Abstract
Cefpodoxime is one of five antimicrobial agents recommended by the National Committee for Clinical Laboratory Standards for screening isolates of Klebsiella spp. and Escherichia coli for extended-spectrum β-lactamase (ESBL) production. In a prior study, we noted that among 131 E. coli isolates for which the MIC of at least one extended-spectrum cephalosporin (ESC) or aztreonam was ≥2 μg/ml (suggesting the presence of ESBL production), there were 59 isolates (45.0%) for which the MIC of cefpodoxime was 2 to 4 μg/ml (i.e., a positive ESBL screening test), but the MICs of ceftazidime, cefotaxime, and ceftriaxone were ≤1 μg/ml (below the ESBL screening breakpoint). Thus, the results appeared to be false-positive ESBL screening tests. These 59 isolates were divided into five phenotypic groups based on the susceptibility patterns of the organisms to a variety of β-lactam agents and further characterized. The first group (32 isolates) all produced a TEM-1 β-lactamase, and changes in the major outer membrane proteins were detected in representative strains. The second group (18 isolates) lacked blaTEM but showed a number of porin changes; some also showed a modest elevation in production of the AmpC chromosomal β-lactamase. In the third phenotypic group (seven isolates) all expressed an OXA-30 β-lactamase. Some also harbored altered porins. The two remaining phenotypes each had a distinct pattern of porin changes with or without β-lactamase production. These data indicate that several factors are associated with decreased susceptibility to cefpodoxime in E. coli, but none of the mechanisms are related to ESBL production. Current screening methods produced false-positive ESBL results for these isolates. Such isolates should not be classified as containing ESBLs, nor should interpretations of ESCs or aztreonam susceptibility be changed to resistant on test reports for these isolates.
Resistance to β-lactam agents in Escherichia coli is often mediated by the acquisition of plasmid-encoded β-lactamases (3, 14). These may be either broad-spectrum β-lactamases, such as TEM-1 or SHV-1, that confer resistance only to penicillins and narrow-spectrum cephalosporins, or extended-spectrum β-lactamases (ESBLs), including those derived from TEM-1, SHV-1, OXA-10, or CTX-M, that also confer resistance to extended-spectrum cephalosporins and aztreonam (ATM) (3, 14). Mutations in the attenuator or the weak promoter of the chromosomal ampC β-lactamase gene can lead to enhanced production of AmpC, resulting in resistance to penicillins and extended-spectrum cephalosporins (4, 12). Changes in major outer membrane protein (OMP) profiles, such as decreased production of OmpC or OmpF, may also result in low-level β-lactam resistance (9, 15, 29).
Most studies of β-lactam resistance in E. coli focus on strains showing high levels of resistance, particularly to extended-spectrum cephalosporins (4, 22). Few studies have investigated the mechanisms of low-level resistance. However, low-level resistance, while of arguable clinical relevance, does have an impact on the accuracy with which clinical laboratories identify ESBL-producing E. coli strains when using the NCCLS screening tests (21). The screening breakpoint for identifying potential ESBL-producing strains is ≥2 μg/ml for cefpodoxime (CPD), ceftazidime (CAZ), ceftriaxone, cefotaxime (CTX), and ATM, significantly lower than the suggested resistance breakpoints of 32 to 64 μg/ml (21). NCCLS guidelines recommend that organisms with a positive screening test for ESBL production (MIC of ≥2 μg/ml for any one of the five agents listed above) undergo additional testing using both CTX and CAZ, with and without clavulanic acid (the NCCLS recently changed the CPD screening breakpoint to ≥8 μg/ml). If a clavulanic acid effect (MIC reduced by ≥3 dilutions) is observed, the test interpretations for penicillins and cephalosporins should be changed to resistant. However, if confirmation testing is not performed on strains that have a positive screening test, then strains that do not produce ESBLs may be falsely reported as resistant to penicillins and cephalosporins. A survey of laboratories participating in the CDC Active Bacterial Core Surveillance program indicates that less than 50% of laboratories perform ESBL confirmatory tests, suggesting a potentially significant problem in overreporting of resistance (6).
In a previous study (F. C. Tenover, P. M. Raney, P. P. Williams, K. L. Brittain, C. D. Steward, S. K. Fridkin, R. P. Gaynes, and J. E. McGowan, Jr., Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. D1606, 2000) we noted that among 131 E. coli isolates for which the MIC of at least one extended-spectrum cephalosporin was ≥2 μg/ml, 59 (45%) showed reduced susceptibility to CPD (MICs of 2 to 4 μg/ml) but remained below the ESBL screening breakpoint for CAZ, CTX, and ceftriaxone (MICs ≤1 μg/ml). Because of the low MICs of CAZ and CTX, no clavulanic effect could be observed. The objective of this study was to characterize the mechanisms of β-lactam resistance in these 59 strains and to determine whether the results of the ESBL screening test with these organisms should be considered false positives.
MATERIALS AND METHODS
Bacterial isolates.
Fifty-nine E. coli isolates for which the CPD MICs were 2 to 4 μg/ml but the CAZ, CTX, and ceftriaxone MICs were ≤1 μg/ml were selected from strains submitted from 26 Project ICARE (Intensive Care Antimicrobial Resistance Epidemiology) laboratories during a 4-year period (1995 to 1999). Microorganisms were identified using Vitek GNI+ cards (bioMérieux, Hazelwood, Mo.) or standard biochemical reactions (19). Three E. coli isolates from the Centers for Disease Control (CDC) culture collection, two whose MICs of only CPD (CDC10 and CDC110) were increased and one whose MICs to all extended-spectrum cephalosporins (CDC3100) were increased, due to enhanced chromosomal AmpC production, were also included. Control strains for OMPs included E. coli MH225 [MC4100φ (ompC lacZ+)] (8) and E. coli C600, a well-characterized laboratory strain.
Susceptibility testing.
