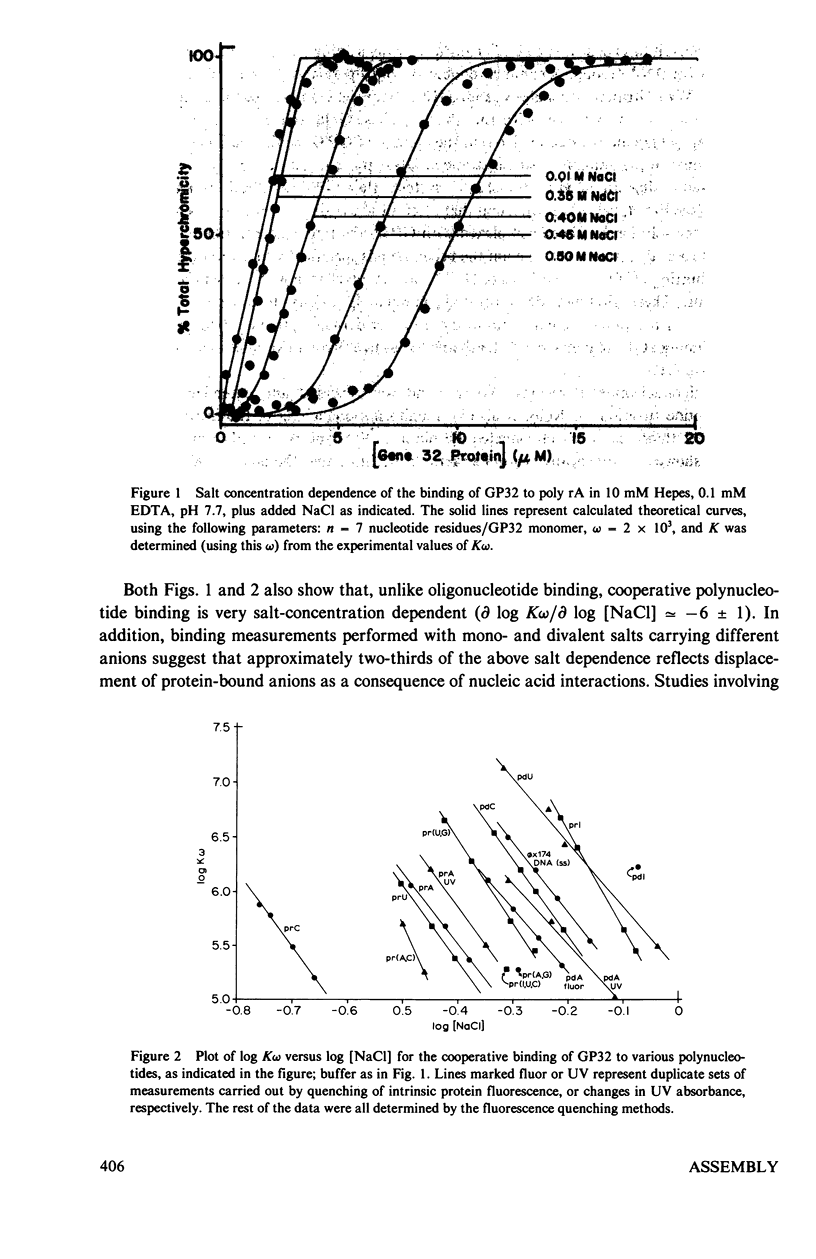
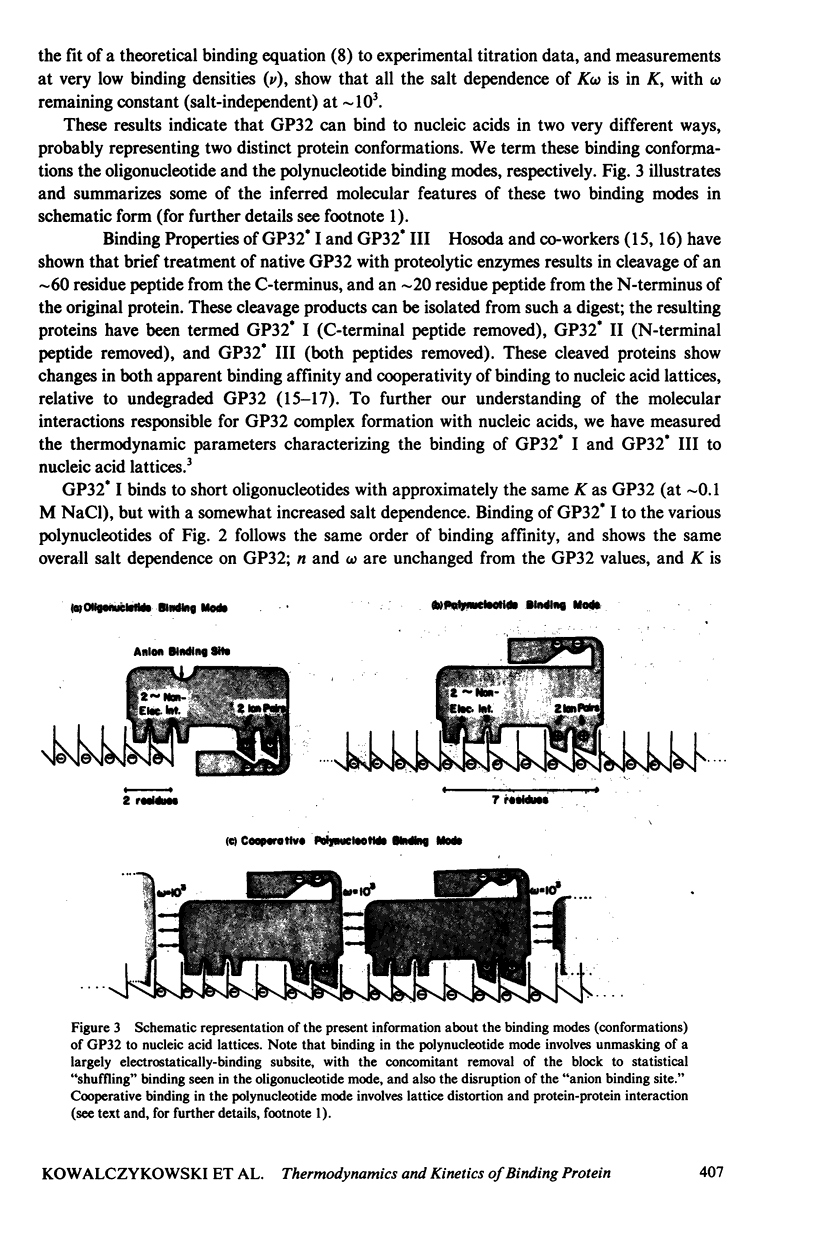
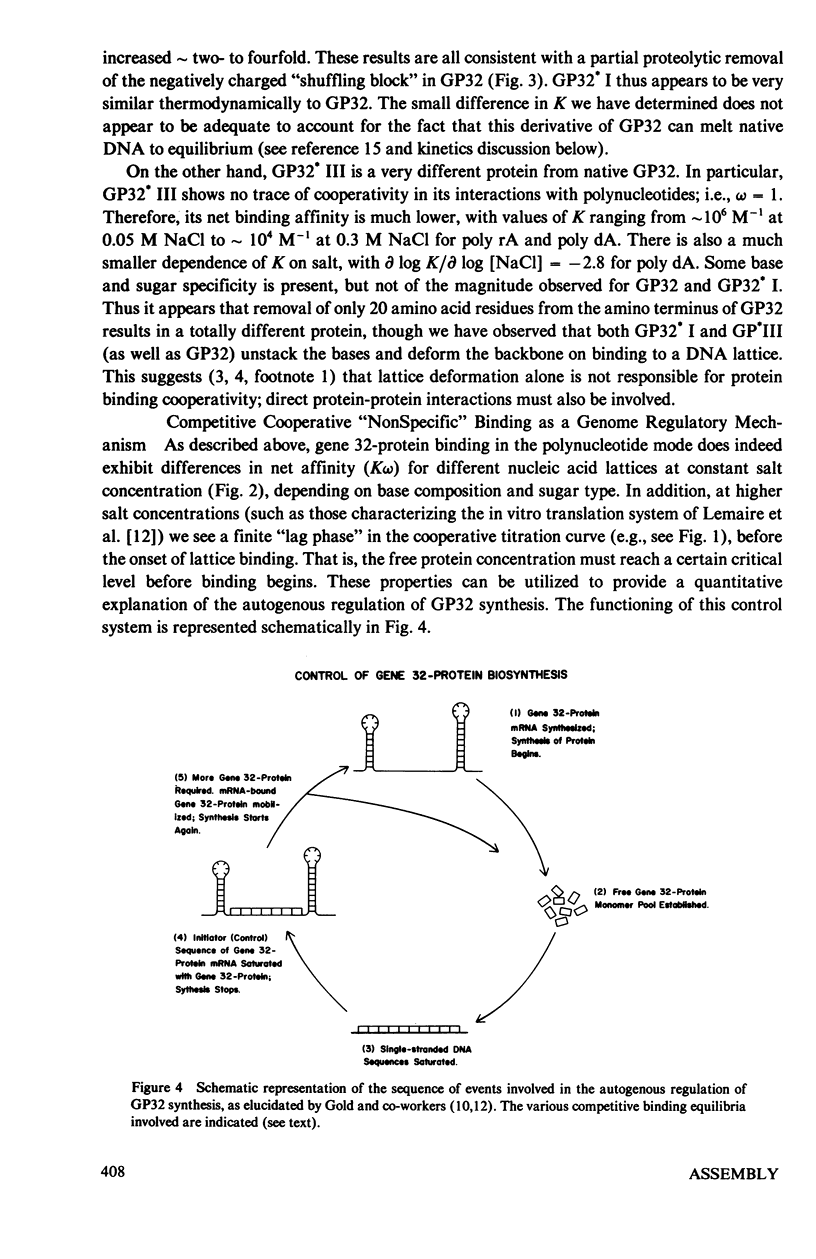
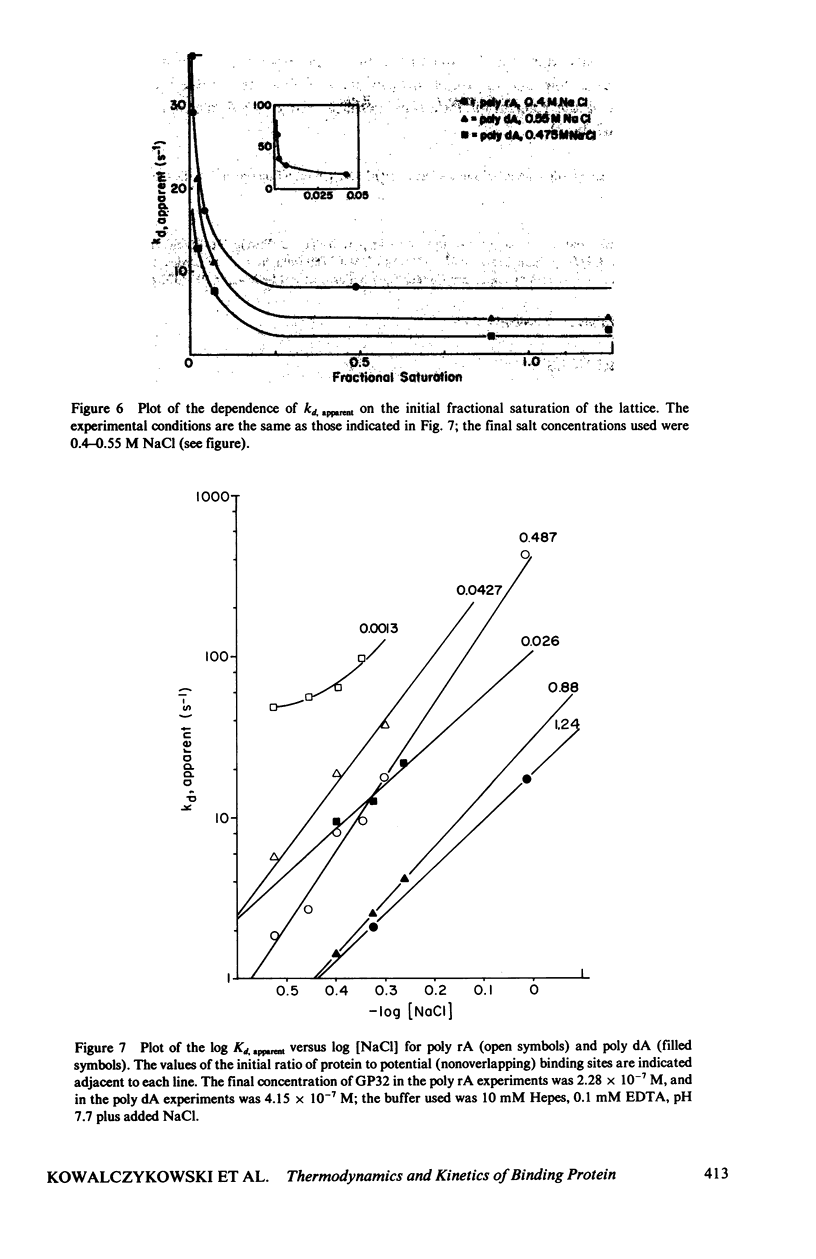
In this paper we summarize a series of thermodynamic, and preliminary kinetic, studies on the molecular details and specificity of interaction of phage T4-coded gene 32-protein (GP32) with nucleic acid lattices. It is shown that the binding of GP32 to short (l = 2--8 residues) oligonucleotides is essentially independent of base composition and sugar-type, as well as of salt concentration. In contrast, cooperative (continuous) or isolated binding of GP32 to single-stranded polynucleotides is base and sugar composition-dependent (binding is tighter to DNA than to RNA) and highly dependent on salt concentrations. Binding constants (K), cooperativity parameters (w), and binding site sizes (n) are determined for binding to various nucleic acid lattices under a variety of environmental conditions. These results are used to show that GP32 can bind to nucleic acid lattices in two different conformations, and to characterize the molecular details of these binding species. Further insight into the molecular origins of binding cooperativity is obtained by determining these thermodynamic parameters also for the specifically proteolytically