Abstract
The relationships between porin deficiency, active efflux of fluoroquinolones, and extended-spectrum β-lactamase (ESBL) production were determined for 53 clinical isolates of Klebsiella pneumoniae. Thirty-two ESBL-positive strains (including 22 strains expressing porins and 10 strains lacking porins) and 21 ESBL-negative strains were evaluated. Active efflux of norfloxacin was defined as a ≥50% increase in the accumulation of norfloxacin in the presence of carbonyl cyanide m-chlorophenylhydrazone (CCCP) in comparison with the corresponding basal value in the absence of CCCP. The quinolone resistance-determining regions of both gyrA and parC from 13 strains, representing all isolates with different porin profiles and with or without active efflux, were determined. Porin loss was significantly more common among ESBL-positive strains (10 of 32 [31.2%]) than among ESBL-negative strains (0 of 2 [0%]) (P < 0.01). Active efflux was observed in 7 of 10 (70%) strains lacking porins and in 4 of 43 (9.3%) strains producing porins (P < 0.001). The 11 strains showing active efflux corresponded to 3 of 21 (14.3%) ESBL-negative strains and 8 of 32 (25.5%) ESBL-positive strains (P > 0.05). Basal values of norfloxacin accumulation were higher in strains lacking active efflux than in those that had this mechanism (P < 0.05). In the absence of topoisomerase changes, the contribution of either porin loss or active efflux to fluoroquinolone resistance in K. pneumoniae was negligible. It is concluded that among K. pneumoniae strains of clinical origin, porin loss was observed only in those producing ESBL, and that a significant number of porin-deficient strains also expressed active efflux of norfloxacin. In terms of fluoroquinolone resistance, both mechanisms are significant only in the presence of topoisomerase modifications.
Klebsiella pneumoniae strains producing extended-spectrum β-lactamases (ESBL) are more frequently resistant to fluoroquinolones than K. pneumoniae strains lacking these enzymes (22). ESBL-producing K. pneumoniae strains intermediate or resistant to ciprofloxacin contain gyrA mutations, alone or combined with parC mutations (4, 5, 6, 12, 25). It is also possible that the plasmid coding for the ESBL, or an additional plasmid within the same strain, may contain a qnr-like determinant as described for organisms expressing plasmid-mediated AmpC type enzymes (14). Qnr is responsible for low-level resistance to quinolones and favors increased resistance to these drugs (14).
An alternative explanation for β-lactam-fluoroquinolone coresistance in ESBL-producing K. pneumoniae is a decrease in the permeability of the outer membrane to both classes of agents because of porin alterations. Non-ESBL-producing strains usually express the two major porins (OmpK35 and OmpK36) of the species, while ESBL-producing strains commonly express either only one of these (normally OmpK36) or no porin (9). Porin loss in ESBL-producing K. pneumoniae causes resistance to cefoxitin and increased resistance or decreased susceptibility to all oxyimino cephalosporins, zwitterionic cephalosporins, and β-lactam-β-lactamase combinations (1, 11, 13). Loss of these channels causes a moderate (two- to fourfold) increase in the level of resistance to fluoroquinolones when modifications of topoisomerase II (alone or combined with changes in topoisomerase IV) are present (5, 12). The direct role of either OmpK35 or OmpK36 in the resistance of K. pneumoniae strains to fluoroquinolones and to β-lactams has been demonstrated. Expression of either porin from a vector containing wild-type ompK35 or ompK36 in strains lacking both OmpK35 and OmpK36 decreases the level of resistance (1, 13; A. Doménech-Sánchez, S. Hernández-Allés, L. Martínez-Martínez, and V. J. Benedí, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C186, 1998).
In addition to decreased permeability, it is possible that active efflux also contributes to fluoroquinolone resistance in K. pneumoniae. A few clinical isolates of ESBL-producing K. pneumoniae that have porin deficiencies and express energy-dependent efflux of fluoroquinolones have been described (5, 12). In Escherichia coli and Salmonella enterica serovar Typhimurium, pumps for efflux of quinolones may also export β-lactams from the cells (19-21). Expression of the ramA locus from K. pneumoniae in E. coli causes resistance to norfloxacin and is related to both increased efflux of antimicrobial agents and loss of the OmpF porin (8). These activities are similar to those caused by the transcriptional activator marA of E. coli, which contributes to multiple antimicrobial resistance by increasing the activity of the AcrAB-TolC efflux system and by decreasing the permeability of the outer membrane due to increased transcription of micF, a transcriptional repressor of the porin gene ompF (3, 21).
The objectives of this study were (i) to determine the activities of several quinolones against clinical isolates of K. pneumoniae with characterized mechanisms of resistance, (ii) to establish whether porin loss and active efflux of fluoroquinolones are more frequently observed in clinical isolates of K. pneumoniae producing ESBL than in those not producing ESBL, (iii) to evaluate the potential relationship between porin expression and active efflux, and (iv) to analyze the roles of porin loss and active efflux in the quinolone resistance levels of clinical strains of K. pneumoniae with or without mutations in topoisomerase-encoding genes.