The MICs of amoxicillin (AMX) (Sigma), ticarcillin (TIC) (GlaxoSmithKline, Collegeville, Pa.), piperacillin (PIP)(Sigma), cefazolin (CFZ) (Eli Lilly and Company, Indianapolis, Ind.), cefuroxime (CXM) (Eli Lilly and Co.), CPD (Pharmacia Upjohn, Peapack, N.J.), cefoxitin (FOX) (Sigma), CTX (Sigma), CAZ (Eli Lilly and Co.), cefepime (FEP) (Bristol-Myers Squibb, Wallingford, Conn.), and ATM (Bristol-Myers Squibb) were determined by broth microdilution according to NCCLS guidelines (20, 21) using cation-adjusted Mueller-Hinton broth (BD Biosciences, Sparks, Md.). Broth microdilution MICs also were determined for several β-lactam-β-lactamase inhibitor combinations, including AMX and clavulanate (CLAV) (GlaxoSmithKline, Philadelphia, Pa.) in a 2:1 ratio; CFZ, CXM, CPD, CTX, CAZ, and FEP each with 4-μg/ml CLAV (fixed concentration); and PIP with a fixed 4-μg/ml concentration of tazobactam (TZ) (Wyeth-Ayerst Pharmaceuticals, Pearl River, N.Y.). E. coli ATCC 25922, Staphylococcus aureus ATCC 29213, Pseudomonas aeruginosa ATCC 27853, and Klebsiella pneumoniae ATCC 700603 were used as control strains.
Pulsed-field gel electrophoresis.
Pulsed-field gel electrophoresis was performed as previously described using the enzyme BlnI (35). Fragment patterns were interpreted as previously described by Tenover et al. (36).
IEF and specific β-lactamase activity determination.
Isoelectric focusing (IEF) was performed using crude cell extracts as previously described (16). Hydrolysis rates (in micromoles of substrate hydrolyzed per minute and per microliter of crude cell extract) were determined using the SoftmaxPRO software for enzyme kinetics (Molecular Devices Corporation, Sunnyvale, Calif.) and a SpectraMAX PLUS spectrophotometer (Molecular Devices Corp.) equipped with a microplate reader. Activity was measured as a decrease in absorbance at 240 nm for a 100 μM solution of cephaloridine. The experiments were performed in triplicate at 25°C, and the mean value was recorded. Specific cephalosporinase activity (in micromoles of substrate hydrolyzed per minute and per microgram of protein) was determined by normalizing hydrolysis rates against the concentration of total proteins in the crude cell extract. Total protein concentration was determined with the bicinchoninic acid method (BCA protein assay kit; Pierce, Rockford, Ill.) (32). Specific β-lactamase activity in isolates harboring TEM β-lactamases was performed by measuring the hydrolysis of nitrocephin (observed as a decrease in absorbance at 482 nm) instead of cephaloridine.
Conjugation, cloning, and DNA sequence analysis of β-lactamase genes.
Conjugation experiments were performed using a rifampin- and streptomycin-resistant variant of E. coli strain HB101 as the recipient. Transconjugants were selected on Mueller-Hinton agar (Difco) containing 100-μg/ml ampicillin and a 100-μg/ml concentration of either rifampin or streptomycin. β-Lactamase genes were cloned using plasmid pBGS18−, which contains a kanamycin resistance marker (33). Plasmid DNA, including pBGS18− DNA, was purified from E. coli hosts using the QIAfilter plasmid midi kit (QIAGEN, Chatsworth, Calif.). The DNA was digested with EcoRI or BamHI as described below, ligated overnight with T4 DNA ligase, and transformed into CaCl2-treated E. coli DH5α competent cells. DH5α transformants harboring the pBGS18− plasmid with the inserted β-lactamase gene were selected on Mueller-Hinton agar containing 100-μg/ml ampicillin and 50-μg/ml kanamycin. β-Lactamase genes were subcloned by digesting transformant plasmids with EcoRI, BamHI, or HindIII, reducing the size of the insert containing the β-lactamase gene to 2 to 4 kb. The resulting DNA inserts from the subclones were sequenced in both directions using universal primers, and with primers specific for the coding region. DNA sequencing reactions were performed using ABI Prism dRhodamine terminator cycle sequencing (PE Applied Biosystems, Foster City, Calif.). Products from sequencing reactions were purified on Centri-Sep spin columns (Princeton Separations, Adelphia, N.J.) before analysis on an ABI Prism 377 DNA sequencer (PE-Applied Biosystems).
PCR amplification of ampC promoter and attenuator regions.
Primers AB1 and ampC2 (Table 1) were used to amplify the E. coli ampC promoter and attenuator regions as previously described (5). The forward and reverse sequences were determined using products from two independent PCRs as templates.
TABLE 1.
Primers used for PCR amplification of β-lactamase genes
| Gene | Primer | Sequence (5′→3′) | PCR product size (pb) |
|---|---|---|---|
| E. coli ampC promoter | AB1 | GATCGTTCTGCCGCTGTG | 271 |
| AmpC2 | GGGCAGCAAATGTGGAGCAA | ||
| TEM | Forward | ATGAGTATTCAACATTTCCG | 867 |
| Reverse | CTGACAGTTACCAATGCTCTC | ||
| SHV | Forward | GGTTATGCGTTATATTCGCC | 867 |
| Reverse | TTAGCGTTGCCAGTGCTC | ||
| OXA-1 | OXA-1F | ACACAATACATATCAACTTCGC | 814 |
| OXA-1R | AGTGTGTGTTTAGAATGGTGATC | ||
| OXA-2 | OXA-2F | TTCAAGCCAAAGGCACGATAG | 704 |
| OXA-2R | TCCGAGTTGACTGCCGGGTTG | ||
| OXA-10 | OXA-10F | CGTGCTTTGTAAAAGTAGCAG | 651 |
| OXA-10R | CATGATTTTGGT GGGAATGG |
PCR amplification of β-lactamases.
PCR amplification of blaTEM and blaSHV β-lactamase genes was performed as previously described (28). Analysis of the DNA sequences of oxacillin-hydrolyzing β-lactamases (OXAs) in GenBank were used to design specific primers for amplification of the most common subgroups in the OXA family of β-lactamases (Table 1). Primers OXA-1F and OXA-1R were designed to amplify an 814-bp fragment of the gene encoding the OXA-1 β-lactamase and the related OXA-4 and OXA-30 genes. Primers OXA-2F and OXA-2R amplify a 704-bp fragment of the OXA-2 gene and the closely related genes OXA-3, OXA-15, and OXA-21. Primers OXA-10F and OXA-10R amplify a 651-bp fragment of the OXA-10 gene and the related genes OXA-7, OXA-11, OXA-13, OXA-14, OXA-16, OXA-17, OXA-19, and OXA-28. Each PCR mixture contained 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, a 0.1 μM concentration of each primer, a 200 μM concentration of each deoxynucleoside triphosphate, 2.5 U of Taq polymerase, and 1 μl of crude cell lysate in a final volume of 100 μl. The PCR conditions were 96°C for 5 min, 35 cycles of 96°C for 1 min, 60°C for 1 min, and 72°C for 1 min, followed by a final extension cycle of 72°C for 10 min. The specificity of each set of primers was confirmed using DNA templates from a laboratory collection of E. coli strains harboring OXA-1, OXA-2, OXA-3, OXA-4, or OXA-7 β-lactamases.
