Abstract
To determine how imipenem inhibits the class C β-lactamase AmpC, the X-ray crystal structure of the acyl-enzyme complex was determined to a resolution of 1.80 Å. In the complex, the lactam carbonyl oxygen of imipenem has flipped by approximately 180° compared to its expected position; the electrophilic acyl center is thus displaced from the point of hydrolytic attack. This conformation resembles that of imipenem bound to the class A enzyme TEM-1 but is different from that of moxalactam bound to AmpC.
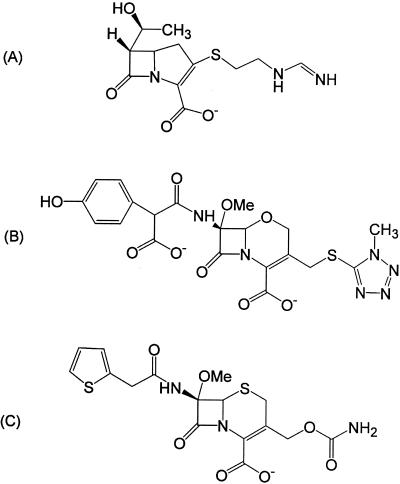
Why do some β-lactams act as substrates and others act as inhibitors of serine β-lactamases? Previous studies have shown that β-lactams that contain large substituents in the 6(7)α position such as imipenem (Fig. 1A), moxalactam (Fig. 1B), and cefoxitin (Fig. 1C) inhibit both class A and class C β-lactamases (9, 10) and actually destabilize these enzymes upon binding (1, 17). These compounds are thought to inhibit both class A and class C enzymes because of conformational changes induced by their large 6(7)α substituents. However, crystallographic data suggest that these inhibitors have different mechanisms of action.
FIG. 1.
Structures of β-lactams that contain large 6(7)α substituents. (A) Imipenem; (B) moxalactam; (C) cefoxitin.
In class A enzymes, crystal structures of the TEM-1 bound to imipenem (PDB code 1BT5) (10) and BS3 bound to cefoxitin (PDB code 1I2W) (6) suggest that the 6(7)α groups force the ligands to adopt a conformation where the carbonyl oxygen of what was the β-lactam ring is “flipped out” from its expected binding location in the oxyanion (11), or electrophilic (16), hole. This dramatic conformational change appears to be a result of steric strain induced in the acyl-enzyme intermediate by the presence of the large substituent in the 6(7)α position. This conformational change, in turn, precludes deacylation.
In class C enzymes, the crystal structure of AmpC bound to moxalactam (PDB code 1FCO) suggests a different mechanism of inhibition (14). In this complex, moxalactam appears to bind as expected for typical substrates, with the carbonyl oxygen of what was the β-lactam ring bound in the oxyanion, or electrophilic, hole. However, the presence of the 6(7)α group in this ligand forces the oxacephem ring into a position that destabilizes the formation of the tetrahedral deacylation transition state. Hence, moxalactam resists deacylation and acts as an inhibitor.
This set of crystal structures raises two questions: is there a fundamental difference between class A and class C enzymes that results in the different binding modes (in or out of the oxyanion hole), or do cefoxitin and imipenem interact differently in the acyl state than does moxalactam? To answer these questions, the X-ray crystal structure of AmpC β-lactamase from Escherichia coli in complex with imipenem has been determined to a resolution of 1.80 Å.
Crystals of wild-type AmpC were grown by vapor diffusion in 6-μl hanging drops in 1.7 M potassium phosphate at pH 8.7 (16). Crystals were harvested and soaked for 40 min in a solution of saturated imipenem in crystallizing buffer. The crystals were then cryoprotected in 25% sucrose in crystallizing buffer with saturated imipenem and flash cooled in liquid nitrogen. X-ray diffraction data were measured with the DuPont-Northwestern-Dow Collaborative Access Team beamline 5IDB apparatus at the Advanced Photon Source research facility by using a Mar charge-coupled device detector with a 162-mm-diameter objective at 100 K. The HKL software suite (13) was used to index, integrate, and scale all data (Table 1). The structure was determined by molecular replacement using a native AmpC wild-type structure (PDB code 1KE4) with all solvent atoms removed. The model was positioned using rigid body refinement and then refined by using simulated annealing, positional minimization, and individual B-factor techniques with the maximum likelihood target, a bulk solvent correction, and a 2-σ cutoff using CNS (2). Sigma A-weighted electron density maps were also calculated with CNS (2). Manual rebuilding and placement of water molecules were performed with the program O (7) and alternated with rounds of positional and B-factor refinement with CNS. The atomic coordinates and structure factors have been deposited in the Protein Data Bank with ID code 1LL5.