MATERIALS AND METHODS
Bacterial strains.
Fifty-three K. pneumoniae strains of clinical origin were studied. Three groups of strains were selected for evaluation in the present study: (i) 10 clonally unrelated strains of ESBL-producing K. pneumoniae deficient in both OmpK35 and OmpK36 expression (see below) isolated either in our laboratory or in another geographical zone (porin-deficient strains), (ii) 22 clonally unrelated ESBL-producing strains expressing porins in their outer membranes (these include 3 strains clonally related to those of the previous group), and (iii) 21 clonally unrelated non-ESBL-producing K. pneumoniae strains. We lack, and are not aware of, ESBL-negative clinical strains of K. pneumoniae deficient in the two major porins of the species.
The 32 ESBL-producing K. pneumoniae strains included strains C1 and C2 (14), strain CSUB10R (1), and strains MCQ-102, MCQ-121, and MCQ-122, kindly provided by P. Bradford (2); strains GAJ-3, GAJ-18, and GAJ-20, kindly provided by G. A. Jacoby (G. A. Jacoby, P. Han, M. Alvarez, and F. Tenover, Abstr. 35th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C40, 1995); six strains (LTZ205, LTZ-3273, RL-1, RL-2, RL-5, and RL-5X) from Greece, kindly provided by L. Tzouvelekis (24); and 17 strains from the University Hospital Virgen Macarena, Seville, Spain, isolated between 1996 and 1998. ESBL-deficient organisms were isolated between 1996 and 1998 at the University Hospital Virgen Macarena, Seville, Spain (HUS strains; n = 19) or the Ciudad Sanitaria Universitaria de Bellvitge, Barcelona, Spain (CSUB strains; 2 strains from the same patient).
Organisms were identified with the API 20E system (bioMérieux, La Balme Les Grottes, France) and maintained in tryptic soy broth (Difco, Detroit, Mich.) with 20% glycerol (Difco) at −80°C until use.
Clonal relationship of strains was determined by pulsed-field gel electrophoresis (PFGE) of genomic DNA digested with SmaI as previously described (1). All ESBL-deficient strains from Seville represented different clones, while the two CSUB strains showed the same PFGE pattern (data not shown).
E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as control strains in susceptibility testing assays.
Susceptibility testing.
MICs of antimicrobial agents were determined by microdilution according to NCCLS guidelines (17). The following agents were tested: ciprofloxacin (Bayer, Leverkusen, Germany), clinafloxacin (Parke-Davis, Ann Arbor, Mich.), moxifloxacin (Bayer), nalidixic acid (Sigma, Madrid, Spain), norfloxacin (Sigma), pefloxacin (Rhône-Poulenc, St. Antoine, France), pipemidic acid (Sigma), and trovafloxacin (Pfizer, Groton, Conn.). When available, clinical categories were determined according to NCCLS guidelines (18).
ESBL production was detected by microdilution, and strains were considered ESBL positive when clavulanic acid (fixed concentration, 2 μg/ml) caused a ≥8-fold reduction of the MICs of ceftazidime, cefotaxime, and/or aztreonam.
Analysis of OMPs.
Bacterial cells grown to logarithmic phase were lysed by sonication. Outer membrane proteins (OMPs) were obtained after treatment of cell membranes recovered by ultracentrifugation with sodium lauryl-sarcosynate (2%; Sigma) and subsequent ultracentrifugation. OMP profiles were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 11% acrylamide, 0.345% bisacrylamide, and 0.1% sodium dodecyl sulfate in the running gel. After electrophoresis, proteins were stained with Coomassie brilliant blue (Sigma) (11).
Accumulation of norfloxacin.
Bacteria grown in nutrient broth were suspended in phosphate-buffered saline (PBS) (ca. 0.650 mg of bacteria [dry weight]/ml), incubated with 10 μg of norfloxacin/ml at 37°C for 30 min, and centrifuged through a silicone oil barrier (ρ, 1.029 g/cm3) to eliminate extracellular quinolone. For the 11 strains of 53 evaluated that did not traverse the silicon oil layer, extracellular norfloxacin was eliminated by a wash with 1 ml of PBS (at 12,800 × g and 4°C for 1 min) (16). The cell pellet obtained by either method was placed in 2 ml of 0.1 M glycine-HCl buffer (pH 3.0), vortexed, and centrifuged for 5 min at 12,800 × g. The amount of norfloxacin in the supernatant (basal accumulation) was measured spectrophotometrically (12). The effect of the energy inhibitor carbonyl cyanide m-chlorophenylhydrazone (CCCP) (0.1 mM) was evaluated in parallel by addition of CCCP to bacterial suspensions 10 min after the addition of norfloxacin and reincubation of the cells for 20 min. Experiments were done in duplicate on three different days. An organism was considered to express energy-dependent accumulation of norfloxacin when CCCP enhanced basal accumulation by at least 50%, a threshold which corresponds to twice the standard deviation of the mean basal accumulation for the strains tested.
