Abstract
We searched for cell-surface-associated proteins overexpressed on B cell chronic lymphocytic leukemia (CLL) to use as therapeutic antibody targets. Antibodies binding the immunosuppressive molecule CD200 were identified by cell panning of an antibody phage display library derived from rabbits immunized with primary CLL cells. B cells from 87 CLL patients exhibited 1.6- to 5.4-fold cell-surface up-regulation of CD200 relative to normal B cells. An effect of increased CD200 expression by CLL cells on the immune system was evaluated in mixed lymphocyte reactions. Addition of primary CLL but not normal B cells to macrophages and T cells downregulated the Th1 response, as seen by a 50–95% reduction in secreted IL-2 and IFN-γ. Antibodies to CD200 prevented downregulation of the Th1 response in most B cell CLL samples evaluated, indicating abrogation of the CD200/CD200R interaction can be sufficient to restore the Th1 response. A disease-progression-associated shift of the immune response from Th1 to Th2 has been observed in numerous cancers. Because this cytokine shift is also believed to promote the induction of regulatory T cells, reverting the immune response to Th1 through direct targeting of the cancer cells may provide therapeutic benefits in CLL by encouraging a cytotoxic T cell response.
Keywords: CD200, chronic lymphocytic leukemia, immune evasion, immunotherapy
Chronic lymphocyte leukemia (CLL), the most common leukemia in Western countries, with ≈7,000 new cases and 4,500 deaths per year in the U.S. (1), is an incurable chronic disease caused by the clonal expansion of mature B lymphocytes with the immunophenotype CD5+CD19+CD23+sIglowCD79b–/low. CLL is a difficult disease to treat with chemotherapeutics, because apoptotic control in B lymphocytes is deregulated (2), and most cells are growth-arrested in early G1 phase with small compartments of proliferating cells present in microenvironments of the lymph nodes, spleen, and bone marrow.
Patients with CLL can be subdivided into a population with stable disease (median survival of 26 years) and a population progressing to aggressive disease (median survival of 8 years) (3, 4). Risk stratification is based on tests examining CD38, p53, zeta associated protein-70 (ZAP-70), variable heavy (VH) mutational status, and FISH (5) profiles.
Despite advances in the ability to diagnose high-risk patients earlier in the disease, CLL remains incurable. Chemotherapeutic approaches have largely failed to result in major advances, but with the development of therapeutic antibodies, a novel form of targeted cancer therapy has emerged either as monotherapy or in combination with chemotherapy. Hematopoietic malignancies are generally considered to be particularly amenable to antibody therapy based on relatively good accessibility of the antibody to target cells. However, despite the success of Rituximab (Genentech, South San Francisco, CA) (anti-CD20) treatment in a number of lymphomas and leukemias, clinical trials in CLL with single-agent Rituximab have demonstrated only partial responses in the majority of patients, with little to no effect on long-term survival. Ongoing clinical studies are evaluating the use of Rituximab in combination chemoimmunotherapy treatment regimens (6). Alemtuzumab (Berlex Laboratories, Richmond, CA) (anti-CD52) was the first approved antibody treatment of CLL, but infusion reactions and opportunistic infections limit its use (7).
We used antibody phage-display technology to identify cell-surface proteins that are up-regulated on CLL cells. Such proteins may be useful as targets for monoclonal antibody therapy or as diagnostic markers for CLL. Our results show that the immunosuppressive protein CD200 is up-regulated on B cells from all patients with CLL. CD200 is a type 1a membrane protein, related to the B7 family of costimulatory receptors, with two extracellular IgSF domains, a single transmembrane region, and a short cytoplasmic tail with no known signaling motifs. It is normally expressed on thymocytes, T and B lymphocytes, some dendritic cells, neurons, and endothelial cells. CD200 binds to its receptor, CD200R, expressed on cells of the monocyte/macrophage lineage; on T lymphocytes; and on monocyte-derived dendritic cells. Knockout of the CD200 gene in mice and studies with blocking antibodies and recombinant Fc fusion proteins containing the CD200 or CD200R extracellular domains have shown that CD200 is a potent immunosuppressant (8, 9).
