Abstract
Streptococcus mutans is resistant to bacitracin, which is a peptide antibiotic produced by certain species of Bacillus. The purpose of this study was to clarify the bacitracin resistance mechanism of S. mutans. We cloned and sequenced two S. mutans loci that are involved in bacitracin resistance. The rgp locus, which is located downstream from rmlD, contains six rgp genes (rgpA to rgpF) that are involved in rhamnose-glucose polysaccharide (RGP) synthesis in S. mutans. The inactivation of RGP synthesis in S. mutans resulted in an approximately fivefold-higher sensitivity to bacitracin relative to that observed for the wild-type strain Xc. The second bacitracin resistance locus comprised four mbr genes (mbrA, mbrB, mbrC, and mbrD) and was located immediately downstream from gtfC, which encodes the water-insoluble glucan-synthesizing enzyme. Although the bacitracin sensitivities of mutants that had defects in flanking genes were similar to that of the parental strain Xc, mutants that were defective in mbrA, mbrB, mbrC, or mbrD were about 100 to 120 times more sensitive to bacitracin than strain Xc. In addition, a mutant that was defective in all of the mbrABCD genes and rgpA was more sensitive to bacitracin than either the RGP or Mbr mutants. We conclude that RGP synthesis is related to bacitracin resistance in S. mutans and that the mbr genes modulate resistance to bacitracin via an unknown mechanism that is independent of RGP synthesis.
Cariogenic Streptococcus mutans is known to be resistant to bacitracin. This property is often exploited in the isolation of this bacterium from the highly heterogeneous oral microflora (6). Bacitracin is a cyclic polypeptide antibiotic that is produced by certain species of Bacillus. The primary mechanism of action of this antibiotic is thought to be the inhibition of peptidoglycan synthesis (28). During peptidoglycan synthesis, C55-isoprenyl phosphate (IP) serves as a lipid carrier (24). After the translocation of sugar-peptide units to the ends of the linear peptidoglycan strands, the C55-isoprenyl pyrophosphate (IPP) is detached and dephosphorylated to IP by a membrane-bound pyrophosphatase, thus recycling IP for subsequent peptidoglycan synthesis (24, 28). Bacitracin binds tightly to IPP and prevents pyrophosphatase from interacting with IPP, thus reducing the amount of IP that is available for carrying sugar-peptide units.
In Escherichia coli, increased phosphorylation of IP, due to elevated intracellular levels of the lipid kinase encoded by bacA, appeared to confer resistance to bacitracin (1). Alternatively, E. coli mutants lacking membrane-derived oligosaccharides had reduced sensitivity to bacitracin because of reduced IP utilization (5). Pollock et al. (20) reported that certain gram-negative bacteria that synthesized exopolysaccharides acquired resistance to bacitracin by shutting down the synthesis of exopolysaccharides. On the other hand, Podlesek et al. (19) suggested that an ABC-type efflux system, which consisted of the BcrA, BcrB, and BcrC proteins, might be involved in the resistance of Bacillus licheniformis to bacitracin. However, the exact mechanism by which this transporter system mediates resistance is still unknown.
The clinical use of bacitracin by oral administration is getting much attention for its ability to eradicate vancomycin-resistant enterococci (VRE) from the gastrointestinal tracts of patients (2, 15, 25). Since S. mutans is found in human feces, the bacitracin-resistant phenotype of S. mutans could presumably be transferred to VRE. Despite increasing fears that VRE might acquire tolerance to bacitracin from S. mutans, the mechanism of bacitracin resistance in S. mutans remains a mystery.
In the present study, we isolated two bacitracin-sensitive mutants of S. mutans using random mutagenesis of an S. mutans genomic library that was constructed in an integration vector. One of the mutants had an inactivated rgpA gene that was previously shown to be involved in glucose-rhamnose polysaccharide (RGP) formation in the cell wall. The second mutant was disrupted in an unknown gene, and we characterized the plasmid-inserted chromosomal region of this mutant. The two mutants differed in their sensitivities to bacitracin. We discussed the relationship between bacitracin resistance and RGP synthesis in S. mutans.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The S. mutans strains and plasmids used in this study are listed in Tables 1 and 2. S. mutans strains Xc, Xc13, Xc26R, Xc41, Xc42, Xc43, and Xc44 were selected from the stock culture collection in the Department of Preventive Dentistry, Kyushu University Faculty of Dental Science, Fukuoka, Japan. Strains of S. mutans were grown at 37°C in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.). For transformants of S. mutans, erythromycin or tetracycline was added at a final concentration of 10 or 5 μg per ml, respectively.
TABLE 1.
S. mutans strains used in this study
| Strain | Relevant characteristicsa | Reference |
|---|---|---|
| Xc | Serotype c wild-type strain | 11 |
| Xc13 | Emr; strain Xc carrying Emr gene inserted into gtfC | 35 |
| Xc26R | Emr; strain Xc carrying P15A replicon and Emr gene both inserted into rmlD | 31 |
| Xc41 | Emr; strain Xc carrying P15A replicon and Emr gene both inserted into rgpA | 37 |
| Xc42 | Emr; strain Xc carrying P15A replicon and Emr gene both inserted into rgpB | 37 |
| Xc43 | Emr; strain Xc carrying P15A replicon and Emr gene both inserted into rgpC | 37 |
| Xc44 | Emr; strain Xc carrying P15A replicon and Emr gene both inserted into rgpD | 37 |
| XcB1 | Emr; strain Xc carrying P15A replicon and Emr gene both inserted into rgpA | This study |
| XcB2 | Emr; strain Xc carrying P15A replicon and Emr gene both inserted into mbrD | This study |
| Xc101 | Emr; strain Xc carrying P15A replicon and Emr gene both inserted into mbrA | This study |
| Xc102 | Emr; strain Xc carrying P15A replicon and Emr gene both inserted into mbrB | This study |
| Xc103 | Emr; strain Xc carrying P15A replicon and Emr gene both inserted into mbrC | This study |
| Xc104 | Emr; strain Xc carrying P15A replicon and Emr gene both inserted into mbrD | This study |
| Xc105 | Emr; strain Xc carrying P15A replicon and Emr gene both inserted into ORF5 | This study |
| Xc106 | Emr; strain Xc carrying Emr gene instead of mbrA, mbrB, mbrC, and mbrD | This study |
| Xc146 | Emr and Tetr; strain Xc106 carrying Tetr gene inserted into rgpA | This study |
Emr, erythromycin resistance; Tetr, tetracycline resistance.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| pResEmBBN | Emr; P15A replicon | 23 |
| pBluescriptII SK(+) | Apr; phagemid cloning vector | Stratagene |
| pResYT10 | Emr; P15A replicon with the unique PvuII site immediately downstream of erythromycin resistance gene | 23 |
| pHT1 | Emr; marker rescue plasmid of 7.0-kb PstI fragment of XcB1 chromosomal DNA which includes downstream region of rmlD gene and four intact rgp genes | This study |
| pHT2 | Emr; marker rescue plasmid of 9.8-kb HindIII fragment of XcB2 chromosomal DNA which includes downstream region of gtfC gene and four intact mbr genes | This study |
| pBSHT1 | pBluescriptII SK+ containing 4.3-kb PstI fragment which carries of Xc rgp genes | This study |
| pBSHT2 | pBluescriptII SK+ containing 7.7-kb HindIII fragment which carries of Xc mbr genes | This study |
DNA manipulation.