TABLE 1.
Crystallographic data collection and refinement statistics
| Parameter (unit) | Value |
|---|---|
| Space group | C-2 |
| Molecules per asymmetric unit | 2 |
| Cell constants | |
| a (Å) | 118.46 |
| b (Å) | 76.91 |
| c (Å) | 98.01 |
| β (degree) | 116.16 |
| Resolution (Å)a | 1.80 (1.84-1.80) |
| Total observations | 230,232 |
| Unique reflections | 64,997 |
| Rmerge (%) | 6.2 (17.9) |
| Completeness (%) | 88.9 (96.4) |
| 〈I〉c/〈σ〉 | 29.1 (7.5) |
| Resolution range for refinement (Å) | 20-1.80 |
| No. of protein residues | 709 |
| No. of water molecules | 254 |
| Rmsd bond lengths (Å) | 0.015 |
| Rmsd bond angles (degree) | 1.77 |
| R factor (%) | 20.5 |
| Rfree (%)b | 25.1 |
| Average B factor, protein atoms (Å2) | 31.9 |
| Average B factor, ligand atoms (Å2) | 43.2 |
| Average B factor, solvent atoms (Å2) | 36.1 |
Values in parentheses are for the highest resolution shell.
Rfree was calculated with a percentage of 4.0 of reflections set aside randomly.
〈I〉, average intensity.
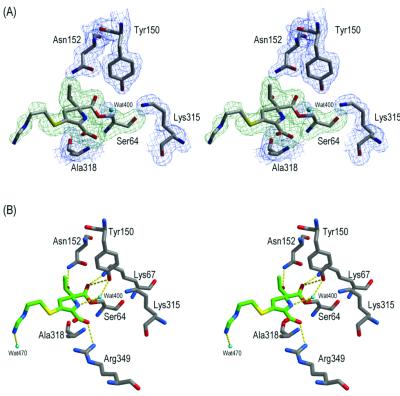
The electron density within each active site revealed an imipenem molecule covalently bound to Ser64 (Fig. 2A). Because the ligands in each active site bind similarly, except for the distal portion of the R2 tail, molecule 1 will be the subject of all further discussion. The electron density reveals that imipenem binds such that its carbonyl oxygen O-7 of what was the β-lactam ring is not bound in the oxyanion, or electrophilic, hole, which is made up of the backbone nitrogens of Ser64 and Ala318 (Fig. 2B; Table 2). Instead, it has flipped approximately 180° from its expected location, a movement of 2.9 Å (Fig. 3). In its new position, the carbonyl oxygen O-7 of imipenem interacts with Lys67 and Tyr150. The hydroxyethyl group in the 6(7)α position hydrogen bonds with Asn152, a residue typically involved in binding of the R1 amide group of β-lactam substrates (8, 12, 14). These interactions of the carbonyl oxygen and the hydroxyethyl group are similar to those observed in the TEM-1-imipenem complex (10). The carbapenem ring is oriented outward such that its C-3 carboxylate interacts with Arg349, displacing a conserved water molecule (Wat401 in molecule 1 and Wat403 in molecule 2) that has been observed in several structures (3, 14, 15). Surprisingly, the putative deacylating water (Wat400 in molecule 1 and Wat402 in molecule 2) is observed in the complex, hydrogen bonding with both Ser64Oγ and the ring nitrogen N-4 of imipenem. The extended R2 tail of imipenem makes few interactions within the site.
FIG. 2.
The active site for molecule 1 of the AmpC-imipenem complexed structure. (A) Stereoview of the electron density of the active site. The 2Fo-Fc electron density of the refined protein model is shown in blue, contoured at 1 σ, and the simulated annealing omit electron density of the refined ligand covalently bound to Ser64 is shown in green, contoured at 3.3 σ. Carbon atoms are colored gray, oxygen atoms are colored red, and nitrogen atoms are colored blue. Cyan spheres indicate water molecules. The figure was generated using SETOR software (4). (B) Stereoview of the interactions observed in the AmpC-imipenem complexed structure. Atoms are colored as described for panel A, except the carbon atoms of the ligand are colored green. Dashed yellow lines indicate hydrogen bonds. Figures 2B and 3 were generated using MidasPlus software (5).