In this report, we demonstrate that up-regulation of CD200 on CLL cells is sufficient to down-regulate a Th1 immune response, including cytokines such as IL-2 and IFN-γ. Th1 cytokines are required for efficient cytotoxic T cell function. Because progression of many cancer types has been correlated with a shift from Th1 to Th2 cytokines (10–14), strategies to reverse this shift are believed to be therapeutically beneficial. Our data indicate that up-regulation of CD200 may be a mechanism used by CLL tumors to evade eradication by the immune system. We demonstrate that blocking CD200 on B cell CLL (B-CLL) cells by monoclonal antibody restores a Th1 cytokine profile, suggesting that blocking the interaction of CD200 with its receptor might be therapeutically useful. CD200 has also been implicated in the induction of regulatory T cells, which are thought to hamper tumor-specific effector T cell immunity (15–17). Blocking the induction of regulatory T cells in CLL by anti-CD200 therapy might reinstate potent tumor-associated antigen-specific T cell production. The combination of direct targeting of the cancer cells, as well as driving a Th1 response against the tumor through a single anti-CD200 treatment regimen, has the potential to be a unique and very efficient therapy for CLL with long-lasting effects.
Results
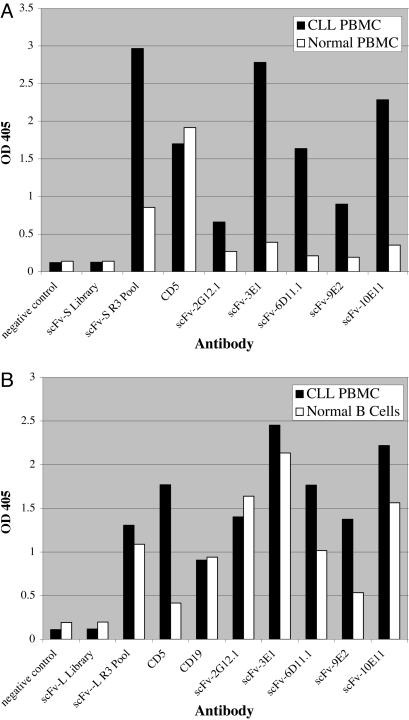
Antibody Selection by Cell Panning. To identify cell-surface antigens commonly associated with CLL, rabbits were immunized with a pool of primary CLL cells from 10 patients. Two single-chain variable region fragment (scFv) phage antibody libraries, each containing >109 independent clones, were constructed. The libraries were enriched for CLL-specific cell surface antigens by multiple rounds of negative selection on the human erythroleukemia cell line TF-1 and positive selection on primary CD19+ B-CLL cells. Analysis of pools of clones obtained after three rounds of positive-negative selection showed ≈200-fold enrichment, indicating that we successfully selected clones recognizing CLL cells (see Table 2, which is published as supporting information on the PNAS web site). Enrichment of the selected pools for scFv antibodies that bind to primary CLL cells was confirmed by flow cytometry (data not shown) and cell ELISA (Fig. 1). The percentage of CLL-positive antibodies in the round three pools was ≈64% and 34% for the two libraries (see Table 2). To identify antibodies recognizing targets that are up-regulated on CLL cells, CLL-positive clones were rescreened by cell ELISA for binding to CLL compared with normal peripheral blood mononuclear cell (PBMC), normal B cells, and three human leukemia cell lines: Ramos (Burkitt's lymphoma), RL (non-Hodgkin's lymphoma), and TF-1 (erythroleukemia). Results from five of six clones chosen for further analysis based on differential binding to CLL cells are shown in Fig. 1 (data for clone 4E5 not shown). Clones 2G12.1, 3E1, 4E5, 6D11.1, 9E2, and 10E11 all bound strongly to PBMC isolated from CLL donors and had minimal binding to PBMC from healthy donors (Fig. 1 A). Five of these clones, 3E1, 4E5, 6D11.1, 9E2, and 10E11, also consistently exhibited significantly higher binding to PBMC isolated from CLL donors relative to normal primary B cells (Fig. 1B) but little or no binding to the human B cell lines Ramos and RL (data not shown). In contrast, clone 2G12.1 consistently showed slightly higher binding to normal primary B cells and B cell lines than to CLL cells (Fig. 1B). None of the six clones bound to TF-1 cells.