Transformation of S. mutans and E. coli was carried out as previously described (34). Chromosomal DNA of S. mutans strains was prepared by the method of Perry et al. (17). Standard DNA recombinant procedures such as DNA isolation, endonuclease restriction, and ligation were carried out according to the methods of Sambrook et al. (22).
Isolation of bacitracin-sensitive transformants of S. mutans.
A complete Sau3AI digest of the chromosomal DNA of S. mutans strain Xc was ligated to BamHI- and BglII-digested pResEmBBN. The pResEmBBN integration plasmid, whose erythromycin resistant gene is able to work in S. mutans, cannot duplicate in S. mutans. This plasmid was produced by Shiroza and Kuramitsu in the process of constructing pResEmMCS11 (23) and is equivalent to pResEmMCS11 except that it lacks the restriction sites between XbaI and NotI in the multicloning site. The S. mutans strain Xc was randomly mutated by transformation with the S. mutans genomic library in pResEmBBN, as described previously (33). The insert fragment of the integration plasmid should be located within the target gene to disrupt the gene by Campbell-type recombination. Transformants were selected on BHI agar plates that contained 10 μg of erythromycin per ml. About 12,000 transformants were transferred to both BHI agar plates with or without 1 unit of bacitracin (Wako Pure Chemical Industries, Ltd., Osaka, Japan) per ml. Bacitracin sensitivity was confirmed on bacitracin-containing (1 U/ml) plates, and then the corresponding transformants on plates containing no drug were picked up.
DNA sequence analysis.
DNA sequencing of insertion fragments in pBluescript SKII(+) was performed by the primer-walking strategy (22), using the −21 M13 primer (5′-TGTAAAACGACGGCAGT-3′) or the M13RP primer (5′-CAGGAAACAGCTATGACC-3′) as an initiation primer, a BigDye Terminator cycle sequencing kit (PE Biosystems, Urayasu, Japan), and an ABI PRISM 310 genetic analyzer automated sequencer (PE Biosystems). To identify the pResEmBBN insertion site in the S. mutans transformants, the DNA sequences of marker-rescued plasmids were determined with the BBN-B primer (5′-GTTACACGTTACTAAAGGGA-3′) for the region downstream from the erythromycin resistance gene and with the BBN-N primer (5′-GATTTGAGCGTCAGATTTCG-3′) for the region upstream from the p15A replicon. The nucleotide sequences were assembled using the DNASIS sequence analysis program (Hitachi Software Engineering Co., Yokohama, Japan). Database searching was performed with the FASTA program of the DDBJ server at the National Institute of Genetics, Mishima, Japan. Multiple alignments of the amino acid sequences were generated with the CLUSTAL W program (29).
Nonpolar insertions in open reading frames (ORFs) flanking the plasmid integration site.
To study the potential involvement of specific ORFs in bacitracin resistance, each ORF was insertionally inactivated with pResYT10. When this plasmid was introduced into the target gene, the inactivated gene and the erythromycin resistance gene were oriented in the same direction, so that the likelihood of polar effects on downstream gene transcription was avoided. It was previously shown that the promoter of the erythromycin resistance gene could direct the transcription of a gene that was located downstream of an inactivated gene in S. mutans (37). Constructions of mbr-inactivated mutants were performed as follows. Briefly, PCR fragments including the four mbr genes, which were amplified using the primer set of 5′-GTAAGCTACGATTCTTTAAG-3′ and 5′-CTTTAGCGGATGATTACGCA-3′, were cloned into pGEM-T PCR-cloning vector (Promega, Madison, Wis.). Four mbr genes on the resultant plasmid were interrupted by linearized pResYT10 at the respective restriction sites indicated (see Fig. 2). The plasmids whose erythromycin resistance gene and mbr gene were oriented in the same direction were selected, digested both sides of inserted site with the appropriate restriction enzymes, and introduced into the chromosome of S. mutans Xc by a double crossover recombination.
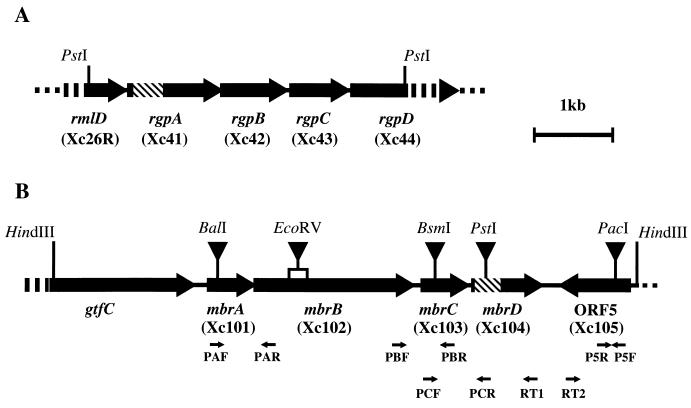
FIG. 2.
Restriction maps of the 4.3-kb PstI fragment (A) in pBSHT1 and the 7.7-kb HindIII fragment (B) in pBSHT2. Arrows indicate ORF locations. Hatched bars indicate the regions corresponding to a single crossover event in strains XcB1 and XcB2. Names of S. mutans mutants whose rml, rgp, or mbr gene was inactivated are indicated in the parentheses below the respective genes' name. Xc26R, Xc41, Xc42, Xc43, and Xc44 were previously constructed as described in Table 1. The pResYT10 insertion sites for inactivation of the mbr genes and ORF5 are indicated by the inverted closed triangles. The construction of mutants of the mbr gene and ORF5 was carried out as described in the text. Primers used in the RT-PCR are indicated as small arrows.
RT-PCR analysis.