TABLE 2.
Key interactions within the AmpC-imipenem complexed structure
| Interaction | Distance (Å)
|
|
|---|---|---|
| AmpC-imipenema | AmpC apob | |
| Y150OH-K315Nζ | 2.8 | 2.8 |
| Y150OH-S64Oγ | NPc | 2.9 |
| Y150OH-K67Nζ | 3.0 | 3.3 |
| K67Nζ-A220O | 3.0 | 2.9 |
| K67Nζ-S64Oγ | NP | 2.8 |
| N152Oδ1-K67Nζ | 2.7 | 2.6 |
| N152Nδ2-Q120Oɛ1 | NP | 2.8 |
| K315Nζ-T316O | 3.1 | 2.8 |
| Y150OH-Wat400/402d | 3.1 | NP |
| S640γ-Wat400/402 | 3.1 | NP |
| ImiN4-Wat400/402 | 2.9 | NP |
| ImiO7-K67Nζ | 2.9 | NP |
| ImiO7-Y150OH | 3.1 | NP |
| ImiO62-N152Nδ2 | 2.9 | NP |
| ImiO31-R349Nη1 | 2.9 | NP |
| ImiO31-N346Oδ1 | NP | NP |
| ImiN26-Wat470 | 2.7 | NP |
Distances are given for molecule 1, in which the density for imipenem was better defined. Similar distances were observed in molecule 2.
From PDB entry 1KE4 molecule 2 (15).
NP, not present.
Wat400 and Wat402 are equivalent waters in molecule 1 and molecule 2, respectively.
FIG. 3.
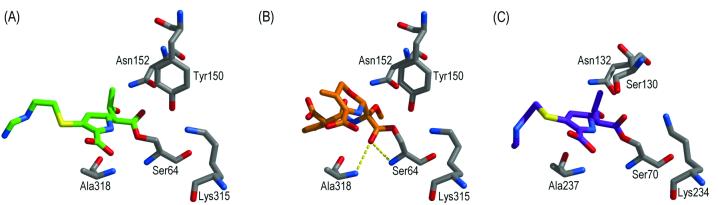
Comparison of the acyl adducts of AmpC and imipenem (PDB entry 1LL5) (A), AmpC and moxalactam (PDB code 1FCO) (14) (B), and TEM-1 and imipenem (PDB code 1BT5) (10) (C), showing that AmpC binds imipenem differently than it does moxalactam but similarly to TEM-1. Atoms are colored as described for Fig. 2A, except the carbon atoms of imipenem in panel A are colored green, the carbon atoms of moxalactam in panel B are colored orange, and carbon atoms of imipenem in panel C are colored purple. Dashed yellow lines indicate hydrogen bonds of the oxyanion, or electrophilic, hole.
Imipenem appears to act as an inhibitor of AmpC β-lactamase because it can react with the enzyme to form an acyl adduct, but once in this form, its bulky 6α-hydroxyethyl side chain forces its electrophilic acyl center to rotate away from the point of hydrolytic attack. The failure of the oxyanion, or electrophilic, hole residues to position the electrophilic center for hydrolytic attack accounts for the effective inhibition of class C enzymes by imipenem as well as the thermodynamic destabilization observed upon complex formation (1). This structure suggests that class C enzymes recognize imipenem differently than they do β-lactams that contain both a large 6(7)α substituent and an R1 side chain, such as moxalactam (14) and cefmetazole (R. A. Powers, B. Beadle, and B. Shoichet, unpublished data) (Fig. 3). Although class A enzymes bind imipenem and cefoxitin similarly, with their carbonyl oxygens forced out of the oxyanion hole, the less-constrained active-site geometry of class C enzymes allows two different binding modes and hence two different mechanisms of inhibition.