Analysis of mutations in topoisomerase-encoding genes.
Thirteen strains, representative of all possible combinations among level of susceptibility, porin profile, and expression of active efflux, were tested. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV were determined by sequencing the quinolone resistance-determining regions (QRDRs) of the gryA and parC genes by use of primers previously described by Deguchi et al. (4).
Statistical analyses.
Relationships between ESBL production, loss of porins, and expression of active efflux were determined by the chi-square test and were considered statistically significant at a P of <0.05. Norfloxacin accumulation values were expressed as means ± standard deviations, and differences among groups were compared by analysis of variance, which was used to assess statistical significance at a P of <0.05.
RESULTS
Susceptibility testing.
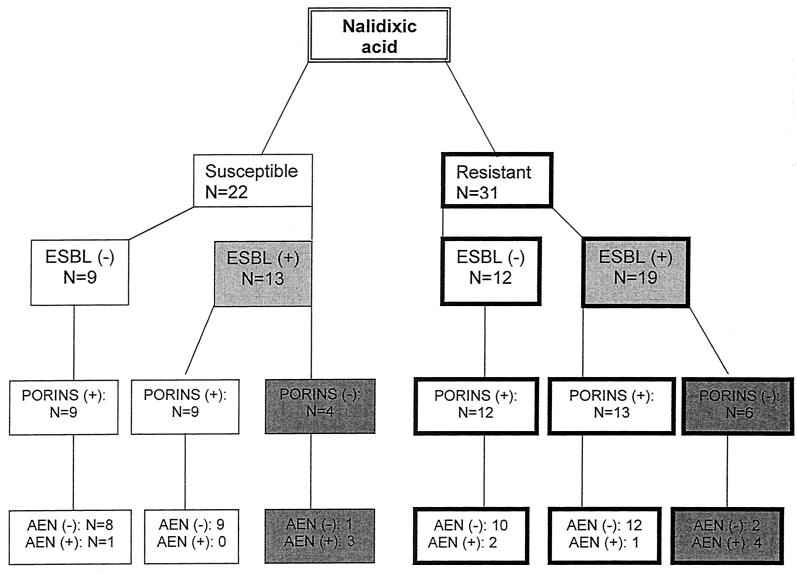
Twenty-two out of the 53 strains studied were susceptible to nalidixic acid (NAL-S), and 31 were resistant to nalidixic acid (NAL-R; MICs, in all cases, were ≥64 μg/ml). NAL-S strains included 13 strains producing ESBL and 9 strains not producing ESBL. NAL-R strains included 19 and 12 organisms producing and not producing ESBL, respectively (Fig. 1).
FIG. 1.
Relationship between susceptibility to nalidixic acid, ESBL-production, porin expression, and active efflux of norfloxacin (AEN) in 53 clinical isolates of K. pneumoniae.
All NAL-S strains were inhibited by pipemidic acid at 8 μg/ml, and all NAL-R strains were inhibited by ≥16 μg of pipemidic acid/ml, except in one case (MIC of pipemidic acid, 4 μg/ml). All NAL-S strains were inhibited by ≤0.06 μg of ciprofloxacin or clinafloxacin/ml. MICs of both norfloxacin and pefloxacin for NAL-S strains were ≤0.06 μg/ml (16 strains) and 0.125 μg/ml (6 strains), while MICs of trovafloxacin and moxifloxacin for NAL-S strains were ≤0.06 μg/ml (20 strains), 0.125 μg/ml (1 strain), and 0.25 μg/ml (1 strain). The MIC90s (MICs at which 90% of isolates were inhibited) of norfloxacin and pefloxacin for NAL-S strains were 0.125 μg/ml, and MIC90s of the remaining four fluoroquinolones for this group of strains were ≤0.06 μg/ml.
Distributions of MICs of fluoroquinolones against NAL-R strains are shown in Table 1. It can be observed that MICs of norfloxacin and pefloxacin for NAL-R strains were always ≥0.5 μg/ml, and those of ciprofloxacin were always ≥0.125 μg/ml. On the other hand, MICs of clinafloxacin were ≤2 μg/ml for all 32 NAL-R strains. As shown in Table 1, MIC90s of all fluoroquinolones for which the NCCLS establishes breakpoints fall within the resistant category.
TABLE 1.
Distribution of MICs, MIC50s, and MIC90s of six fluoroquinolones against 31 NAL-R strains of K. pneumoniae
| Drug | No. of strains inhibited at the following MIC (μg/ml):
|
MIC50 (μg/ml) | MIC90 (μg/ml) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | ≥64 | |||
| Ciprofloxacin | 1 | 1 | 5 | 1 | 5 | 6 | 4 | 4 | 2 | 2 | 4 | 32 | |
| Clinafloxacin | 10 | 2 | 4 | 9 | 4 | 2 | 0.25 | 1 | |||||
| Moxifloxacin | 4 | 3 | 6 | 6 | 8 | 2 | 1 | 1 | 2 | 8 | |||
| Norfloxacin | 4 | 4 | 2 | 2 | 5 | 8 | 2 | 4 | 8 | 64 | |||
| Pefloxacin | 4 | 2 | 3 | 3 | 6 | 8 | 2 | 3 | 8 | 32 | |||
| Trovafloxacin | 2 | 3 | 7 | 4 | 8 | 4 | 2 | 1 | 2 | 8 | |||
Analysis of OMPs.