Total RNA was prepared from S. mutans strains by using the FastPrep device (Savant Instruments, Holbrook, N.Y.) in combination with FastRNA kit-BLUE (Bio 101, Vista, Calf.) with minor modifications of the manufacturer's protocol. Briefly, bacterial cells that were grown to middle log phase in 15 ml of BHI broth were harvested and suspended in 300 μl of diethylpyrocarbonate-treated distilled water. The suspension was mixed with 900 μl of ISOGEN-LS (Nippon Gene Inc., Tokyo, Japan) and transferred to a FastPrep BLUE tube. The tube was processed for 40 s at speed level 6 in the FastPrep device, 200 μl of chloroform was added, and the mixture was shaken. Total RNA in the resultant supernatant was precipitated with 1 volume of isopropanol and washed with 500 μl of 75% ethanol. The dried pellet was suspended in an appropriate amount of diethylpyrocarbonate-treated distilled water. Before the solution was used for reverse transcriptase PCR (RT-PCR), contaminating DNA was eliminated by digestion with RNase-free DNase (DNase I; Gibco BRL Life Technologies, Grand Island, N.Y.). Reverse transcription was performed with SuperScript II RNase H− reverse transcriptase (Gibco BRL). The relevant primers for RT-PCR are shown in Table 3. Primers RT1 and RT2 were used to synthesize cDNA in the mbr and ORF5 regions, respectively (Table 3). The reaction was carried out at 42°C for 50 min. The cDNA was used directly for PCR amplification with the sets of primers listed in Table 3. The PCR procedure performed with Ex Taq DNA polymerase (TAKARA BIO Inc., Otsu, Japan) consisted of a denaturation step at 94°C for 2 min followed by 25 cycles that comprised the following steps: denaturation at 94°C for 30 s, annealing at an appropriate temperature for 30 s, and extension at 72°C for 1 min. A final extension step was performed at 72°C for 10 min. Negative control reactions were performed that left out the first reverse transcriptase treatment.
TABLE 3.
Primers used for PCR and RT-PCR
| Primer | Sequence (5′ to 3′) | Positionsa |
|---|---|---|
| PAF | CTTGCAAGACGTTGACTTCA | 2120-2139 |
| PAR | GGTAGCCAAACCTAGAGCAT | 2960-2979 |
| PBF | GACTCCTCTATGGCGATGATGAT | 4346-4368 |
| PBR | GCCCTAAAGTTATCAACGC | 4945-4963 |
| PCF | GGTTGAAGATGATACAACC | 4876-4895 |
| PCR | CAAGATCACTTGGTGCAG | 5763-5780 |
| P5F | AGGCATACGCCGTGATACTA | 7570-7589 |
| P5R | CAATGACTGATGCTTGATC | 6970-6988 |
| RT1 | GTAACATAAACCTGTGTTCCTTG | 6437-6459 |
| RT2 | ACCAGTCACCAGACTTGTTGGTTCCTC | 6876-6892 |
The numbers give the positions on a 7.7-kb HindIII fragment sequenced in this study (EMBL/GenBank/DDBJ data bank accession no. AB078507).
Antibiotics sensitivity tests.
On the broth assay, S. mutans mid-log-phase cultures were adjusted to 5 × 106 CFU/ml. One hundred microliters of CFU-adjusted bacterial suspensions were inoculated in 2.9 ml of BHI broth containing various concentrations of antibiotics. After 20 h of incubation at 37°C, the cultures were sonicated, and the optical densities at 550 nm (OD550) of the test cultures were measured. Cultures of all strains without antimicrobial agents could attain stationary phase within the 20 h at 37°C. The relative cell densities were calculated as (OD550 of culture in the presence of each concentration of antibiotics)/(OD550 of culture in the absence of antibiotics) × 100. The MIC was determined as the minimum antibiotic needed to ensure that culture did not grow to over 10% of the relative cell density.
In the plating assay, the tested strains were grown to mid-log phase in BHI broth. Approximately 50 CFU of bacterial cells were plated on the BHI agar plates containing various concentrations of antibiotics. After 5 days of incubation at 37°C, colonies were counted. MICs were determined from the highest concentration showing complete inhibition of the tested strains.
Immunological analysis of cell wall sugar components.
Lyophilized S. mutans cells were resuspended in phosphate-buffered saline (pH 7.3; 100 mg/ml) and autoclaved at 121°C for 30 min. The suspensions were then centrifuged at 10,000 ×g for 20 min, and the supernatants were collected and used as autoclaved extracts. Immunodiffusion was performed in 1% (wt/vol) Noble agar in phosphate-buffered saline (pH 7.3) (16) with rabbit antisera to whole cells of S. mutans strains MT8148 (serotype c) and Xc31, whose cell wall polysaccharides contain only rhamnan backbones.
Chemical analysis of cell wall sugars.
The sugar compositions of the cell wall preparations from S. mutans strains were analyzed by high-performance liquid chromatography, as described by Tsukioka et al. (32).
Nucleotide sequence accession number.
The 7,733-bp nucleotide sequence presented in this paper has been submitted to the EMBL/GenBank/DDBJ data bank under accession number AB078507.
RESULTS
Cloning and nucleotide sequencing of the S. mutans genes involved in resistance to bacitracin.
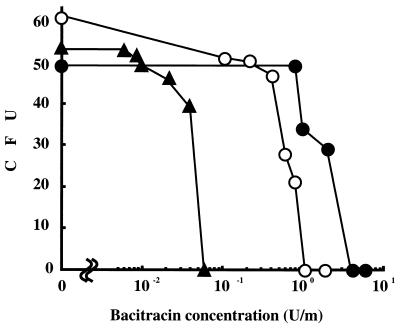
Among the 12,000 transformants, only two were sensitive to bacitracin, and these were designated as strains XcB1 and XcB2. The dose dependent bacitracin sensitivities of strains XcB1 and XcB2 are displayed in Fig. 1. Neither of the mutants grew in the presence of 1 U of bacitracin per ml, and both were clearly sensitive to bacitracin compared to the wild-type strain Xc. In addition, strain XcB2 was distinctly more sensitive to bacitracin than strain XcB1. Southern blotting with a digoxigenin (DIG)-labeled PCR probe that was specific for the erythromycin resistance gene revealed that the probe hybridized with a 7.0-kb PstI fragment from strain XcB1 and a 9.8-kb HindIII fragment from strain XcB2 but did not hybridize with any fragments from the wild-type strain Xc (data not shown). Using the marker rescue strategy (14), fragments that hybridized with the probe were recovered from PstI-digested XcB1 and HindIII-digested XcB2 chromosomal DNA. The plasmids recovered from strains XcB1 and XcB2 were designated pHT1 and pHT2, respectively. The 4.3-kb PstI fragment of the strain Xc chromosome hybridized with pHT1 that had been labeled with DIG-dUTP via random primer labeling. In contrast, the 7.7-kb HindIII fragment of the strain Xc chromosome hybridized with DIG-labeled pHT2. Both fragments were cloned in pBluescript SKII(+) by colony hybridization using DIG-labeled pHT1 and pHT2 as probes. The recombinant plasmids hybridized with pHT1 and pHT2 and were designated pBSHT1 and pBSHT2, respectively. The fragments inserted in these plasmids were sequenced. Restriction maps of the two cloned fragments are shown in Fig. 2.