OMP profile determination.
OMPs were isolated from selected strains as previously described (9, 28). Briefly, cells were grown to mid-log phase in Luria-Bertani broth or nutrient broth and harvested by centrifugation. Cell pellets were washed and resuspended in sodium phosphate buffer, pH 7.0, and the cell walls were disrupted by treatment with lysozyme and sonication. Total membrane proteins were separated from cell debris by differential centrifugation. Sarkosyl-insoluble OMPs were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis through 10% polyacrylamide-8 M urea gels. E. coli strains C600 and MH225 (OmpC−) were used as controls.
RESULTS
Fifty-nine E. coli isolates for which the CPD MICs were 2 to 4 μg/ml were divided into five phenotypic groups (groups A to E) based on β-lactam susceptibility profiles, IEF results, and PCR assays for blaSHV, blaTEM, and blaOXA. Characteristics of phenotypic groups A to E are presented in Table 2. Representative isolates from each group were selected for further study.
TABLE 2.
β-Lactam resistance phenotypes for 59 E. coli isolates for which the MIC of cefpodoxime is ≥2 and that of ceftazidime is ≤1 μg/mla
| MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|
| Major phenotypes
|
Minor phenotypes
|
||||
| A | B | C | D | E | |
| n strains (%) | 32 (54.2%) | 18 (30.5%) | 7 (11.9%) | 1 (1.7%) | 1 (1.7%) |
| β-Lactamase pI | 5.4 | ≥8.5 | 7.2 | 5.4, 5.7, 6.8 | |
| AMX | >64 | 32->64 | >64 | 4 | >64 |
| AMX-CLAV | 16-32 | 16-32 | 32->64 | 4 | 32 |
| TIC | >128 | 8-32 | >128 | 8 | >128 |
| PIP | >128 | <4-8 | >128 | 16 | >128 |
| PIP-TZ | 8->128 | <4-8 | ≥128 | 16 | 16 |
| CFZ | 16->256 | 4-64 | 16-32 | 2 | 16 |
| CFZ-CLAV | 2-4 | 2-16 | 4-8 | 1 | 2 |
| CXM | 16-32 | 8-32 | 32-128 | 16 | 128 |
| CPD | 2-4 | 2-4 | 4-8 | 4 | 2 |
| FOX | 16->32 | 8-32 | 8-32 | 16 | 16 |
| CAZ | 0.5-1 | 0.5-1 | 0.25-1 | 0.5 | 1 |
| CAZ-CLAV | 0.5 | 0.5-1 | 0.12-0.5 | 0.5 | 0.5 |
| CTX | 0.5 | 0.25-1 | 1 | 1 | 0.5 |
| CTX-CLAV | 0.25-0.5 | 0.12-0.5 | 0.25 | 0.5 | 0.5 |
| FEP | 0.12-0.5 | 0.06-0.25 | 2-4 | 0.25 | 0.5 |
| FEP-CLAV | 0.12 | 0.06-0.12 | 0.12-0.5 | 0.12 | 0.12 |
NCCLS breakpoints (S/I/R): AMX, CAZ, CXM, CFZ, FEP, FOX, ≤8/16/≥32; CPD, ≤2/4/≥8; CTX, ≤8/16-32/≥64; PIP, TIC, ≤16/32-64/≥128
Characterization of isolates with phenotype A.
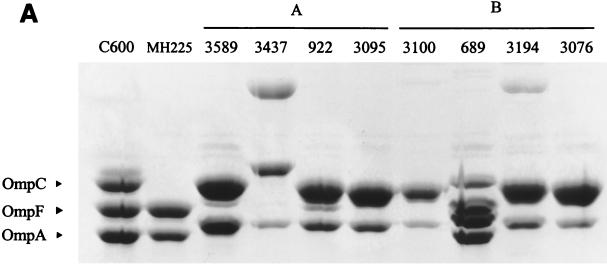
The 32 isolates in this group were obtained from 11 hospitals with a maximum of eight isolates from a single hospital. No more than two isolates from any hospital had the same pulsed-field gel electrophoresis (PFGE) profile (data not shown). Isolates in phenotype A were identified by the following characteristics: (i) positive for blaTEM by PCR and positive for a β-lactamase with pI 5.4, consistent with a TEM-type enzyme; (ii) high MICs for penicillin-type agents (AMX, TIC, and PIP); (iii) high MICs for FOX; (iv) low-level resistance to β-lactam-β-lactamase-inhibitor combinations (AMX-CLAV and PIP-TZ); and (v) low-level resistance to CFZ and CXM. Analysis of OMPs from six strains showed a variety of patterns, including isolates with modifications in both OmpC and OmpF. Two strains, EC410 and EC3437 (from two different hospitals), were investigated further. OmpF was missing in both EC410 (data not shown) and EC3437 (Fig. 1). An additional OMP, larger than OmpC, was produced by EC3437.
FIG. 1.
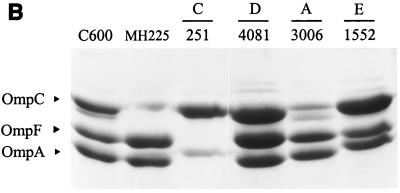
OMP profiles of selected E. coli isolates and control strains grown in nutrient broth. Proteins were separated on 10% polyacrylamide-8 M urea gels and stained with Coomassie blue. (A) Comparison of isolates with phenotypes designated as A (EC3589, EC3437, EC922, and EC3095) or B (EC689, EC3194, EC3076) and control strains E. coli C600 (wild type), E. coli MH225 (OmpC−), and CDC3100 (enhanced AmpC producer). (B) Comparison of isolates with phenotypes designated as C (EC251), D (EC4081), A (EC3006), or E (EC1552) and E. coli C600 and E. coli MH225 (OmpC−).