degraded GP32 fragments GP32 I (C-terminal peptide removed) and GP32 III (C- and N-terminal peptides removed). It is also shown that these GP32-nucleic acid binding measurements can be used to provide a quantitative molecular interpretation of the sequential (competitive) binding equilibria involved in the autogenous translational regulation of GP32 synthesis (Lemaire et al., 1978, J. Mol. Biol. 126:73, 1978), and to illustrate some general principles of the development of interactional specificity in cooperatively binding protein-nucleic acid complexes. Preliminary experiments have also been carried out on the kinetics of GP32 association to, and dissociation from, single-stranded nucleic acid lattices. In particular, fluorescence stopped-flow measurements of the dissociation of GP32 from such lattices as a function of lattice saturation (and protein cluster size) can be interpreted to suggest that the protein may translocate ("slide") on the lattice before dissociation, These studies permit an approach to possible rates and mechanisms of such translocation events.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Amodio F. J., Jenkins M., Gutmann E. D., Ferris F. L. Studies with DNA-cellulose chromatography. I. DNA-binding proteins from Escherichia coli. Cold Spring Harb Symp Quant Biol. 1968;33:289–305. doi: 10.1101/sqb.1968.033.01.033. [DOI] [PubMed] [Google Scholar]
- Alberts B. M., Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970 Sep 26;227(5265):1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- Berg O. G., Blomberg C. Association kinetics with coupled diffusional flows. Special application to the lac repressor--operator system. Biophys Chem. 1976 Jul;4(4):367–381. doi: 10.1016/0301-4622(76)80017-8. [DOI] [PubMed] [Google Scholar]
- Epstein I. R. Kinetics of nucleic acid-large ligand interactions: exact Monte Carlo treatment and limiting cases of reversible binding. Biopolymers. 1979 Aug;18(8):2037–2050. doi: 10.1002/bip.1979.360180815. [DOI] [PubMed] [Google Scholar]
- Gold L., O'Farrell P. Z., Russel M. Regulation of gene 32 expression during bacteriophage T4 infection of Escherichia coli. J Biol Chem. 1976 Nov 25;251(22):7251–7262. [PubMed] [Google Scholar]
- Hosoda J., Moise H. Purification and physicochemical properties of limited proteolysis products of T4 helix destabilizing protein (gene 32 protein). J Biol Chem. 1978 Oct 25;253(20):7547–7558. [PubMed] [Google Scholar]
- Hosoda J., Takacs B., Brack C. Denaturation of T4 DNA by an in vitro processed gene 32-protein. FEBS Lett. 1974 Oct 15;47(2):338–342. doi: 10.1016/0014-5793(74)81043-4. [DOI] [PubMed] [Google Scholar]