FIG. 1.
Bacitracin sensitivities of S. mutans strain Xc and its derivatives. Approximately 50 CFU of mid-log-phase bacterial cells were plated on the BHI agar plates containing various concentrations of antibiotics. After 5 days incubation at 37°C, colonies were counted. The experiments were performed three times, and similar results were obtained in each experiment. Symbols: •, strain Xc; ○, strain XcB1; ▴, strain XcB2.
Nucleotide sequence analysis revealed the presence of three complete and two truncated ORFs in the 4.3-kb PstI fragment of pBSHT1 (Fig. 2A). The sequences of these ORFs are identical to previously characterized ORFs of the rmlD, rgpA, rgpB, rgpC, and rgpD genes, which are involved in RGP synthesis (37). Sequencing of the region flanking the pResEmBBN insertion in pTH1 revealed that rgpA was insertionally inactivated by pResEmBBN in strain XcB1 (Fig. 2A).
Six ORFs were found in the 7.7-kb HindIII fragment of pBSHT2 (Fig. 2B). The first ORF represents a truncated form of the gtfC gene, which codes for a glucosyltransferase-SI and which is responsible for the synthesis of water-insoluble glucan (8). With the exception of the last ORF, the other five ORFs are in the same orientation. Since inactivation of the second to fifth ORFs in S. mutans resulted in bacitracin sensitivity, these four ORFs were designated as mutans bacitracin resistance (mbr) genes, i.e., mbrA, mbrB, mbrC, and mbrD, respectively (Fig. 2B). The remaining ORF (ORF5) was not involved in bacitracin resistance (Table 4). The mbrA gene is located 273 bp downstream from gtfC. In contrast, the intergenic regions between mbrA and mbrB, mbrB and mbrC, and mbrC and mbrD are only −8, 41, and −4 bp, respectively. Potential transcription terminators, in the form of stem-loop structures followed by a poly(T) sequence (positions 1847 to 1866 and 6522 to 6542) are present in the regions between gtfC and mbrA and downstream of mbrD. Putative Shine-Dalgarno ribosome binding sequences lie just upstream of the potential initiation codons in all of the mbr genes and in ORF5.
TABLE 4.
MICs of bacitracin for S. mutans strains used in this study
| Straina | MICb |
|---|---|
| Xc | 4.0 ± 0.0d,f |
| Xc13 (gtfC) | 4.0 ± 0.0d,f |
| Xc26R (rmlD) | 0.80 ± 0.00c,f |
| Xc41 (rgpA) | 0.73 ± 0.09c,f |
| Xc42 (rgpB) | 0.73 ± 0.09c,f |
| Xc43 (rgpC) | 0.80 ± 0.00c,f |
| Xc44 (rgpD) | 0.73 ± 0.09c,f |
| XcB1 (rgpA) | 0.73 ± 0.09c,f |
| XcB2 (mbrD) | 0.033 ± 0.009c,d |
| Xc101 (mbrA) | 0.033 ± 0.009c,d |
| Xc102 (mbrB) | 0.033 ± 0.009c,d |
| Xc103 (mbrC) | 0.040 ± 0.000c,d |
| Xc104 (mbrD) | 0.040 ± 0.000c,d |
| Xc105 (ORF5) | 3.3 ± 0.9e,g |
| Xc106 (mbrA, mbrB, mbrC, and mbrD) | 0.033 ± 0.009c,d |
| Xc146 (mbrA, mbrB, mbrC, mbrD, and rgpA) | 0.020 ± 0.000c,d |
For all strains besides Xc, the genes were inactivated.
Results are expressed in international units per milliliter. Each value represents the mean ± standard deviation for assays performed three times.
Differences from strain Xc (P < 0.05 [Welch's t test]).
Differences from strain Xc41 (P < 0.01 [Welch's t test]).
Differences from strain Xc41 (P < 0.05 [Welch's t test]).
Differences from strain Xc106 (P < 0.01 [Welch's t test]).
Differences from strain Xc106 (P < 0.05 [Welch's t test]).
The deduced amino acid sequences encoded by mbrA and mbrB genes show strong similarities to the components of other bacterial ABC transporters. The mbrA-encoded protein contains four highly conserved motifs that are characteristic of ATP-binding proteins: a Walker A site at amino acid positions 41 to 48, a Walker B site at positions 165 to 169, a linker peptide at positions 145 to 152, and a switch region at the positions 193 to 202 (9, 12). Secondary structure predictions for the MbrB protein (performed with the SOSUI program; http://sosui.proteome.bio.tuat.ac.jp/welcomeE.html) revealed 10 putative transmembrane helices, suggesting that this protein may be membrane localized.
The MbrC and MbrD proteins exhibit high levels of identity with the response regulators and the histidine sensor kinases, respectively, of two-component regulatory systems in certain bacteria. MbrC protein contains a region that is characteristic of the helix-turn-helix motif (at amino acid positions 36 to 55), which is associated with DNA binding. The N terminus of MbrC is highly conserved and includes several aspartate residues (at amino acid positions 12, 13, 49, 54, 98, and 99) that may form the phosphorylation site of the protein. The histidine kinase domain of MbrD contains four highly conserved amino acid sequences, termed the H, N, D/F, and G boxes (27). These motifs presumably form a nucleotide-binding surface within the active site. Moreover, secondary structure prediction for the MbrD protein using the SOSUI program revealed that this protein possesses two putative transmembrane helices (at amino acid positions 12 to 34 and 38 to 60) (10). The flanking region of the pResEmBBN inserted in pTH2 was sequenced and we confirmed that mbrD was insertionally inactivated by pResEmBBN in XcB2 (Fig. 2B).
Transcription of the mbr genes in S. mutans.
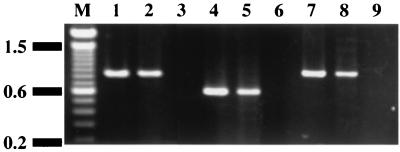
The mbr genes are located close to each other, suggesting polycistronic transcription of these genes. RT-PCR analysis of total RNA from the wild-type strain Xc was performed to demonstrate that all of the mbr genes were included in a single polycistronic transcript. RT-PCR using the following primer sets: PAF and PAR; PBF and PBR; and PCF and PCR (Fig. 2), produced 0.9-, 0.6-, and 0.9-kb fragments, respectively (Fig. 3). Total RNA preparations that had not undergone reverse transcription did not give amplified fragments, suggesting that the RT-PCR products were derived from mRNA and not from contaminating chromosomal DNA (Fig. 3).
FIG. 3.