Conjugation experiments between EC410 or EC3437 and E. coli HB101 revealed a transferable β-lactamase with a pI of 5.4 that was associated with a carriage of a plasmid of approximately 150 kb. The antimicrobial resistance profiles of transconjugants from both isolates (Table 3) were typical of strains expressing a TEM-1 β-lactamase. The transconjugants were resistant to penicillins, but the MICs of cephalosporins (including those of CFZ, CXM, and FOX) and β-lactam-β-lactamase inhibitor combinations remained low. The MICs of CAZ and CTX remained close to the baseline levels (0.5 and 0.12 μg/ml, respectively) for the host strain. Thus, the mechanisms associated with the isolates of phenotype A appear to be production of TEM-1 β-lactamase in conjunction with modification of porins.
TABLE 3.
MICs for EC410 and EC3437 and transconjugants
| Antibiotic | MIC (μg/ml) for:
|
||||
|---|---|---|---|---|---|
| HB101 | EC410 | HB101/410TC | EC3437 | HB101/3437TC | |
| AMX | 4 | >64 | >64 | >64 | >64 |
| AMX-CLAV | 4 | 16 | 8 | 16 | 4 |
| TIC | ≤4 | >128 | >128 | >128 | >128 |
| PIP | ≤4 | >128 | >128 | >128 | >128 |
| PIP-TZ | ≤4 | ≤4 | ≤4 | 32 | ≤4 |
| CFZ | 2 | 16 | 4 | 32 | 4 |
| CFZ-CLAV | 1 | 4 | 1 | 4 | 1 |
| CXM | 4 | 32 | 4 | 16 | 4 |
| CXM-CLAV | 2 | 16 | 2 | 16 | 2 |
| FOX | 4 | 32 | 4 | 32 | 4 |
| CPD | 1 | 2 | 1 | 4 | 1 |
| CPD-CLAV | 0.5 | 2 | 0.5 | 2 | 0.5 |
| CAZ | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| CAZ-CLAV | 0.25 | 0.25 | 0.25 | 0.5 | 0.25 |
| CTX | 0.12 | 0.5 | 0.12 | 0.25 | 0.12 |
| CTX-CLAV | 0.12 | 0.25 | 0.06 | 0.25 | 0.12 |
| ATM | 0.25 | 0.5 | 0.25 | 0.5 | 0.25 |
| ATM-CLAV | 0.12 | 0.5 | 0.12 | 0.25 | 0.25 |
| FEP | 0.06 | 0.12 | 0.06 | 0.12 | 0.12 |
| FEP-CLAV | 0.06 | 0.06 | 0.06 | 0.12 | 0.06 |
Strains EC410, EC3437 and their respective transconjugants harbored a pI 5.4 TEM beta-lactamase (see characterization of phenotype A).
Strain EC3006 had an antimicrobial resistance pattern similar to those of the other strains from phenotype A, but with a higher resistance to CFZ. The CFZ MIC was markedly reduced by CLAV (Table 2). PCR was positive for blaTEM, and IEF revealed a TEM-type β-lactamase with a pI of 5.4. Penicillin and narrow-spectrum cephalosporin resistance was transferred to E. coli HB101 by conjugation; however, the MICs of several β-lactams, including CFZ, CXM, FOX, and CPD (Table 4), were significantly lower than those of the EC3600 parent, suggesting that alteration of the porins in the parent strain contributed to the higher level of resistance. A significant CLAV effect was observed for CFZ. A 25-kb BamHI fragment harboring a β-lactamase gene was cloned from the HB101 transconjugant into pBGS18− plasmid, and an approximately 3-kb EcoRI-HindIII fragment from this plasmid was isolated and subcloned. DNA sequence analysis revealed an open reading frame with a nucleotide sequence identical to the TEM-1 β-lactamase (data not shown). Further analysis revealed a blaTEM-1B promoter (2, 13) which is characterized by a C-to-T nucleotide substitution at position 32 (177 bp upstream of the coding region). This substitution is found in TEM-2 β-lactamase and its derivatives and is known to create two overlapping promoter sequences, resulting in a 10-fold increase in penicillin resistance (7) Assays of specific enzymatic activity demonstrated a significant increase in β-lactamase production of EC3006 compared to an isolate producing a classic TEM-1 β-lactamase (350.9 ± 39.5 versus 41.9± 4.5 nmol min−1 mg−1, respectively). As previously demonstrated (24, 34, 37), TEM-1 hyperproduction is likely responsible for the higher resistance levels for AMX-CLAV and CFZ found in the EC3006 transconjugant compared with expected MICs of an unmodified TEM-1 β-lactamase. Additionally, the porin profile for EC3006 indicated that the production of OmpC was significantly decreased, and an additional OMP intermediate in size between OmpC and OmpF, was present. The relative quantity of OmpF did not appear to be altered when compared with the OMP profiles of the control strain, E. coli C600.
TABLE 4.