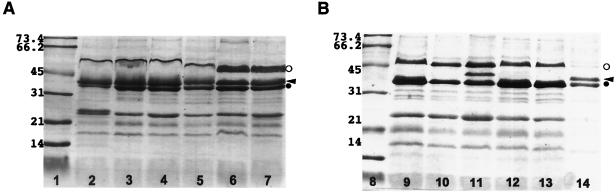
All 53 strains tested produced an OmpA-like protein of about 32 kDa. All 21 ESBL-negative strains (either NAL-S or NAL-R) produced OMPs in the range of 35 to 37 kDa, compatible with the hypothesis that they are porins. Among ESBL-producing strains, porin expression was observed in 22 strains (68.8%), and not in the remaining 10 strains (31.2%) (Fig. 1). These data indicate that porin loss is significantly (P < 0.01) more frequent among ESBL-producing strains. Representative results of OMP analysis are presented in Fig. 2.
FIG. 2.
OMP profiles of ESBL-producing (A) and non-ESBL-producing (B) K. pneumoniae strains. (A) Lane 2, strain HUS 76-96; lane 3, strain HUS 56-96; lane 4, strain HUS 33-96; lane 5, strain HUS 71-96; lane 6, strain HUS 20-97; lane 7, strain HUS 91-96. (B) Lane 9, strain RL-1; lane 10, strain RL-2; lane 11, strain RL-5X; lane 12, strain RL-5b; lane 13, strain GAJ-3; lane 14, strain HUS 8-98. Lanes 1 and 8, molecular size markers (in kilodaltons). Symbols: open circle, LamB-like protein; arrowhead, porin; solid circle, OmpA-like protein.
Accumulation of norfloxacin.
In a set of independent experiments in which the absolute value of norfloxacin accumulation was determined for 10 strains (representative of the different accumulation models observed) by both the silicon oil method and the PBS washing method, lower absolute values were obtained with the latter method, but the percentages of increase in norfloxacin accumulation caused by CCCP determined by the two methods were not significantly different (data not shown). For this reason we have considered the percentage of increase in norfloxacin accumulation caused by CCCP to be an indicator of efflux, regardless of whether this was determined by the silicon oil method (42 strains) or the PBS washing method (11 strains).
Only for 11 of 53 strains did CCCP enhance the concentration of accumulated norfloxacin more than 50% over the corresponding basal accumulation value. Active efflux was significantly more frequent (P < 0.001) among porin-deficient strains (7 of 10 [70.0%]) than among porin-producing strains (4 of 43 [9.3%]). A higher percentage of strains showing active efflux of norfloxacin was noted among ESBL-positive strains (8 of 32 [25.0%]) than among ESBL-negative strains (3 of 21 [14.3%]), but this difference was not statistically significant (P > 0.05). For seven strains the increase in norfloxacin accumulation due to CCCP was 210 to 275%, and for four strains the increase was 56 to 75%. Addition of CCCP caused a <50% increase in the accumulation of norfloxacin in 19 strains: 1 to 20% in 6 strains, 11 to 20% in 8 strains, and 31 to 49% in 5 strains. Interestingly, CCCP caused decreased norfloxacin accumulation in the remaining 23 strains: the percentages of decrease were 62% (n = 1), 36% (n = 1), 20 to 26% (n = 3), 10 to 19% (n = 10), and 1 to 9% (n = 8).
Among the 42 strains for which accumulation of norfloxacin was assayed by the silicon oil method, basal accumulation values were higher in strains lacking active efflux of norfloxacin than in strains showing active efflux of norfloxacin (P < 0.05). No such comparison was attempted for strains assayed by the centrifugation method because none of them expressed active efflux as previously defined.
Accumulation of norfloxacin in strains lacking active efflux after cells were treated with CCCP ranged from 219 to 589 ng/mg of bacteria (dry weight), while the corresponding range in active-efflux-positive strains was 273 to 505 ng/mg of bacteria (dry weight) (P > 0.05).
Norfloxacin accumulation in 11 strains with active efflux of norfloxacin is presented according to porin expression pattern in Table 2. Norfloxacin accumulation in three strains deficient in porin expression and lacking active efflux of norfloxacin (Table 3) was not significantly different (P > 0.05) from that observed for strains that do not express active efflux but express porins.
TABLE 2.