RT-PCR analysis of mRNA from the mbr operon. Lanes 1, 4, and 7, PCR using the chromosomal DNA of strain Xc as the template (positive controls); lanes 2, 5, and 8, RT-PCR using total RNA from strain Xc as the template; lanes 3, 6, and 9, PCR using total RNA from strain Xc as the template (negative controls). The RT1 primer was used for reverse transcription. PCR amplification was performed with the following primer sets: PAF and PAR (lanes 1, 2, and 3), PBF and PBR (lanes 4, 5, and 6), and PCF and PCR (lanes 7, 8, and 9).
The effect of insertional inactivation of mbr genes on downstream genes.
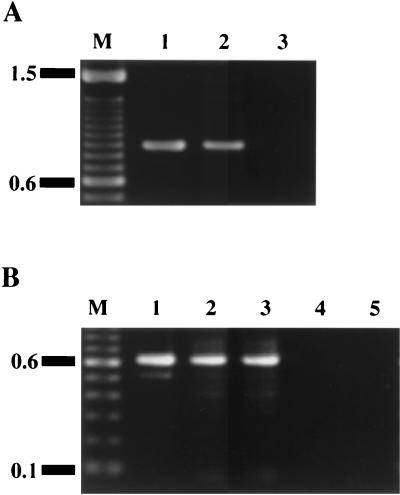
We used pResYT10 to insertionally inactivate each mbr gene in order to reduce the likelihood of a polar effect on the transcription of the downstream genes. RT-PCR was performed to confirm mbrD gene transcription in strain Xc102 (an mbrB-defective mutant) and ORF5 transcription in strains Xc102 and Xc104 (an mbrD-defective mutant). A 0.9-kb RT-PCR fragment was amplified from the total RNA of strain Xc102 (Fig. 4A), whereas 0.6-kb RT-PCR products were obtained from the total RNA of strains Xc102 and Xc104 (Fig. 4B). Total RNA preparations that were not first reverse transcribed give no PCR products (Fig. 4). We thus concluded that insertional inactivation of the mbr genes with pResYT10 did not interrupt the transcription of downstream genes.
FIG. 4.
RT-PCR analysis of the transcription of the region located downstream of the mbrC gene and upstream of the mbrD gene in strain Xc102 (A), and of ORF5 transcription in strains Xc102 and Xc104 (B). (A) Lane 1, PCR using the chromosomal DNA of strain Xc as the template (positive control); lane 2, RT-PCR using total RNA from strain Xc102 as the template; lane 3, PCR using total RNA from strain Xc102 as the template (negative control). The RT1 primer was used for reverse transcription. PCR amplification was performed with primers PCF and PCR. (B) Lane 1, PCR using the chromosomal DNA of strain Xc as the template (positive control); lanes 2 and 3, RT-PCR using total RNA from strains Xc102 and Xc104, respectively, as the templates; lanes 4 and 5, PCR using total RNA from strains Xc102 and Xc104, respectively, as the templates (negative controls). The primer RT2 was used for reverse transcription. PCR amplification was performed with primers P5F and P5R.
Bacitracin sensitivity tests.
To determine which genes were responsible for resistance to bacitracin, S. mutans mutants that were defective in mbrA, mbrB, mbrC, mbrD, or ORF5 were constructed by insertional inactivation with pResYT10, as described in the Materials and Methods section (Table 1 and Fig. 2B). The strains in which mbrA, mbrB, mbrC, mbrD, and ORF5 were inactivated were designated Xc101, Xc102, Xc103, Xc104, and Xc105, respectively. S. mutans mutants that were defective in each of the rgp genes or in rmlD were constructed as described previously (31, 37). The previously constructed strain Xc13 (35) was used as the gtfC mutant. The bacitracin sensitivity of gtfC-deficient strain Xc13 was the same as that of the wild-type strain Xc (Table 4). Similarly, insertional inactivation of ORF5 (strain Xc105) did not significantly change the bacitracin resistance of S. mutans (Table 4). On the other hand, strains Xc101, Xc102, Xc103, and Xc104 exhibited about 100- to 120-fold-higher sensitivity to bacitracin than did strain Xc; the enhanced sensitivities to bacitracin were not significantly different from that of strain XcB2 (Table 4). In addition, we constructed an S. mutans mutant in which chromosomal region corresponding to all four mbr genes was replaced by erythromycin resistance gene (strain Xc106). The bacitracin resistance of strain Xc106 was not significantly different from that of strains Xc101, Xc102, Xc103, and Xc104 (Table 4).
On the other hand, strains Xc26R, Xc41, Xc42, Xc43, and Xc44, which are defective in rmlD, rgpA, rgpB, rgpC, and rgpD, respectively, were about fivefold more sensitive to bacitracin than strain Xc (Table 4). Strain XcB1 was similar in sensitivity to bacitracin as strains Xc26R, Xc41, Xc42, Xc43, and Xc44 (Table 4). In addition, we constructed a double mutant that was defective in all of the mbr genes and the rgpA gene (strain Xc146), to examine the relationship between mbr genes and rgp genes functions in bacitracin resistance of S. mutans. Strain Xc146 was more sensitive to bacitracin than strains XcB1 and XcB2 and exhibited an approximately 200-fold-higher sensitivity to bacitracin than strain Xc (Table 4). Similar results were obtained from the experiments by plating assay (Fig. 1).
Analysis of cell wall polysaccharides of the mbr mutant.
To examine whether the inactivation of mbr genes influenced RGP synthesis, cell wall polysaccharides of strain Xc106 were analyzed immunologically and chemically. Immunodiffusion analysis was carried out with both serotype c-RGP-specific antiserum and rhamnan-backbone-specific rabbit antiserum (36). The serotype c-specific antiserum formed a single precipitin line between the antiserum and the autoclaved extracts of strains Xc or Xc106, while the rhamnan-specific antiserum did not react with autoclaved extracts of either strain Xc or Xc106 (data not shown). In addition, the sugar composition of the cell wall preparations of strains Xc and Xc106 were analyzed by high-performance liquid chromatography. The amounts of rhamnose, glucose, and N-acetylglucosamine per milligram (dry weight) of cell wall preparation of strain Xc106 did not differ from the corresponding values for strain Xc (Table 5).
TABLE 5.
Sugar composition of cell wall preparations
| Strain | Sugar contenta (μg/mg)
|
||
|---|---|---|---|
| Glucose | Rhamnose | N-Acetylglucosamine | |
| Xc | 161 | 357 | 163 |
| Xc106 | 126 | 253 | 121 |
Values are micrograms per 1 mg (dry weight) of the purified cell wall preparation.
Effect of the culture supernatants of S. mutans strains Xc and Xc106 on the bacitracin sensitivity of strain Xc106.