Resistance profiles of EC251 and EC3006 and their respective transconjugants and clonesa
| Antibiotic | MIC (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| HB101 | DH5α | EC251 | HB101 251TC | DH5α (pBG251) | EC3006 | HB101 3006TC | DH5α(pBG3006) | |
| AMX | 4 | ≤2 | >64 | >64 | >64 | >64 | >64 | >64 |
| AMX-CLAV | 4 | ≤2 | 32 | 16 | 32 | 32 | 16 | 32 |
| TIC | ≤4 | ≤4 | >128 | >128 | >128 | >128 | >128 | >128 |
| PIP | ≤4 | ≤4 | >128 | 64 | >128 | >128 | >128 | >128 |
| PIP-TZ | ≤4 | ≤4 | 128 | 16 | >128 | >128 | 128 | >128 |
| CZ | 2 | 1 | 16 | 4 | 32 | >256 | 32 | 256 |
| CZ-CLAV | 1 | 1 | 4 | 1 | 1 | 2 | 1 | 1 |
| CXM | 4 | 2 | 32 | 8 | 64 | 32 | 8 | 16 |
| CXM-CLAV | 2 | 1 | 16 | 4 | 4 | 16 | 4 | 2 |
| FOX | 4 | 2 | 32 | 4 | 2 | 16 | 4 | 2 |
| CPD | 1 | 0.5 | 4 | 1 | 16 | 4 | 1 | 2 |
| CPD-CLAV | 0.5 | 0.5 | 2 | 0.5 | 0.5 | 2 | 0.5 | 0.25 |
| CAZ | 0.5 | 0.25 | 0.5 | 0.25 | 0.25 | 1 | 0.5 | 0.5 |
| CAZ-CLAV | 0.25 | 0.25 | 0.25 | 0.25 | 0.12 | 0.5 | 0.25 | 0.25 |
| CTX | 0.12 | 0.06 | 1 | 0.5 | 4 | 0.5 | 0.12 | 0.12 |
| CTX-CLAV | 0.12 | 0.06 | 0.25 | 0.12 | 0.12 | 0.5 | 0.06 | 0.06 |
| ATM | 0.25 | 0.06 | 0.5 | 0.12 | 0.5 | 0.5 | 0.25 | 0.12 |
| ATM-CLAV | 0.12 | 0.06 | 0.5 | 0.12 | 0.06 | 0.25 | 0.12 | 0.12 |
| FEP | 0.06 | 0.06 | 2 | 1 | >32 | 0.5 | 0.12 | 0.5 |
| FEP-CLAV | 0.06 | 0.03 | 0.12 | 0.12 | 0.25 | 0.12 | 0.06 | 0.12 |
EC251 and its corresponding transconjugant (HB101 251TC) and clone [DH5α (pBG251)] harbored an OXA-30 β-lactamase (see characterization of phenotype C) and EC3006 and its corresponding transconjugant (HB101 3006TC) and clone [DH5α (pBG3006)] harbored a TEM-1 β-lactamase.
Characterization of isolates with phenotype B.
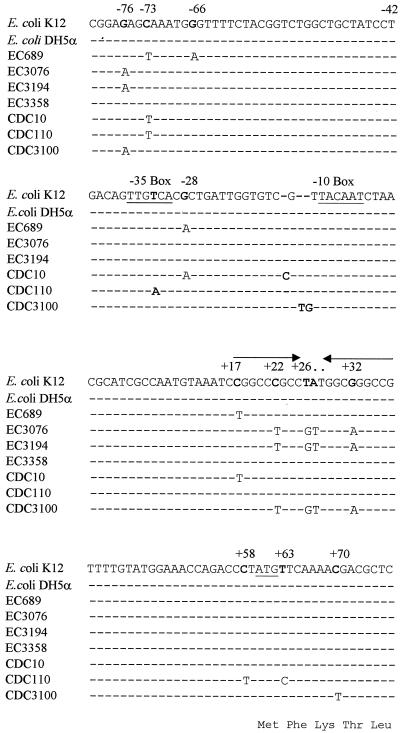
The second group of 18 isolates (phenotype B) also showed multiple PFGE patterns (data not shown). These strains, isolated from nine different hospitals, were characterized by antimicrobial susceptibility profiles that included resistance to AMX but susceptibility or intermediate resistance to TIC and PIP (Table 2). Susceptibility to CFZ, CPD, and FOX varied among the isolates, and no significant CLAV effect was observed. Since a pI band of ≥8.5 was observed in the IEF, enhanced AmpC production was initially suspected as a β-lactam resistance mechanism. The cephalosporinase-specific activity of four isolates from four different hospitals showed only a modest 2- to 2.5-fold increase when compared with E. coli strain DH5α (Table 5). DNA sequence analysis of the ampC promoter and attenuator regions revealed identical mutations in two isolates at position −76 (upstream from the start codon) and at positions +22, +26, +27, and +32 in the attenuator loop. Nucleotide substitutions were noted at positions −73, −66, −28, and +17 (attenuator loop) of the third isolate, and no mutations were detected in the fourth isolate (Fig. 2).
TABLE 5.
Mutations in ampC promoter and attenuator regions and cephalosporinase specific activity of selected E. coli isolates
| Strain | Mutations in promoter and attenuatora | Cephalo- sporinase sp actb |
|---|---|---|
| DH5α | Same as E. coli K-12 control | 1.00 |
| EC689 | −73, −66, −28, +17 | 2.59 ± 0.26 |
| EC3076 | −76, +22, +26, +27, +32 | 2.04 ± 0.13 |
| EC3194 | −76, +22, +26, +27, +32 | 2.67 ± 0.12 |
| EC3358 | No mutations detected | 2.40 ± 0.09 |
| Control strains | ||
| CDC10 | −73, −28, 1-bp insert between −35 and −10, +17 | 13.6 ± 2.2 |
| CDC110 | −73, −32, +58 | 14.6 ± 1.3 |
| CDC3100 | −76, 2-bp insert between −35 and −10, +22, +26, +27, +32 | 17.2 ± 3.7 |
Key mutations associated with enhanced AmpC production are indicated by bold type.
Specific cephalosporinase activities are relative values based on the activity in the wild type strain DH5α.
FIG. 2.
DNA sequence of the ampC promoter and attenuator regions from selected E. coli isolates compared with analogous sequences of K12 and DH5α (4, 5). The −35 and −10 consensus promoter sequences and the translation initiation codon for AmpC β-lactamase are underlined. Arrows indicate the transcription attenuator region. The amino acid sequence of the initial segment of the AmpC leader region is shown below the corresponding nucleotide sequence.
To investigate whether these nucleotide substitutions contribute to CPD resistance by increasing the production of AmpC, the study isolates were compared with control strains CDC10, CDC110, and CDC3100, which express increased levels of AmpC (Table 5). Increased production of AmpC was associated with MICs of CPD of 8 to 16 μg/ml (data not shown) and cephalosporinase activity that was increased 14- to 17-fold when compared with that observed in E. coli DH5α. DNA sequence analysis revealed the following mutations that are known to result in increased transcription of ampC: CDC110, a nucleotide substitution at the −32 position; CDC10 and CDC3100; and a 1-bp and a 2-bp insertion between the −35 and the −10 promoter consensus sequences, respectively. Each of the CDC isolates and the study isolates also had several additional nucleotide substitutions when compared with the E. coli K-12 sequence (Fig. 2).