- Jensen D. E., Kelly R. C., von Hippel P. H. DNA "melting" proteins. II. Effects of bacteriophage T4 gene 32-protein binding on the conformation and stability of nucleic acid structures. J Biol Chem. 1976 Nov 25;251(22):7215–7228. [PubMed] [Google Scholar]
- Kelly R. C., Jensen D. E., von Hippel P. H. DNA "melting" proteins. IV. Fluorescence measurements of binding parameters for bacteriophage T4 gene 32-protein to mono-, oligo-, and polynucleotides. J Biol Chem. 1976 Nov 25;251(22):7240–7250. [PubMed] [Google Scholar]
- Kelly R. C., von Hippel P. H. DNA "melting" proteins. III. Fluorescence "mapping" of the nucleic acid binding site of bacteriophage T4 gene 32-protein. J Biol Chem. 1976 Nov 25;251(22):7229–7239. [PubMed] [Google Scholar]
- Krisch H. M., Bolle A., Epstein R. H. Regulation of the synthesis of bacteriophage T4 gene 32 protein. J Mol Biol. 1974 Sep 5;88(1):89–104. doi: 10.1016/0022-2836(74)90296-4. [DOI] [PubMed] [Google Scholar]
- Lemaire G., Gold L., Yarus M. Autogenous translational repression of bacteriophage T4 gene 32 expression in vitro. J Mol Biol. 1978 Nov 25;126(1):73–90. doi: 10.1016/0022-2836(78)90280-2. [DOI] [PubMed] [Google Scholar]