Accumulation of norfloxacin without and with 100 μM CCCP in 11 strains of K. pneumoniae showing energy-dependent accumulation of norfloxacin and expressing porins or not
| Strain | Accumulation of norfloxacina (ng/mg of bacteria [dry wt])
|
|
|---|---|---|
| Basalb | With CCCPc | |
| Porin deficient | ||
| RL-1 | 103 ± 25 | 273 ± 53 |
| RL-5b | 128 ± 25 | 352 ± 34 |
| LTZ-3273 | 253 ± 41 | 437 ± 59 |
| C1 | 247 ± 61 | 505 ± 55 |
| C2 | 220 ± 35 | 410 ± 81 |
| CSUB10R | 188 ± 19 | 429 ± 50 |
| MCQ-122 | 269 ± 39 | 430 ± 32 |
| Porin expressing | ||
| CSUB-61A | 298 ± 22 | 445 ± 13 |
| HUS 71-96 | 245 ± 59 | 436 ± 73 |
| HUS 298-98 | 182 ± 80 | 417 ± 56 |
| GAJ-18 | 161 ± 63 | 331 ± 32 |
Data are means ± standard deviations from three independent duplicate experiments.
Tested in the absence of CCCP.
Tested in the presence of 100 μM CCCP.
TABLE 3.
Accumulation of norfloxacin without and with CCCP in three strains of K. pneumoniae deficient in porins and lacking energy-dependent accumulation of norfloxacin
| Strain | Accumulation of norfloxacina (ng/mg of bacteria [dry wt])
|
|
|---|---|---|
| Basal | With 100 mM CCCP | |
| RL-2 | 441 ± 80 | 387 ± 90 |
| GAJ-3 | 386 ± 33 | 386 ± 33 |
| MCQ-102 | 454 ± 119 | 372 ± 88 |
Data are expressed as means ± standard deviations from three independent duplicate experiments.
Target modifications.
As shown in Table 4, the QRDRs of both GyrA and ParC of the four NAL-S strains evaluated were identical to the sequences reported for K. pneumoniae strain ATCC 13833: codons 83 and 87 of gyrA coded for Ser and Asp, respectively, while codons 78, 80, and 84 of parC coded for Gly, Ser, and Glu, respectively. The same results were obtained for two strains with low-level resistance to nalidixic acid: MICs of nalidixic acid were 64 and 256 μg/ml for strains HUS 33-99 and HUS 298-98, respectively. All strains with high-level resistance to nalidixic acid (MIC ≥ 1,024 μg/ml) had mutations at gyrA, either alone or combined with mutations in parC. No cases of mutation in parC were observed in the absence of mutations in gyrA. Mutations in codon 83 of gyrA caused a change from Ser to either Phe or Tyr, while mutations in codon 87 caused a change from Asp to either Tyr or Asn. Mutations in parC caused changes at position 78 (Gly to Cys), 80 (Ser to Arg), or 84 (Glu to Gly). There were no obvious relationships between the types of mutations in topoisomerase genes and the level of resistance to fluoroquinolones.
TABLE 4.
Mutations at the QRDRs of gyrA and parC in 13 clinical strains of K. pneumoniae
| Strain | Presence or absencea of:
|
MIC
|
Mutationd
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ESBL | Porin | Efflux | NALb | CIPc | GyrA
|
ParC
|
||||
| Ser83 | Asp87 | Gly78 | Ser80 | Glu84 | ||||||
| CSUB-61A | − | + | − | 0.5 | 0.004 | — | — | — | — | — |
| CSUB-61B | − | + | + | 0.5 | 0.008 | — | — | — | — | — |
| HUS 76-96 | − | + | − | 2 | ≤0.06 | — | — | — | — | — |
| LTZ-3273 | + | − | + | 16 | ≤0.06 | — | — | — | — | — |
| HUS 33-96 | − | + | − | 64 | 0.125 | — | — | — | — | — |
| HUS 298-98 | − | + | + | 256 | 0.5 | — | — | — | — | — |
| C2 | + | − | + | >1,024 | 0.5 | Phe | — | — | — | — |
| MCQ-102 | +e | − | − | >1,024 | 0.5 | Tyr | — | — | — | — |
| C1 | + | − | + | >1,024 | 4 | Tyr | — | — | — | |
| HUS 22-97 | + | + | − | >1,024 | 4 | Phe | Tyr | — | — | Gly |
| HUS 20-97 | − | + | − | >1,024 | 8 | Phe | Tyr | — | — | Gly |
| MCQ-121 | +e | + | − | >1,024 | 64 | Phe | Asn | Cys | Arg | — |
| MCQ-122 | +e | − | + | >1,024 | 128 | Phe | Asn | Cys | Arg | — |
+, presence; −, absence.
NAL, nalidixic acid.
CIP, ciprofloxacin.
—, no change with respect to wild-type sequence.
AmpC type β-lactamase producer.
Norfloxacin accumulation and porin expression in pairs of clonally related strains (presenting the same PFGE pattern) are presented in Table 5.
TABLE 5.