The OMP profiles were altered for most of the selected isolates. EC689 showed a decrease in OmpC and an additional OMP between OmpC and OmpF, whereas EC3076 and EC3194 appeared to have lost OmpF (Fig. 1). AmpC-hyperproducing strains from the CDC collection for which the MICs of only CPD (8 to 16 μg/ml) among the extended-spectrum cephalosporins were increased (CDC10 and CDC110) showed no apparent alterations of OMPs. On the other hand, strain CDC3100, for which MICs to all extended-spectrum cephalosporins were increased, appeared to have lost OmpF in addition to the AmpC hyperproduction (Fig. 1). Altogether these results suggest that the β-lactam resistance phenotype in this group of strains is mainly due to the loss of porin expression and not to AmpC hyperproduction.
Characterization of phenotype C.
Seven isolates from five hospitals, each with a unique PFGE pattern, were grouped in phenotype C. Each isolate expressed a β-lactamase with a pI of 7.1 to 7.2. PCR assays were positive for blaOXA and negative for blaTEM and blaSHV. Phenotypically, these strains showed high-level resistance to penicillins that was not significantly decreased by the addition of CLAV (Table 2). Resistance to CFZ, CXM, and FOX was variable. CTX MICs of 0.25 to 1 μg/ml were below the resistant breakpoint but were slightly above those for other E. coli isolates. A modest CLAV effect was observed with CTX but was more apparent with FEP. Conjugation of E. coli strain EC251 with HB101 resulted in the transfer of a β-lactamase gene that was associated with a plasmid of approximately 160 kb. The β-lactamase gene was localized to a 2.3-kb HindIII restriction fragment and subcloned into E. coli DH5α(pBG251B3H1). A search of the GenBank database indicated sequence similarity with a fragment of Tn2603 that encodes the OXA-1 β-lactamase (26, 38). PCR amplification with the blaOXA-1 primers and DNA sequence analysis indicated that each of the isolates in the phenotype C contained the OXA-30 enzyme, which differs from OXA-1 by an Arg131-to-Gly modification (31). In addition to the OXA β-lactamase, analysis of the OMPs from strain EC251 revealed that OmpF was missing (Fig. 1). These data suggest that the combination of an OXA β-lactamase and alteration of porin proteins results in the phenotype C susceptibility profile.
Characterization of phenotype D.
For EC4081, the MICs of the penicillins, narrow-spectrum cephalosporins, and FOX were increased but remained below the breakpoint for resistance (Table 2); no CLAV effect was observed. β-Lactamase activity was not detected by either IEF or hydrolysis experiments. The porin profile was unchanged when compared with that of the control strain, E. coli C600. Thus, the mechanism of decreased CPD susceptibility is unclear. Efflux mechanisms, point mutations in the genes coding for the OMP, or penicillin-binding protein modification could account for the low level β-lactam resistance found in this strain.
Characterization of phenotype E.
IEF analysis of EC1552 representing phenotype E indicated the presence of three β-lactamases with pIs of 5.4, 5.7, and 6.8. The resistance pattern was similar to phenotype A, except that EC1552 showed high-level resistance to CXM. A modest CLAV effect was observed with AMX, but a more significant effect was noted with PIP. PCR assays were positive for blaTEM but negative for blaSHV and blaOXA. Plasmid analysis revealed two plasmids of approximately 180 and 120 kb. HB101 transconjugants of strain EC1552 had a resistance pattern typical of a broad-spectrum β-lactamase conferring resistance only to penicillins (data not shown). IEF of transconjugants revealed two β-lactamases of pI 5.4 and 6.8. Plasmid analysis of 10 transconjugants indicated that only the smaller plasmid was transferred, suggesting that the third β-lactamase may be encoded on the larger plasmid. The 180-kb plasmid may be either nontransferable or transferred with lower frequency than the smaller plasmid. Analysis of the OMP profile for EC1552 indicated that production of OmpC was significantly increased when compared with OmpF. Although the pI 5.7 β-lactamase of strain EC1522 could reflect a false-negative ESBL screening test for CAZ and CTX, this is highly unlikely since the low level resistance to CTX and CAZ found in EC1552 was not inhibited by clavulanic acid (Table 2).
DISCUSSION
The NCCLS established a supplemental set of MIC and disk diffusion screening breakpoints for CTX, ceftriaxone, CAZ, CPD, and ATM and added new CLAV-based confirmation tests to improve the accuracy of detecting ESBLs in E. coli, K. pneumoniae, and Klebsiella oxytoca (21). A recent study of K. pneumoniae isolates showed that approximately 84% of isolates with positive results from the ESBL screening test proved to be ESBL-producing strains by the confirmatory tests (35). In sharp contrast, a study of 131 E. coli strains for which the MICs of at least one extended-spectrum cephalosporin were ≥2 μg/ml showed that only 14% were confirmed as ESBL-producing strains (Tenover et al., 40th ICAAC). Fifty-nine (45%) of the E. coli isolates (i.e., those isolates studied here) showed reduced susceptibility to CPD (MIC = 2 to 4 μg/ml; i.e., a positive ESBL screening test), but the MICs for the other extended-spectrum cephalosporins remained below the ESBL screening breakpoint (MICs ≤ 1 μg/ml). These data suggested that the results of the ESBL screening test were false positives, and indicated that laboratories that did not perform the CLAV-based confirmation tests may be inappropriately reporting resistant results for extended-spectrum cephalosporins and ATM for a potentially high percentage of E. coli isolates. The objective of the present study was to characterize the mechanisms of reduced susceptibility to CPD to determine if these ESBL screening test results were, in fact, false positives.
In general, susceptibility to β-lactam agents in E. coli depends on the concentration of antimicrobial agent available in the periplasmic space and, therefore, on the amount of the antimicrobial agent capable of binding the penicillin-binding proteins. In wild-type strains, the intrinsic level of resistance depends on the diffusion rate of the drug across the outer membrane; activity of efflux pumps, such as the AcrAB-TolC system; and the hydrolysis of the drug by the chromosomal AmpC β-lactamase (17, 23). Although present at very low levels, the AmpC β-lactamase plays a key role in the intrinsic resistance of E. coli to β-lactam agents (17, 23). Mutations leading to a stronger promoter (12); the acquisition of a stronger promoter by horizontal DNA transfer from other species, such as Shigella sonnei (25); or mutations in the transcriptional attenuator (11) of the E. coli AmpC chromosomal β-lactamase lead to enhanced production of AmpC which increases the level of resistance to β-lactam agents, including both penicillins and cephalosporins. Acquisition of plasmid-mediated β-lactamases also plays a major role in β-lactam resistance (14). However, porin alterations, as shown in this study, although frequently overlooked as a mechanism of β-lactam resistance, clearly play a key role in β-lactam resistance, especially when present in strains containing β-lactamases (9, 15, 29).