- Lohman T. M. Kinetics of the t4 gene 32 protein-single-stranded nucleic Acid interaction. Biophys J. 1980 Oct;32(1):458–460. doi: 10.1016/S0006-3495(80)84982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., von Hippel P. H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974 Jun 25;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Peterman B. F., Wu C. W. Ionic strength perturbation kinetics of gene 32 protein dissociation from its complex with single-stranded DNA. Biochemistry. 1978 Sep 5;17(18):3889–3892. doi: 10.1021/bi00611a033. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Lohman M. L., De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976 Oct 25;107(2):145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- Richter P. H., Eigen M. Diffusion controlled reaction rates in spheroidal geometry. Application to repressor--operator association and membrane bound enzymes. Biophys Chem. 1974 Oct;2(3):255–263. doi: 10.1016/0301-4622(74)80050-5. [DOI] [PubMed] [Google Scholar]
- Russel M., Gold L., Morrissett H., O'Farrell P. Z. Translational, autogenous regulation of gene 32 expression during bacteriophage T4 infection. J Biol Chem. 1976 Nov 25;251(22):7263–7270. [PubMed] [Google Scholar]
- Williams K. R., Konigsberg W. Structural changes in the T4 gene 32 protein induced by DNA polynucleotides. J Biol Chem. 1978 Apr 10;253(7):2463–2470. [PubMed] [Google Scholar]