Fig. 1.
Identification of scFv antibodies that bind preferentially to CLL cells by cell ELISA. Microtiter plates were coated with 105 PBMC from patients diagnosed with CLL, PBMC from a normal donor, or purified normal human B lymphocytes. The cells were incubated with bacterial supernatants containing HA-tagged scFv antibodies, and unbound antibodies were removed by washing with PBS. Bound scFv antibodies were detected with mouse anti-HA IgG, AMDEX alkaline phosphatase-conjugated sheep anti-mouse IgG, and PNPP p-Nutrophenyl phosphate substrate. (A) Cell ELISA comparing binding of selected scFv antibodies with CLL PBMC and normal PBMC. Negative control, cells incubated with bacterial supernatant not containing any scFv; ScFv-S library, supernatant from bacteria infected with the entire phage scFv-S library; ScFv-S R3 Pool, supernatant from bacteria infected with the entire pool of scFv-S phage after three rounds of selection by cell panning; CD5, mouse anti-human CD5 IgG. (B) Cell ELISA comparing binding of scFv antibodies with CLL PBMC and purified normal B lymphocytes. ScFv-L Library, supernatant from bacteria infected with the entire phage scFv-L library; ScFv-L R3 Pool, supernatant from bacteria infected with the entire pool of scFv-L phage after three rounds of selection by cell panning; CD19, mouse anti-human CD19 IgG.
Antigen Identification. Antigens recognized by those scFv clones that appeared to bind to epitopes up-regulated on CLL cells relative to normal B cells were identified by immunoprecipitation (IP) from CLL-AAT cells, a stable B cell line derived from a patient with CLL (described in Supporting Text, which is published as supporting information on the PNAS web site) and reverse-phase HPLC nanoelectrospray tandem MS. Representative IP results are shown in Fig. 4, which is published as supporting information on the PNAS web site. Clone 2G12.1 bound to the pan B cell marker CD19. Of the five clones that recognized epitopes up-regulated on CLL, 3E1 and 6D11.1 bound to CD23, a well known marker for CLL; and 9E2, 10E11, and 4E5 bound to CD200. Specificity of these three clones for CD200 was confirmed by testing the ability of the scFvs to bind specifically to CD200-transfected 293-EBNA cells by flow cytometry (Fig. 5A, which is published as supporting information on the PNAS web site). On normal lymphocytes, all three antibodies bound to B cells (Fig. 5Bd) and to a subset of CD4+ T cells (Fig. 5Bb and data not shown), a pattern consistent with that previously reported for CD200 (18).
To examine the up-regulation of CD200 in CLL in more detail, as well as correlation with other cell surface markers, CD200 levels on peripheral blood lymphocytes from CLL donors and normal donors were compared by flow cytometry (Table 1 and Fig. 5C). The geometric mean fluorescence intensity of anti-CD200 mAb staining was consistently higher on B-CLL lymphocytes (CD19+CD5+) relative to normal B lymphocytes (CD19+) with a mean B-CLL/normal B ratio of 3.3 ± 1.1 (n = 16; Table 1). In this study, the extent of CD200 up-regulation did not correlate with the prognostic marker CD38 (Table 1). A separate larger study was carried out to evaluate whether the extent of CD200 up-regulation correlated with ZAP-70 status. This study encompassed 23 additional patients in one center and 48 additional patients in another center (Fig. 6, which is published as supporting information on the PNAS web site). CD200 levels were again consistently up-regulated in B-CLL cells relative to normal B cells, but no correlation was found between the extent of CD200 up-regulation and ZAP-70. To evaluate a potential therapeutic effect of targeting CD200 for CLL, we isolated a panel of chimeric murine anti-human CD200 antibodies containing murine V regions and human constant regions and evaluated these for their ability to bind to human CD200 and compete with scFv-9E2 (unpublished results). One antibody, d1B5, was characterized in more detail for antagonist properties, as described below.