Since MbrA and MbrB represent a putative ABC transporter, they may function to export specific molecules that inactivate bacitracin. In order to examine this possibility, spent culture supernatants from strains Xc and Xc106 were added, in various proportions, to cultures of strain Xc106, and the bacitracin sensitivity of strain Xc106 was measured. Briefly, S. mutans strains Xc and Xc106 were grown to stationary phase in BHI broth. After centrifugation, culture supernatants were collected and filtered through 0.2-μm-pore-size filters. The collected supernatants were mixed with fresh BHI broth in proportions that ranged from 1:28 to 15:14 to give a total volume of 2.9 ml for each mixture. Aliquots (100 μl) of overnight cultures of strain Xc106 were inoculated in the supernatant mixtures in BHI broth that contained bacitracin at 0.1 U per ml. The cultures were incubated at 37°C for 20 h, and the OD550 was measured. There were no significant differences between the culture supernatants of strains Xc and Xc106 in terms of their effects on the bacitracin sensitivity of Xc106.
Sensitivities to other antibiotics.
In addition to its resistance to bacitracin, the S. mutans wild-type strain Xc is resistant to kanamycin (MIC, 60 μg/ml [broth assay]) and spectinomycin (MIC, 100 μg/ml [broth assay]) (Table 6). To investigate whether the mbr- and RGP-mediated bacitracin resistance mechanisms participated in resistance to kanamycin or spectinomycin, we examined the sensitivities to some antibiotics of strains Xc, Xc106, Xc41, and Xc146 by the broth assay. Strain Xc106 was resistant to kanamycin, spectinomycin, and nisin to the same extent as strain Xc (Table 6). In addition, the sensitivity to ampicillin of strain Xc106 was as high as that of the strain Xc (Table 6). In contrast, strain Xc41, which is defective in RGP synthesis, was more sensitive to these antibiotics than strain Xc (Table 6). The sensitivities of strain Xc146 to kanamycin, spectinomycin, nisin, and ampicillin were not significantly different from those of strain Xc41, although they differed in their sensitivities to bacitracin (Table 6). No difference was found between the sensitivities to tetracycline and ofloxacin of strains Xc, Xc106, and Xc41 (data not shown). The double mutant Xc146 had the same sensitivity to ofloxacin of strain Xc, Xc106, and Xc41 (data not shown). As strain Xc146 has tetracycline resistance gene, this mutant had strong resistance to tetracycline (data not shown).
TABLE 6.
MICs of antibiotics for S. mutans strains
| Antibiotic | MICa
|
|||
|---|---|---|---|---|
| Xc | Xc106 | Xc41 | Xc146 | |
| Kanamycin | 60 ± 0d | 53.3 ± 9.4e | 20 ± 0b,g | 16.7 ± 4.7b,f |
| Spectinomycin | 100 ± 0d | 93.3 ± 9.4 | 80 ± 0b | 73.3 ± 9.4c,h |
| Bacitracin | 4.0 ± 0.0d,f | 0.033 ± 0.009b,d | 0.73 ± 0.09b,f | 0.02 ± 0.00b,d |
| Ampicillin | 0.06 ± 0.00d | 0.06 ± 0.00d | 0.04 ± 0.00b,f | 0.04 ± 0.00b,f |
| Nisin | 100 ± 0d | 100 ± 0d | 0.25 ± 0.00b,f | 0.23 ± 0.02b,f |
Results are expressed in micrograms per milliliter, except for those with bacitracin, which are in international units per milliliter. Each value represents the mean ± standard deviation for assays performed three times.
Differences from strain Xc (P < 0.01 [Welch's t test]).
Differences from strain Xc (P < 0.05 [Welch's t test]).
Differences from strain Xc41 (P < 0.01 [Welch's t test]).
Differences from strain Xc41 (P < 0.05 [Welch's t test]).
Differences from strain Xc106 (P < 0.01 [Welch's t test]).
Differences from strain Xc106 (P < 0.05 [Welch's t test]).
Differences from strain Xc106 (P < 0.1 [Welch's t test]).
DISCUSSION
In this study, we identified and sequenced genes that were involved in resistance of S. mutans to bacitracin and examined the bacitracin sensitivity of mutants that were defective in the rml, rgp, and mbr genes. We found that strains Xc26R, Xc41, Xc42, Xc43, and Xc44, which are defective in rmlD, rgpA, rgpB, rgpC, and rgpD, respectively, were about five- to six-fold more sensitive to bacitracin than the wild-type strain Xc (Table 4). The RGPs of S. mutans have a backbone structure that is composed of α 1,2- and α 1,3-linked rhamnosyl polymer with glucose side chains (13, 21). We previously reported that the rgpA, rgpB, rgpC, and rgpD genes were required for the assembly of RGP from dTDP-l-rhamnose and for the export of RGP across the cytoplasmic membrane and that mutants defective in these genes did not incorporate RGP into their cell walls (37). The rmlD gene is involved in the synthesis of dTDP-l-rhamnose, which is an immediate precursor for RGP-backbone production, and RGP was not found in the cell wall of rmlD mutant Xc26R (31). All of these mutants exhibited similar sensitivities to bacitracin (Table 4). These findings suggest that the presence of RGP in the cell wall may confer resistance to bacitracin in S. mutans. Pollock et al. (20) reported that exopolysaccharide-synthesizing gram-negative bacteria acquired resistance to bacitracin by repressing the synthesis of exopolysaccharide. Exopolysaccharide synthesis requires the same carrier IP that is needed for the synthesis of peptidoglycan. By repressing the synthesis of exopolysaccharide, bacteria can use the excess IP for peptidoglycan synthesis and thus become resistant to bacitracin. If IP is required for RGP synthesis, mutants defective in rmlD, rgpA, rgpB, rgpC, or rgpD genes should be more resistant to bacitracin than the wild-type strain Xc. We previously suggested that the transfer of N-acetylglucosamine to a lipid carrier, such as IP, was required for the RGP synthesis (33). However, the mutants defective in rmlD, rgpA, rgpB, rgpC, or rgpD (strains Xc26R, Xc41, Xc42, Xc43, or Xc44, respectively) exhibited significantly enhanced sensitivity to bacitracin compared with strain Xc (Table 4). These results appear to contradict the model outlined above. The lipid carrier for N-acetylglucosamine transfer during RGP synthesis in S. mutans might not be IP. Putative mechanisms for RGP-mediated resistance of S. mutans to bacitracin are discussed below.