Accumulation of norfloxacin without and with CCCP in five pairs of clonally related K. pneumoniae strains with different mechanisms of resistance to fluoroquinolones
| Strain | PFGE pattern | Presence or absencea of:
|
Norfloxacin accumu- lationsb (ng/mg of bacteria [dry wt])
|
Mutation(s)c
|
MIC (μg/ml)
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ESBL | Porin | Basal | With CCCP | GyrA | ParC | NALd | CIPe | NORf | CLINg | ||
| CUSB-61A | 1 | − | + | 570 ± 27 | 219 ± 78 | wt | wt | 0.5 | 0.004 | 0.03 | ≤0.06 |
| CUSB-61B | 1 | − | + | 298 ± 22 | 445 ± 13 | wt | wt | 0.5 | 0.008 | 0.06 | ≤0.06 |
| RL-5X | 2 | + | + | 470 ± 25 | 409 ± 79 | wt | wt | 8 | ≤0.06 | 0.25 | ≤0.06 |
| RL-2 | 2 | + | − | 441 ± 80 | 387 ± 90 | wt | wt | 4 | ≤0.06 | 0.5 | ≤0.06 |
| LB2h | 3 | + | + | 286 ± 24 | 385 ± 65 | Ser83Tyr | wt | >1,024 | 0.5 | 2 | ≤0.06 |
| LB4h | 3 | + | − | 116 ± 21 | 263 ± 38 | Ser83Tyr | wt | >1,024 | 1 | 4 | ≤0.06 |
| CSUB10S | 4 | + | + | 313 ± 6 | ND | Ser83Phe | wt | >1,024 | 0.5 | 1 | ≤0.06 |
| CSUB10R | 4 | + | − | 188 ± 19 | 429 ± 50 | Ser83Phe | wt | >1,024 | 4 | 8 | ≤0.06 |
| MCQ-121 | 5 | +i | + | 434 ± 53 | 405 ± 79 | Ser83Phe, Asp87Asn | Gly78Cys, Ser80Arg | >1,024 | 64 | 256 | 1 |
| MCQ-122 | 5 | +i | − | 298 ± 22 | 445 ± 13 | Ser83Phe, Asp87Asn | Gly78Cys, Ser80Arg | >1,024 | 128 | >256 | 2 |
+, presence; −, absence.
Basal, without CCCP; with CCCP, in the presence of 100 μM CCCP. Data are means ± standard deviations from three independent duplicate experiments. ND, not determined.
wt, wild type.
NAL, nalidixic acid.
CIP, ciprofloxacin.
NOR, norfloxacin.
CLIN, clinafloxacin.
Data for LB2 and LB4 have been reported previously (11) and are included for comparison.
Also producing the AmpC type ACT-1 β-lactamase.
DISCUSSION
Resistance to fluoroquinolones in K. pneumoniae is more commonly found among ESBL-producing strains than among non-ESBL producers (22). In this study the possible associations between altered permeability, active efflux of norfloxacin, and ESBL production in clinical isolates of K. pneumoniae were evaluated. Additionally, topoisomerase changes were also determined for some strains in order to evaluate in more detail the relevance of the above-mentioned mechanisms. The possible protection of DNA gyrase by a Qnr-like protein (14, 23) has been ruled out in a parallel study (data not shown). Figure 1 shows the distribution of the 53 strains tested in this study according to quinolone susceptibility, ESBL production, porin expression, and active efflux expression.
Our results indicate that in clinical isolates of K. pneumoniae, porin loss is more frequent among ESBL-positive than among ESBL-negative strains and that in this organism active efflux of norfloxacin occurs more commonly among porin-deficient strains. Although the percentage of strains showing active efflux was higher among ESBL-positive than among ESBL-negative isolates, this trend was not statistically significant. It is possible that the method we used for detecting active efflux is not sensitive enough and/or that the breakpoint for defining it (a 50% increase in accumulation in the presence of CCCP) is too strict (note that for five strains, CCCP enhanced accumulation by 31 to 49%). It would be interesting to evaluate a larger number of strains, to measure accumulation of other substrates, or to use other methodological approaches (an assay with radiolabeled fluoroquinolone, or immunological detection of the efflux system proteins) to confirm that active efflux is not related to ESBL production.
The data in Table 1 indicate the existence of cross-resistance to nalidixic acid and fluoroquinolones in K. pneumoniae, although the effects of the mechanisms of resistance are different for different compounds. Clinafloxacin was, in terms of MIC90s, the most active fluoroquinolone of those tested here, followed by moxifloxacin, trovafloxacin, ciprofloxacin, pefloxacin, and norfloxacin.
K. pneumoniae ATCC 13833 and many other quinolone-susceptible clinical isolates contain serine instead of threonine at position 83, as previously reported for K. pneumonaie M5a1 (7). A recent report has shown that Klebsiella oxytoca 13182 contains threonine (25). Our data with four NAL-S strains also indicate that serine is normally found at position 83 of GyrA in K. pneumoniae, while Phe or Tyr is present in K. pneumoniae strains with high-level resistance to NAL. Changes in GyrA Asp87 to Tyr or to Asp have been observed in our NAL-R strains. These have also been the most frequent substitutions found in other studies (4, 5, 6, 25). Mutations in parC (coding for Ser80Arg and Glu84Gly) were observed in four out of five NAL-R and ciprofloxacin-resistant strains. In the two clonally related strains MCQ-121 and MCQ-122, a mutation in parC that is responsible for a Gly78Cys change and that has not been rpeviously reported for K. pneumoniae was noted. The relevance of this change awaits further study.