Acknowledgments
This work was supported by National Institutes of Health grant GM63815 (to B.K.S.). Crystallography data were collected by the DuPont-Northwestern-Dow Collaborative Access Team at the APS, which is supported by E.I. DuPont, deNemours & Co., the Dow Chemical Company, the National Science Foundation, and the State of Illinois.
We thank Xiaojun Wang and John Irwin for reading the manuscript.
REFERENCES
- 1.Beadle, B. M., S. L. McGovern, A. Patera, and B. K. Shoichet. 1999. Functional analyses of AmpC beta-lactamase through differential stability. Protein Sci. 8:1816-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 3.Crichlow, G. V., M. Nukaga, V. R. Doppalapudi, J. D. Buynak, and J. R. Knox. 2001. Inhibition of class C beta-lactamases: structure of a reaction intermediate with a cephem sulfone. Biochemistry 40:6233-6239. [DOI] [PubMed] [Google Scholar]
- 4.Evans, S. V. 1993. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph. 11:127-128, 134-138. [DOI] [PubMed] [Google Scholar]
- 5.Ferrin, T. E., C. C. Huang, L. E. Jarvis, and R. Langridge. 1988. The MIDAS display system. J. Mol. Graph. 6:13-27. [Google Scholar]
- 6.Fonze, E., M. Vanhove, G. Dive, E. Sauvage, J. M. Frere, and P. Charlier. 2002. Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the acyl-enzyme adduct formed with cefoxitin. Biochemistry 41:1877-1885. [DOI] [PubMed] [Google Scholar]
- 7.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 8.Lobkovsky, E., E. M. Billings, P. C. Moews, J. Rahil, R. F. Pratt, and J. R. Knox. 1994. Crystallographic structure of a phosphonate derivative of the Enterobacter cloacae P99 cephalosporinase: mechanistic interpretation of a beta-lactamase transition-state analog. Biochemistry 33:6762-6772. [DOI] [PubMed] [Google Scholar]
- 9.Matagne, A., J. Lamotte-Brasseur, G. Dive, J. R. Knox, and J. M. Frere. 1993. Interactions between active-site-serine beta-lactamases and compounds bearing a methoxy side chain on the alpha-face of the beta-lactam ring: kinetic and molecular modelling studies. Biochem. J. 293:607-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maveyraud, L., L. Mourey, L. P. Kotra, J. Pedelacq, V. Guillet, S. Mobashery, and J. Samama. 1998. Structural basis for clinical longevity of carbapenem antibiotics in the face of challenge by the common class A beta-lactamases from the antibiotic-resistant bacteria. J. Am. Chem. Soc. 120:9748-9752. [Google Scholar]
- 11.Murphy, B. P., and R. F. Pratt. 1988. Evidence for an oxyanion hole in serine beta-lactamases and DD-peptidases. Biochem. J. 256:669-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oefner, C., A. D'Arcy, J. J. Daly, K. Gubernator, R. L. Charnas, I. Heinze, C. Hubschwerlen, and F. K. Winkler. 1990. Refined crystal structure of beta-lactamase from Citrobacter freundii indicates a mechanism for beta-lactam hydrolysis. Nature 343:284-288. [DOI] [PubMed] [Google Scholar]
- 13.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 14.Patera, A., L. C. Blaszczak, and B. K. Shoichet. 2000. Crystal structures of substrate and inhibitor complexes with AmpC beta-lactamase: possible implications for substrate-assisted catalysis. J. Am. Chem. Soc. 122:10504-10512. [Google Scholar]
- 15.Powers, R. A., and B. K. Shoichet. 2002. Structure-based approach for binding site identification on AmpC beta-lactamase. J. Med. Chem. 32:3222-3234. [DOI] [PubMed] [Google Scholar]
- 16.Usher, K. C., L. C. Blaszczak, G. S. Weston, B. K. Shoichet, and S. J. Remington. 1998. Three-dimensional structure of AmpC beta-lactamase from Escherichia coli bound to a transition-state analogue: possible implications for the oxyanion hypothesis and for inhibitor design. Biochemistry 37:16082-16092. [DOI] [PubMed] [Google Scholar]
- 17.Wang, X., G. Minasov, and B. K. Shoichet. 2002. Noncovalent interaction energies in covalent complexes: TEM-1 beta-lactamase and beta-lactams. Proteins Struct. Funct. Genet. 47:86-96. [PubMed] [Google Scholar]