All of the mbr-defective mutants were approximately 100- to 120-fold more sensitive to bacitracin than the parental strain Xc. Analysis of the deduced amino acid sequences of MbrA and MbrB strongly suggested that these proteins represented the components of an ABC transporter. Assuming that this is true, what is the target of this transporter? We suggest two possibilities: (i) the transporter exports a molecule that inactivates bacitracin, and indeed, it has been reported that some metabolites inhibit bacitracin activity at very low concentrations (18); or (ii) the transporter modulates the movement of bacitracin itself. Since Podlesek et al. could not detect any substances that suppressed the bacitracin activity of a strain of B. subtilis that carried cloned bcr genes, they assumed that the Bcr-encoded ABC transporter of B. licheniformis transported bacitracin itself (19). The latter hypothesis is likely to be true in S. mutans, because the culture supernatants of strains Xc and Xc106 had similar effects on the bacitracin sensitivity of strain Xc106. The deduced amino acid sequences of MbrC and MbrD were highly homologous to those of response regulators and histidine-sensor kinases, respectively, which are found in two-component regulatory systems of certain bacteria, and these proteins have some highly conserved motifs. However, based on current information, it is difficult to speculate on the functions of MbrC and MbrD.
The strain Xc146, which is defective in four mbr genes and rgpA, was constructed, and its bacitracin sensitivity was examined to elucidate the relationship between the two mechanisms of resistance to bacitracin. Strain Xc146 was approximately 200 and 37 times more sensitive to bacitracin than strains Xc and Xc41, respectively, and was approximately two times more sensitive to bacitracin than strain Xc106. These results suggest that the mbr-mediated mechanism of bacitracin resistance is independent of RGP synthesis. Indeed, immunological (data not shown) and chemical (Table 5) analysis of the cell-wall components of strain Xc106 confirmed that the mbr genes were not involved in RGP synthesis.
The S. mutans wild-type strain Xc is resistant not only to bacitracin but also to kanamycin and spectinomycin (Table 6). To clarify whether the mbr- and RGP-related bacitracin resistance mechanisms were specific for bacitracin, we examined the sensitivities to kanamycin and spectinomycin of S. mutans strains Xc, Xc106, Xc41, and Xc146. The kanamycin and spectinomycin sensitivities of the mbr-defective strain Xc106 were similar to those of the wild-type strain Xc. In addition, there were no significant differences in sensitivity to ampicillin, ofloxacin, tetracycline, and nisin between strains Xc and Xc 106 (Table 6 and Results section). Furthermore, the kanamycin, spectinomycin, ampicillin, ofloxacin, and nisin sensitivities of strain Xc146 were similar to those of strain Xc41 (Table 6 and Results section). These results suggest that the mbr genes of S. mutans are specific for resistance to bacitracin. On the other hand, RGP-defective strain Xc41 was more sensitive to kanamycin, spectinomycin, bacitracin, ampicillin, and nisin than strain Xc (Table 6). As well as strain Xc41, strain Xc24R (defective in the rmlB gene, which is involved in dTDP-l-rhamnose synthesis) lacked RGP on the cell surface (30, 32) and exhibited a higher sensitivity to bacitracin than strain Xc (data not shown). The bacterial capsule and exopolysaccharide prevent the migration of the antibiotic to its target by prolonging the time to needed to equilibrate the difference in antibiotic concentration between the external medium and the bacterial cell surface and by providing a frictional resistance to diffusion (3, 4, 26). Indeed, electron microscopic observations of cell surface architectures indicated that the cell-wall-like layers of strain Xc24R were thinner than those of strain Xc (30). Thus, we speculate that RGP may act as a barrier that prevents some antibiotics reaching their target molecules and may partly contribute to the resistance to some antibiotics of S. mutans. However, sensitivities of RGP mutant to tetracycline and ofloxacin did not significantly differ from those of wild type strain (data not shown). At present, it is difficult to precisely define the function of RGP in the resistance of S. mutans to antibiotics.
Since the first reports in the late 1980s, VRE have become established pathogens in many hospitals, and the number of cases involving this organism has increased rapidly throughout the world (A. H. Uttley, C. H. Collins, J. Naidoo, and R. C. George, Letter, Lancet i:57-58, 1988). Outbreaks reported in the press have spread alarm in the community. The emergence and widespread incidence of VRE have produced a therapeutic dilemma. Some clinicians have recently suggested that oral administration of bacitracin might be a safe and effective way to eliminate VRE from the gastrointestinal tract of patients, and in this respect bacitracin is receiving a good deal of attention (2, 15, 25). On the other hand, bacitracin-resistant S. mutans colonizes in the oral cavities of most of human beings, and this organism has been detected in feces (7). Oral administration of bacitracin might ensure the predominance of S. mutans in the gastrointestinal tract, thus increasing the potential for contact between VRE and bacitracin-resistant S. mutans. Under these conditions, the mbr genes of S. mutans might be transferred to VRE, thereby conferring the bacitracin resistance phenotype of S. mutans to VRE. Therefore, it is important to understand the bacitracin resistance mechanism of S. mutans, in order to prevent the appearance of bacitracin-resistant VRE. At present, we can only eliminate S. mutans from the oral cavity of VRE-infected patients mechanically, by stringent tooth brushing. Further studies on the bacitracin resistance mechanism of S. mutans are needed in preparation for the emergence of bacitracin-resistant VRE.
Acknowledgments
This work was supported in part by Satoh Fund, Nihon University School of Dentistry, and a grant to promote multidisciplinary research projects and grants-in-aid for developmental scientific research (12357013 and 12557186) from the Ministry of Education, Science, Sports, and Culture and Technology of Japan.
Footnotes
Dedicated to the memory of Toshihiko Koga, esteemed researcher and our mentor.
REFERENCES
- 1.Cain, B. D., P. J. Norton, W. Eubanks, H. S. Nick, and C. M. Allen. 1993. Amplification of the bacA gene confers bacitracin resistance to Escherichia coli. J. Bacteriol. 175:3784-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chia, J. K., M. M. Nakata, S. S. Park, R. P. Lewis, and B. McKee. 1995. Use of bacitracin therapy for infection due to vancomycin-resistant Enterococcus faecium. Clin. Infect. Dis. 21:1520. [DOI] [PubMed] [Google Scholar]
- 3.Costerton, J. W. 1977. Cell envelope as a barrier to antibiotics, p. 151-157. In D. Schlessinger (ed.), Microbiology—1977. American Society for Microbiology, Washington, D. C.