This study and previous studies (4, 5, 6, 12) indicate that the levels of resistance to nalidixic acid and fluoroquinolones in clinical isolates of K. pneumoniae are not completely explained by changes in the QRDRs of gyrA and parC (Table 4; compare strain HUS76-96 with strain LTZ-3273 and strain MCQ-102 with strain C1).
Strains CSUB10R and LB4 are deficient in porins, contain a GyrA change, and in addition exhibit energy-dependent accumulation of fluoroquinolones (11, 12). On the other hand, the clonally related strains CSUB10S and LB2 express OmpK36 and do not show energy-dependent accumulation of fluoroquinolones. This suggests a link between the loss of the two porins and expression of energy-dependent accumulation of fluoroquinolones in clinical isolates of K. pneumoniae. To evaluate this hypothesis, norfloxacin accumulation was determined in a collection of unrelated porin-deficient strains and, for comparison, in a representative number of porin-expressing strains (either producing or not producing ESBL).
We measured norfloxacin accumulation by using a fluorometric assay, eliminating extracellular fluoroquinolone by centrifugation through a silicon oil barrier or by washing cells with PBS. In this study, lower norfloxacin accumulation was obtained when the PBS washing method instead of the silicon oil method was used, in agreement with previous reports (16; unpublished data). Although the actual reasons for these differences have not yet been satisfactorily explained, the approach we used of defining active efflux as a ratio of accumulation in the presence of CCCP to accumulation in the absence of CCCP allows comparison of the results obtained by these two methods.
In some of the strains we studied, CCCP caused decreased (rather than increased) accumulation of norfloxacin. It can be hypothesized that, in these strains, CCCP induces the expression of one or more efflux pumps also involved in fluoroquinolone elimination, but further work is needed to clarify these results.
Seven of the 11 strains expressing active efflux of norfloxacin were porin deficient, while among the 10 porin-deficient strains evaluated, 7 expressed energy-dependent accumulation of fluoroquinolones (Fig. 1).
Accumulations (either in the absence or in the presence of CCCP) in the seven strains expressing efflux of fluoroquinolones and lacking porins were not statistically different (P < 0.05) from those in four strains expressing energy-dependent accumulation of fluoroquinolones and producing porins (Table 2), while accumulations in strains lacking active efflux were not different (P > 0.05) for strains expressing or lacking porins (Table 3). This suggests that when the major porins of K. pneumoniae are lost, norfloxacin (and perhaps other fluoroquinolones) may still accumulate in the cell by other means and that decreased accumulation depends more on elimination by active efflux than on decreased permeability by porin loss. Further studies are required to deduce the independent roles of active efflux and of loss of porins in the final amount of norfloxacin accumulated in strains expressing both mechanisms.
In a previous study it was shown that when other mechanisms of resistance are absent, loss of porins in K. pneumoniae C3 does not contribute significantly to quinolone resistance (10). In this study we have observed, for strains CSUB-61A and CSUB-61B, that expression of energy-dependent accumulation of fluoroquinolones alone does not significantly increase the level of resistance (Table 5). When both porin loss and active efflux are present in the same organism that has a topoisomerase change(s), a moderate increase in the level of fluoroquinolone resistance is observed. This has been described previously for strains LB2 and LB4 (11) and for strains CSUB10S and CSUB10R (1) and has been observed here for strains MCQ-121 and MCQ-122 (with double mutations in both gyrA and parC). This indicates that multiple target mutations are much more important than nonspecific mechanisms for quinolone resistance, but the latter may be a critical step favoring the emergence of mutants with altered targets.
K. pneumoniae may express a basal efflux of fluoroquinolones, as already observed for E. coli and other enterobacteria. The wide range of norfloxacin accumulation in the strains we studied may reflect variation in undefined factors that determine binding of the drug to the cell but may also represent variable expression of active efflux in different organisms. In E. coli, AcrAB-TolC is the more clinically relevant efflux system, since it is involved in the elimination of fluoroquinolones, β-lactams, and other antimicrobial agents (15, 20). Preliminary results (data not shown) indicate that K. pneumoniae contains an AcrAB homologue. New studies are in progress to evaluate the importance of AcrAB-TolC and other efflux systems in the strains included in this report.
Acknowledgments
We thank C. Ardanuy, P. Bradford, G. A. Jacoby, and L. Tzouvelekis for supplying bacterial strains.
This work was supported by the Fondo de Investigación Sanitaria, Ministerio de Sanidad y Consumo of Spain, grants 00/0242 (to L.M.-M.) and 01/0034 (to V.J.B.). A.D.-S. was supported by a predoctoral fellowship from the Ministerio de Educación y Cultura of the Spanish government.