Table 1. FACS analysis of CD200 expression on B-CLL cells in comparison with normal B cells.
| CLL donor
|
Normal donor
|
|||
|---|---|---|---|---|
| Donor ID | B-CLL CD200 (GMFI) | % CD38+ | Normal B CD200 (GMFI) | GMFI ratio (CLL/normal) |
| 011731 | 93 | 27 | 58 | 1.6 |
| 020934 | 659 | 39 | 185 | 3.6 |
| 073031* | 334, 409 | 1.5, 0 | 64, 98 | 5.2, 4.2 |
| 011846* | 156, 190 | 62, 42 | 64, 55 | 2.4, 3.5 |
| 101735 | 420 | 0 | 95 | 4.4 |
| 6988172 | 290 | 67 | 97 | 2.9 |
| 8074020 | 403 | 10 | 97 | 4.2 |
| 8267677 | 300 | 18 | 97 | 3.1 |
| 040439 | 123 | 0 | 41 | 3.0 |
| 700036926 | 145 | 0 | 27 | 5.4 |
| 1325248 | 178 | 4.3 | 77 ± 7† | 2.3 |
| 7029373 | 154 | 14.5 | 77 ± 7† | 2.0 |
| 8942820 | 146 | 26.6 | 77 ± 7† | 1.9 |
| 8451869 | 237 | 3.2 | 77 ± 7† | 3.1 |
| 4539931 | 215 | 1.7 | 77 ± 7† | 2.8 |
| 6787771* | 305, 273 | 2.7, 2.4 | 77 ± 7†, 55 | 4.0, 5.0 |
| Mean = 3.3 | ||||
| SD = 1.1 | ||||
PBMC were isolated from CLL patients or from healthy donors, and CD200 protein expression on the B lymphocyte population was measured as described in Fig. 3C. The geometric mean fluorescent intensity (GMFI) of CD200 staining was used as a measure of CD200 expression and the ratio of GMFIs as a measure of CD200 up-regulation on CLL relative to the normal B cell control. The percentage of CD38+ B cells in the CLL samples was determined by flow cytometry by using CD38-APC, CD19-PE, and CD5-FITC.
Two values are shown for these CLL donors, because they were analyzed in two independent experiments.
Mean ± SD of GMFIs from six normal B cell donors analyzed in the same experiment.
Validation of Potential Antagonistic Properties of the Anti-CD200 Antibody. To evaluate the antagonistic properties of the chimeric antibody, disruption of the interaction of CD200-coated fluorescent beads with CD200 receptor-transfected cells was assessed. The presence of the chimeric antibody at 20 μg/ml completely blocked the interaction of CD200 with its receptor (data not shown). Because it has been demonstrated in murine systems that the presence of CD200 in mixed lymphocyte reactions results in a shift from secretion of Th1 cytokines such as IL-2 and IFN-γ to Th2 cytokines such as IL-4 and IL-10 (19), we evaluated whether these results could be extended to the human setting. As shown in Fig. 2 (IL-2) and Fig. 7, which is published as supporting information on the PNAS web site (IFN-γ), cytokine analysis of culture supernatants from mixed lymphocyte reactions in the presence of CD200-transfected cells, but not untransfected cells, indeed showed lower Th1 cytokine levels. IFN-γ and IL-2 production dropped to below detection in the presence of CD200. IL-4 levels were below our limit of detection of 70 pg/ml. Addition of 20 μg/ml anti-CD200 restored Th1 cytokine profiles, indicating that the anti-CD200 has antagonistic properties.