Alteration of the major porin proteins, OmpF and OmpC, may be due to mutations in the regulatory genes, such as mar (30) or ompB (8), or to alteration of the individual porin genes by insertions, deletions, or point mutations. Interruption of porin genes by insertion sequences is a common cause of porin loss in Enterobacteriaceae (10, 27), and the correlation of antimicrobial resistance and porin loss has been demonstrated in several enterobacterial species (1, 10, 18).
The most common mechanism of reduced susceptibility to CPD among the isolates in this study was production of a TEM-1 β-lactamase associated with the loss or alteration of a major porin protein. While the effect on the MICs of CTX and CAZ was minimal, the changes were sufficient to elevate the CPD MICs above the screening breakpoint for ESBLs set by the NCCLS.
The mechanism responsible for the second most common phenotype among the 59 isolates was initially considered to be enhanced production of the chromosomal AmpC β-lactamase. Data supporting this hypothesis included PCR assays that were negative for blaTEM, blaSHV, and blaOXA and detection of a β-lactamase with a pI of 8.3 to 8.5 by IEF. On the other hand, although nucleotide substitutions in the ampC promoter or attenuator regions were found, they were not those known to be associated with enhanced AmpC production, and only a modestly enhanced β-lactam hydrolysis (2 to 2.5 times the background levels) was detected. Control strains showing ampC activity 14 to 17 times that of wild-type E. coli controls did show known mutations in ampC, including nucleotide insertions between the −35 and −10 promoter sequences, and a nucleotide substitution at the −32 position (4, 12). In addition, the study strains expressed lower levels of resistance to the extended-spectrum cephalosporins than expected for strains with enhanced AmpC production (14). Thus, alteration of porin proteins, as shown for strains EC689, EC3194, and EC3076, is the resistance mechanism most likely to be responsible for this phenotype. Strains CDC10 and CDC110 with high cephalosporinase activity had higher CPD MICs (8 to 16 μg/ml) than the study strains but remained susceptible to other extended-spectrum cephalosporins (CAZ ≤ 1 μg/ml), whereas the only strain with high extended-spectrum cephalosporin MICs, CDC3100, had a combination of AmpC hyperproduction and OmpF loss. These results suggest that many of the previously reported AmpC-producing strains that have higher levels of resistance to extended-spectrum cephalosporins may also have porin alterations. Unfortunately, few investigators determined the OMP profiles of strains that they reported as AmpC-associated resistance (4, 22). Therefore, the significance of this mechanism of resistance in terms of actual challenge for treatment of E. coli infections remains unknown.
The third mechanism of reduced susceptibility involved production of an OXA-30 β-lactamase that was also associated with porin changes in some strains. The isolates in this study came from a variety of hospitals in the United States, suggesting that this β-lactamase is widely distributed. The extended-spectrum cephalosporin most affected by OXA-30 is FEP. MICs of FEP were 2 to 4 μg/ml for isolates in this phenotypic group, in contrast to MICs of 0.06 to 0.12 μg/ml that are typical for wild-type E. coli isolates. In transformants containing the cloned OXA-30 β-lactamase gene, where the enzyme is produced in larger amounts, MICs of FEP and CTX were >32 and 4 μg/ml, respectively. Thus, inoculum effects may be important.
It is clear that CPD MICs are more heavily influenced by porin changes, enhanced TEM production, and OXA-type β-lactamases than are the MICs of CTX or CAZ. This is likely the reason for the high number of false-positive results obtained when CPD is used for screening E. coli isolates for ESBLs. The NCCLS recommends that strains harboring ESBLs be considered resistant to all penicillins and cephalosporins whether the MICs are above the traditionally recommended breakpoints or not (21). We support these recommendations. Recently, the NCCLS changed the CPD screening breakpoint from ≥2 to ≥8 μg/ml based on additional data from our laboratory (P. M. Raney, P. P. Williams, J. E. McGowan, Jr., and F. C. Tenover, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. D-182, 2001). In summary, we have characterized a series of E. coli isolates that display decreased susceptibility to CPD only, producing what we believe to be false-positive ESBL screening test results. Such isolates should not be classified as ESBLs, nor should they be classified as containing ESBLs, nor should the interpretations of extended-spectrum cephalosporins be changed to “resistant” on susceptibility test reports.
Acknowledgments
We thank Gregory Anderson for assistance with DNA sequencing studies and Christine Steward for helpful discussions. We also thank T. Silhavy for the generous gift of E. coli porin mutants.