REFERENCES
- 1.Ardanuy, C., J. Liñares, M. A. Domínguez, S. Hernández-Allés, V. J. Benedí, and L. Martínez-Martínez. 1998. Outer membrane profiles of clonally related Klebsiella pneumoniae isolates from clinical samples and activities of cephalosporins and carbapenems. Antimicrob. Agents Chemother. 42:1636-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen, S. P., L. M. McMurry, and S. B. Levy. 1989. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob. Agents Chemother. 33:1318-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deguchi, T., A. Fukuoka, M. Yasuda, M. Nakano, S. Ozeki, E. Kanematsu, Y. Nishino, S. Ishihara, Y. Ban, and Y. Kawada. 1997. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 41:699-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deguchi, T., T. Kawamura, M. Yasuda, M. Nakano, H. Fukuda, H. Kato, N. Kato, Y. Okano, and Y. Kawada. 1997. In vivo selection of Klebsiella pneumoniae strains with enhanced quinolone resistance during fluoroquinolone treatment of urinary tract infections. Antimicrob. Agents Chemother. 41:1609-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deguchi, T., M. Yasuda, T. Kawamura, M. Nakano, S. Ozeki, E. Kanematsu, Y. Nishino, and Y. Kawada. 1997. Improved antimicrobial activity of DU-6859a, a new fluoroquinolone, against quinolone-resistant Klebsiella pneumoniae and Enterobacter cloacae isolates with alterations in GyrA and ParC proteins. Antimicrob. Agents Chemother. 41:2544-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimri, G. P., and H. K. Das. 1990. Cloning and sequence analysis of gyrA gene of Klebsiella pneumoniae. Nucleic Acids Res. 18:151-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.George, A. M., R. M. Hall, and H. W. Stokes. 1995. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology 141:1909-1920. [DOI] [PubMed] [Google Scholar]
- 9.Hernández-Allés, S., S. Albertí, D. Alvarez, A. Doménech-Sánchez, L. Martínez-Martínez, J. Gil, J. M. Tomás, and V. J. Benedí. 1999. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145:673-679. [DOI] [PubMed] [Google Scholar]
- 10.Hernández-Allés, S., M. C. Conejo, A. Pascual, J. M. Tomás, V. J. Benedí, and L. Martínez-Martínez. 2000. Relationship between outer membrane alterations and susceptibility to antimicrobial agents in isogenic strains of Klebsiella pneumoniae. J. Antimicrob. Chemother. 46:273-277. [DOI] [PubMed] [Google Scholar]
- 11.Martínez-Martínez, L., S. Hernández-Allés, S. Albertí, J. M. Tomás, V. J. Benedí, and G. A. Jacoby. 1996. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 40:342-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez-Martínez, L., I. García, S. Ballesta, V. J. Benedí, S. Hernández-Allés, and A. Pascual. 1998. Energy-dependent accumulation of fluoroquinolones in quinolone-resistant Klebsiella pneumoniae strains. Antimicrob. Agents Chemother. 42:1850-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Martínez, L., A. Pascual, S. Hernández-Allés, D. Alvarez-Díaz, A. I. Suárez, J. Tran, V. J. Benedí, and G. A. Jacoby. 1999. Roles of β-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 43:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez-Martínez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 15.Mazzariol, A., Y. Tokue, T. M. Kanegawa, G. Cornaglia, and H. Nikaido. 2000. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob. Agents Chemother. 44:3441-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mortimer, P. G., and L. J. Piddock. 1991. A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 28:639-653. [DOI] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial susceptibility testing, 11th informational supplement. NCCLS document M100-S11. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.Nikaido, H., M. Basina, V. Nguyen, and E. Y. Rosenberg. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side chains. J. Bacteriol. 180:4686-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikaido, H., and H. I. Zgurskaya. 2001. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:215-218. [PubMed] [Google Scholar]
- 21.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paterson, D. L., L. Mulazimoglu, J. M. Casellas, W. C. Ko, H. Goossens, A. von Gottberg, S. Mohapatra, G. M. Trenholme, K. P. Klugman, J. G. McCormack, and V. L. Yu. 2000. Epidemiology of ciprofloxacin resistance and its relationship to extended-spectrum β-lactamase production in Klebsiella pneumoniae isolates causing bacteremia. Clin. Infect. Dis. 30:473-478. [DOI] [PubMed] [Google Scholar]
- 23.Tran, J. H., and G. A Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 99:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzouvelekis, L., E. Tzelepi, E. Prinarakis, M. Gazouli, A. Katrahoura, P. Giakkoupi, O. Paniara, and N. J. Legakis. 1998. Sporadic emergence of Klebsiella pneumoniae strains resistant to cefepime and cefpirome in Greek hospitals. J. Clin. Microbiol. 36:266-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weigel, L. M., C. D. Steward, and F. C. Tenover. 1998. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob. Agents Chemother. 42:2661-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]