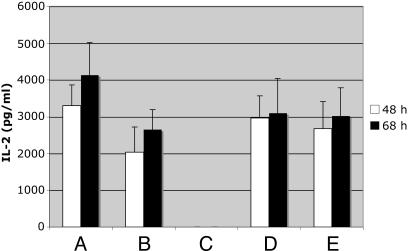
Fig. 2.
Effect of anti-CD200 on IL-2 production in mixed lymphocyte reactions in the presence of CD200 overexpressing cells. Mixed lymphocyte reactions were set up by using 200,000 dendritic cells/macrophages matured as described under Materials and Methods and 1 × 106 T cell-enriched lymphocytes. Two hundred thousand CD200 transfected or untransfected Epstein–Barr nuclear antigen (293-EBNA) cells were added to the dendritic cells in the presence or absence of 20 μg/ml anti-CD200 antibody or 2–4 h before lymphocyte addition. Supernatants were collected after 48 and 68 h and analyzed for the presence of IL-2. (A) Dendritic cells + T cells; (B) dendritic cells + T cells + untransfected EBNA cells; (C) dendritic cells + T cells + CD200-transfected EBNA cells; (D) dendritic cells + T cells + untransfected EBNA cells + anti-CD200; (E) dendritic cells + T cells + CD200-transfected EBNA cells+ anti-CD200. All samples represent the average of duplicates from two experiments. The experiment was repeated four times with similar results. ***, P < 0.0001 as determined by Student's unpaired t test.
Cytokine Profiles of Mixed Lymphocyte Reactions in the Presence of B-CLL Cells. To extend our results to the primary CLL setting, B cells from either healthy donors or CLL patients were added to mixed lymphocyte reactions to determine whether their presence would prevent Th1 cytokine production. Data in Fig. 3 (IL-2) and Fig. 8, which is published as supporting information on the PNAS web site (IFN-γ) demonstrate that the presence of B-CLL cells resulted in substantially lower levels of IL-2 and IFN-γ. Reduction varied between 2- and 20-fold. Addition of cells from donor 1325248 and 4539931 also resulted in production of IL-10 (data not shown). All samples overexpressing CD200 reduced Th1 cytokine levels, whereas B cells from healthy donors did not. Our CD200 antibody (d1B5) prevented that shift in four of five patient samples. In contrast, a control antibody (ALXN-4100) that does not react with CLL cells did not affect the cytokine profile. Because the sample that was not affected by the antibody has equal or higher CD200 expression compared with the other samples, the lack of response to antibody treatment might be due to other factors intrinsic to the CLL cells themselves and contributing to a Th2 response. Alternatively, extrinsic factors that cannot be ruled out include the presence of contaminating regulatory T cells in the B-CLL preparations, which could also alter the cytokine profile.
Fig. 3.
Effect of CLL cells on IL-2 production in mixed lymphocyte reactions Mixed lymphocyte reactions were set up in the presence of either PBMCs from healthy donors or PBMCs from CLL patients. Twenty μg/ml anti-CD200 or ALXN-4100 (nonbinding control antibody) were added to samples, as indicated. All samples represent the average of triplicates of one experiment. The experiment was repeated seven times.
Discussion
We identified potential therapeutic antibodies for CLL by selecting antibodies to tumor-associated proteins from immune repertoire libraries. Previous studies have demonstrated the feasibility of this approach (20–22). Using positive selection of antibody libraries on primary CLL cells and negative selection on an erythroleukemia cell line, we enriched for antibodies to CD19, CD23, and CD200.
We find CD200 protein to be up-regulated 1.6- to 5.4-fold on CLL cells in all disease patients compared with CD200 expression on B cells from healthy volunteers. CD200 appears to be up-regulated early in the disease process of CLL, because we find it to be up-regulated in patients irregardless of their ZAP-70 or variable heavy mutational status, and the degree of upregulation does not correlate with those disease markers. However, continuous elevated expression of CD200 in all CLL patients suggests an important role of this molecule in the disease.