- 4.Costerton, J. W., and K. J. Cheng. 1975. The role of the bacterial cell envelope in antibiotic resistance. J. Antimicrob. Chemother. 1:363-377. [DOI] [PubMed] [Google Scholar]
- 5.Fiedler, W., and H. Rotering. 1988. Properties of Escherichia coli mutants lacking membrane-derived oligosaccharides. J. Biol. Chem. 263:14684-14689. [PubMed] [Google Scholar]
- 6.Gold, O. G., H. V. Jordan, and J. van Houte. 1973. A selective medium for Streptococcus mutans. Arch. Oral Biol. 18:1357-1364. [DOI] [PubMed] [Google Scholar]
- 7.Hamada, S., N. Masuda, and S. Kotani. 1980. Isolation and serotyping of Streptococcus mutans from teeth and feces of children. J. Clin. Microbiol. 11:314-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanada, N., and H. K. Kuramitsu. 1988. Isolation and characterization of the Streptococcus mutans gtfC gene, coding for synthesis of both soluble and insoluble glucans. Infect. Immun. 56:1999-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 10.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 11.Koga, T., H. Asakawa, N. Okahashi, and I. Takahashi. 1989. Effect of subculturing on expression of a cell-surface protein antigen by Streptococcus mutans. J. Gen. Microbiol. 135:3199-3207. [DOI] [PubMed] [Google Scholar]
- 12.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5-13. [DOI] [PubMed] [Google Scholar]
- 13.Linzer, R., M. S. Reddy, and M. J. Levine. 1986. Immunochemical aspects of serotype carbohydrate antigens of Streptococcus mutans, p. 29-38. In S. Hamada, M. Michalek, H. Kiyono, L. Menaker, and J. R. McGhee (ed.), Molecular microbiology and immunology of Streptococcus mutans. Elsevier Science Publisher, Amsterdam, The Netherlands.
- 14.Niaudet, B., A. Goze, and S. D. Ehrlich. 1982. Insertional mutagenesis in Bacillus subtilis: mechanism and use in gene cloning. Gene 19:277-284. [DOI] [PubMed] [Google Scholar]
- 15.O'Donovan, C. A., P. Fan-Havard, F. T. Tecson-Tumang, S. M. Smith, and R. H. Eng. 1994. Enteric eradication of vancomycin-resistant Enterococcus faecium with oral bacitracin. Diagn. Microbiol. Infect. Dis. 18:105-109. [DOI] [PubMed] [Google Scholar]
- 16.Öuchterlony. 1958. Diffusion-in-gel methods for immunological analysis. Prog. Allergy 5:1-78. [PubMed] [Google Scholar]
- 17.Perry, D., L. M. Wondrack, and H. K. Kuramitsu. 1983. Genetic transformation of putative cariogenic properties in Streptococcus mutans. Infect. Immun. 41:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Podlesek, Z., and A. Comino. 1994. Antagonists of bacitracin. Lett. Appl. Microbiol. 19:102-104. [DOI] [PubMed] [Google Scholar]
- 19.Podlesek, Z., A. Comino, B. Herzog-Velikonja, D. Zgur-Bertok, R. Komel, and M. Grabnar. 1995. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol. Microbiol. 16:969-976. [DOI] [PubMed] [Google Scholar]
- 20.Pollock, T. J., L. Thorne, M. Yamazaki, M. J. Mikolajczak, and R. W. Armentrout. 1994. Mechanism of bacitracin resistance in gram-negative bacteria that synthesize exopolysaccharides. J. Bacteriol. 176:6229-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pritchard, D. G., R. L. Gregory, S. M. Michaleck, and J. R. McGhee. 1986. Biochemical aspects of serotype carbohydrate antigens of Streptococcus mutans, p. 39-49. In S. Hamada, M. Michalek, H. Kiyono, L. Menaker, and J. R. McGhee (ed.), Molecular microbiology and immunology of Streptococcus mutans. Elsevier Science Publisher, Amsterdam, The Netherlands.
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Shiroza, T., and H. K. Kuramitsu. 1993. Construction of a model secretion system for oral streptococci. Infect. Immun. 61:3745-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siewert, G., and J. L. Strominger. 1967. Bacitracin: An inhibitor of the dephosphorylation of lipid pyrophosphate, an intermediate in biosynthesis of the peptidoglycan of bacterial cell walls. Proc. Natl. Acad. Sci. USA 57:767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silverblatt, F. J., C. Tibert, D. Mikolich, J. Blazek-D'Arezzo, J. Alves, M. Tack, and P. Agatiello. 2000. Preventing the spread of vancomycin-resistnt enterococci in a long-term care facility. J. Am. Geriatr. Soc. 48:1211-1215. [DOI] [PubMed] [Google Scholar]
- 26.Slack, M. P., and W. W. Nichols. 1982. Antibiotic penetration through bacterial capsules and exopolysaccharides. J. Antimicrob. Chemother. 10:368-372. [DOI] [PubMed] [Google Scholar]
- 27.Stock, J. B., M. G. Srette, M. Levit, and P. Park. 1995. Structure-function relationships and mechanisms of catalysis, p. 25-51. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction systems. American Society for Microbiology, Washington, D.C.
- 28.Storm, D. R. 1974. Mechanism of bacitracin action: a specific lipid-peptide interaction. Ann. N. Y. Acad. Sci. 235:387-398. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuda, H., Y. Yamashita, K. Toyoshima, N. Yamaguchi, T. Oho, Y. Nakano, K. Nagata, and T. Koga. 2000. Role of serotype-specific polysaccharide in the resistance of Streptococcus mutans to phagocytosis by human polymorphonuclear leukocytes. Infect. Immun. 68:644-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsukioka, Y., Y. Yamashita, Y. Nakano, T. Oho, and T. Koga. 1997. Identification of a fourth gene involved in dTDP-rhamnose synthesis in Streptococcus mutans. J. Bacteriol. 179:4411-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukioka, Y., Y. Yamashita, T. Oho, Y. Nakano, and T. Koga. 1997. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J. Bacteriol. 179:1126-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamashita, Y., Y. Shibata, Y. Nakano, H. Tsuda, N. Kido, M. Ohta, and T. Koga. 1999. A novel gene required for rhamnose-glucose polysaccharide synthesis in Streptococcus mutans. J. Bacteriol. 181:6556-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita, Y., T. Takehara, and H. K. Kuramitsu. 1993. Molecular characterization of a Streptococcus mutans mutant altered in environmental stress responses. J. Bacteriol. 175:6220-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamashita, Y., Y. Tsukioka, Y. Nakano, Y. Shibata, and T. Koga. 1996. Molecular and genetic analysis of multiple changes in the levels of production of virulence factors in a subcultured variant of Streptococcus mutans. FEMS Microbiol. Lett. 144:81-87. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita, Y., Y. Tsukioka, Y. Nakano, K. Tomihisa, T. Oho, and T. Koga. 1998. Biological functions of UDP-glucose synthesis in Streptococcus mutans. Microbiology 144:1235-1245. [DOI] [PubMed] [Google Scholar]
- 37.Yamashita, Y., Y. Tsukioka, K. Tomihisa, Y. Nakano, and T. Koga. 1998. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J. Bacteriol. 180:5803-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]