REFERENCES
- 1.Aggeler, R., R. Then, and R. Ghosh. 1987. Reduced expression of outer-membrane proteins in β-lactam-resistant mutants of Enterobacter cloacae. J. Gen. Microbiol. 133:3383-3392. [DOI] [PubMed] [Google Scholar]
- 2.Backman, A., P. Orvelid, J. A. Vazquez, O. Skold, and P. Olcen. 2000. Complete sequence of a β-lactamase-encoding plasmid in Neisseria meningitidis. Antimicrob. Agents Chemother. 44:210-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caroff, N., E. Espaze, I. Berard, H. Richet, and A. Reynaud. 1999. Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyminocephalosporins without extended spectrum β-lactamase production. FEMS Microbiology Lett. 173:459-465. [DOI] [PubMed] [Google Scholar]
- 5.Caroff, N., E. Espaze, D. Gautreau, H. Richet, and A. Reynaud. 2000. Analysis of the effects of −42 and −32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing AmpC. J. Antimicrob. Chemother. 45:783-788. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2000. Laboratory capacity to detect antimicrobial resistance, 1998. Morb. Mortal. Wkly. Rep. 48:1167-1171. [PubMed] [Google Scholar]
- 7.Chen, S. T., and R. C. Clowes. 1984. Two improved promoter sequences for the beta-lactamase expression arising from a single base-pair substitution. Nucleic Acids Res. 12:3219-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall, M. N., and T. J. Silhavy. 1981. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J. Mol. Biol. 146:23-43. [DOI] [PubMed] [Google Scholar]
- 9.Harder, K. J., H. Nikaido, and M. Matsuhashi. 1981. Mutant of Escherichia coli that are resistant to certain beta-lactam compounds lack the ompF porin. Antimicrob. Agents Chemother. 20:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez-Alles, S., V. J. Benedi; L. Martinez-Martinez, A. Pascual, A. Aguilar, J. M. Tomas, and S. Alberti. 1999. Development of resistance during antimicrobial therapy caused by insertion sequence interruption of porin genes. Antimicrob. Agents Chemother. 43:937-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaurin, B., T. Grundstrom, T. Edlund, and S. Normark. 1981. The E. coli β-lactamase attenuator mediates growth rate-dependent regulation. Nature 290:221-225. [DOI] [PubMed] [Google Scholar]
- 12.Jaurin, B., T. Grundstrom, and S. Normark. 1982. Sequence elements determining ampC promoter strength in E. coli. EMBO J. 1:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leflon-Guibout, V., V. Speldooren, B. Heym, and M. H. Nicolas-Chanoine. 2000. Epidemiological survey of amoxicillin-clavulanate resistance and corresponding molecular mechanisms in Escherichia coli isolates in France: new genetic features of blaTEM genes. Antimicrob. Agents Chemother. 44:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livermore, D. M. 1995. β-lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Martinez, L., M. C. Conejo, A. Pascual, S. Hernandez-Alles, S. Ballesta, E. Ramirez de Arellano-Ramos, V. J. Benedi, and E. J. Perea. 2000. Activities of imipenem and cephalosporins against clonally related strains of Escherichia coli hyperproducing chromosomal β-lactamase and showing altered porin profiles. Antimicrob. Agents Chemother. 44:2534-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 17.Mazzariol, A., G. Cornaglia, and H. Nikaido. 2000. Contributions of the AmpC β-lactamase and the AcrAB multidrug efflux system in intrinsic resistance of Escherichia coli K12 to β-lactams. Antimicrob. Agents Chemother. 44:1387-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medeiros, A. A., T. F. O'Brien, E. Y. Rosenberg, and H. Nikaido. 1987. Loss of ompC porin in a strain of Salmonella typhimurium causes increased resistance to cephalosporins during therapy. J. Infect. Dis. 156:751-757. [DOI] [PubMed] [Google Scholar]
- 19.Murray, P. R., E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. American Society for Microbiology. Washington, D.C.
- 20.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing. NCCLS approved standard M100-S12. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.Nelson, E. C., and B. G. Elisha. 1999. Molecular basis of AmpC hyperproduction in clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 43:957-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikaido, H., and S. Normark. 1987. Sensitivity of Escherichia coli to various β-lactams is determined by the interplay of outer membrane permeability and the degradation by periplasmic β-lactamases: a quantitative predictive treatment. Mol. Microbiol. 1:29-35. [DOI] [PubMed] [Google Scholar]
- 24.Oliver, A., Perez-Vazquez, M., M. Martinez-Ferrer, F. Baquero, L. de Rafael, and R. Canton. 1999. Ampicillin-sulbactam and amoxicillin-clavulanate susceptibility testing of Escherichia coli isolates with different β-lactam resistance phenotypes. Antimicrob. Agents Chemother. 43:862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson, O., S. Bergstrom, F. P. Lindberg, and S. Normak. 1983. AmpC β-lactamase hyperproduction in Escherichia coli: natural ampicillin resistance generated by horizontal chromosomal DNA transfer from Shigella. Proc. Natl. Acad. Sci. USA 80:7556-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouellette, M., L. Bissonnette, and P. H. Roy. 1987. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: Nucleotide sequence of the OXA-1 β-lactamase gene. Proc. Natl. Acad. Sci. USA 84:7378-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozawa, Y., S. Mizushima, and T. Mizuno. 1990. Osmoregulatory expression of the ompC gene in Escherichia coli K-12; IS1 insertion in the upstream regulatory region results in constitutive activation of the promoter. FEMS Microbiol. Lett. 68:295-300. [DOI] [PubMed] [Google Scholar]
- 28.Rasheed, J. K., C. Jay, B. Metchock, F. Berkowitz, L. Weigel, J. Crellin, C. Steward, B. Hill, A. A. Medeiros, and F. C. Tenover. 1997. Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 41:647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumacher, H., U. Skibsted, R. Skov, and J. Scheibel. 1996. Cefuroxime resistance in Escherichia coli. APMIS 104:531-538. [DOI] [PubMed] [Google Scholar]
- 30.Seoane, A. S., and S. B. Levy. 1995. Identification of new genes regulated by the marRAB operon in Escherichia coli. J. Bacteriol. 177:530-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siu, L. K., J. Y. C. Lo, K. Y. Yuen, P. Y. Chau, M. H. NG, and P. L. Ho. 2000. β-lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel oxa-1 like β-lactamase, oxa-30. Antimicrob. Agents Chemother. 44:2034-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Garter, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 33.Spratt, B. G., P. J. Hedge, S. te Heesen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41:337-342. [DOI] [PubMed] [Google Scholar]
- 34.Stapleton, P., P. J. Wu, A. King, K. Shanon, G. French, and I. Phillips. 1995. Incidence and mechanism of resistance to the combination of amoxicillin and clavulanic acid in Escherichia coli. Antimicrob. Agents Chemother. 39:2478-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steward, C. D., J. K. Rasheed, S. K. Hubert, J. W. Biddle, P. M. Raney, G. J. Anderson, P. P. Williams, K. L. Brittain, A. Oliver, J. E. McGowan, Jr., and F. C. Tenover. 2001. Characterization of clinical isolates of Klebsiella pneumoniae from 19 laboratories using the National Committee for Clinical Laboratory Standards extended-spectrum β-lactamase detection methods. J. Clin. Microbiol. 39:2864-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, P. J., K. Shanon, and I. Phillips. 1994. Effect of hyperproduction of TEM-1 β-lactamase on in vitro susceptibility of Escherichia coli to β-lactam antibiotics. Antimicrob. Agents Chemother. 38:494-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto, T., M. Tanaka, R. Baba, and S. Yamagishi. 1981. Physical and functional mapping of Tn2603, a transposon encoding ampicillin, streptomycin, sulfonamide, and mercury resistance. Mol. Gen. Genet. 181:464-469. [DOI] [PubMed] [Google Scholar]