Up-regulation of CD200 appears to be another example where tumors have proven adept at concealing their identity from the immune system. CD200 has a demonstrated potent role in suppressing immune responses. Soluble CD200-Fc can prolong allograft and xenograft survival in mice and can suppress autoimmune responses in mouse models (8, 23). Additionally, human herpesvirus and myxoma virus each express CD200-related proteins that result in down-regulation of the human immune response (24). Further, Gorczynski et al. (25) demonstrated in a mouse model that infusion of CD200-Fc suppressed protection of tumor growth by the immune system. Our data suggest that CLL cells themselves have found a direct mechanism to evade eradication by the immune system through up-regulation of CD200. Our data further suggest that even a 2-fold up-regulation is biologically relevant. Interaction of CD200 as a cell-bound ligand with its receptor on macrophages has been shown to perturb immune functioning and shift immune responses from a Th1 toward a Th2 cytokine profile. We demonstrate that primary PBMCs from CLL patients down-regulate Th1 cytokine production in mixed lymphocyte reactions in vitro, whereas B cells from healthy donors do not. The presence of our anti-CD200 antibody restores the profile seen in the absence of CD200, indicating that we have selected an antagonistic antibody that blocks a key mediator in these immunomodulation reactions.
A shift from Th1 to Th2 cytokine production has been observed during the progression of many cancer types, including various lymphomas, renal carcinoma, prostate carcinoma, cervical carcinoma, lung carcinoma, and melanoma, and has been associated with a negative prognosis (10–14). Podhorecka et al. (26) showed that a Th1 cytokine response shifts to a Th2 cytokine response during the progression of CLL. Our observation that CLL cells alter the cytokine profile in mixed lymphocyte reactions suggests that CLL cells themselves can directly affect the cytokine profile of surrounding cells. A shift to Th2 might prevent the support of cytotoxic T cells, thought to be the most important effector cells to eradicate cancer cells, and might favor the production of regulatory T cells. Also, type 2 cytokines have been reported to down-modulate tumor-specific immune responses by reducing tumor-associated MHC expression and by inhibiting tumor antigen presentation by antigen-presenting cells. Furthermore, IL-4 has been shown to prevent apoptosis of CLL cells (27). Although cytokine imbalance is a well known phenomenon in cancer, the underlying mechanisms are not well defined. Certain tumors, including CLL (28), have the capability of producing soluble factors such as IL-10 and IL-6, thereby inhibiting a broad array of immune functions, including Th1 cytokine production. Here, we demonstrate that increased cell surface expression by tumor cells of the costimulatory molecule CD200 is sufficient to prevent a Th1 response, thereby altering the cytokine milieu surrounding the tumor cells.
For therapeutic benefit, targeting CD200 to skew the immune system toward a Th1 response would similarly require that the immune system mounts an anti-CLL response. Autologous cytotoxic T cells capable of killing CLL cells have been generated in vitro by expansion over several weeks, indicating that they are present at some level in patients (29, 30). However, it has been difficult to initiate autologous T cell responses against unstimulated CLL cells, and CLL cells are ineffective stimulator cells in autologous and allogeneic mixed lymphocyte cultures (31). The lack of response has, in part, been explained by low levels of expression of costimulatory molecules on CLL cells, rendering them inefficient antigen-presenting cells (31). Recent data suggest that good-prognosis cytogenetics in CLL is associated with enhanced immunogenicity in vitro (32). Our results point to the importance of the capability of CLL cells to affect the most potent antigen-presenting cells in the body, dendritic cells. By stimulating CD200R on dendritic cells/macrophages, the immune system is driven to a Th2 response and potentially to the production of regulatory T cells. Our observations are consistent with the finding that dendritic cells in patients with CLL are defective and unable to stimulate an effective T cell response (33).
Shifting the cytokine balance has shown some success in preclinical models and patients. It has been demonstrated that arabinomannan, an agent shifting Th2 to Th1 response, suppresses tumor metastasis in a fibrosarcoma model (34), and its effectiveness is currently being tested in patients with head and neck cancer. Cyclophosphamide had a similar effect in a rat metastatic lymphoma model (35). Furthermore, administration of IFN-α in CML patients resulting in remission restored the Th1 cytokine balance (36).
In recent years, it has become clear that the presence of regulatory T cells provides a major obstacle for efficient elimination of tumor cells by the immune system (37). Interestingly, interaction of CD200 with its receptor on macrophages turns on an immunosuppressive pathway, resulting in the induction of regulatory T cells (15–17). Furthermore, Beyer et al. (38) demonstrated higher frequencies of regulatory T cells in CLL patients. Because expression of CD200 on CLL cells most likely contributes to higher regulatory T cell frequencies, anti-CD200 therapy might be very beneficial in preventing regulatory T cell induction, as well as in combination with ONTAK or Fludarabine therapy to eliminate preexisting regulatory T cells.
In future studies, we may evaluate the benefits of blocking immune suppression in CLL by targeting human leukemic cells directly through CD200. In particular, we may address in animal models whether stimulating the immune system with anti-CD200 will allow the eradication of CLL cells from the spleen and lymph nodes through CLL reactive T cells that may have better access to these organs than antibodies. Another crucial question that remains is whether targeting CD200 on CLL cells and blocking its interaction with CD200R will lead to the lysis of tumor cells. The potential effect of direct CLL cell killing through antibody-dependent cellular cytotoxicity activity or other signaling events triggered by anti-CD200 may be explored in future studies. The combination of direct cell killing, driving the immune response toward a Th1 profile and thereby preventing the induction of regulatory T cells, and enhancing immune function may be a particularly powerful approach that could potentially be achieved by treatment with anti-CD200 alone or in combination with other B cell cytotoxins, cancer vaccines, or other immunostimulatory therapies.
Materials and Methods
Patients and Sample Collection. After obtaining Internal Review Board approval and informed consent, blood was drawn from patients diagnosed with CLL at Scripps Clinic (La Jolla, CA), as well as from healthy donors at Alexion Antibody Technologies, San Diego. PBMC were isolated by Ficoll–Paque gradient centrifugation (Histopaque, Sigma-Aldrich) and stored frozen.
Cell Immunizations and scFv Antibody Library Construction. Two rabbits were each immunized three times with 2 × 107 PBMC pooled from five different CLL donors (10 donors total). Five days after the final immunization, spleen, bone marrow, and PBMC were harvested. Total RNA was isolated by using Tri-Reagent (Molecular Research Center, Cincinnati). scFv antibody phage display libraries were constructed as described (39).
Antibody Selection by Cell Surface Panning. Libraries were enriched for CLL cell-surface-specific antibodies by positive-negative selection with a magnetically activated cell sorter, as described (40), with some modifications. Primary CLL cells (107) pooled from several donors were labeled with anti-human CD19 conjugated to microbeads (Miltenyi Biotec, Auburn, CA). The labeled cells were mixed with a 10-fold excess of absorber cells (the human erythroleukemia cell line TF-1), pelleted, and resuspended in 50 μl (1010-1011 plaque-forming units) of phage particles for 30 m at 25°C. CLL cells and bound phage were captured by using a MiniMACS MS+ separation column (Miltenyi Biotec). Phage particles were eluted and amplified in Escherichia coli for the next round of panning, as described (40). For each round of panning, the input and output phage titers were determined. An enrichment factor (E) was calculated by using the formula E = (Rn output/Rn input)/(R1 output/R1 input).
Antibody Production and Purification. Pools of hemagglutinin (HA)-tagged scFv antibodies were produced in E. coli strain TOP10F′ and used as culture supernatants. Periplasmic fractions from individual clones expressing scFv antibodies were prepared as described (39). scFv antibodies were affinity-purified by using rat anti-HA affinity columns (Roche Molecular Biochemicals).
Flow Cytometry. Binding of HA-tagged scFv antibodies to cells was assayed by flow cytometry by using rat anti-HA (clone 3F10, Roche Molecular Biochemicals) and phycoerythrin (PE)-conjugated anti-rat IgG. Alternatively, biotinylated rat anti-HA was used as secondary antibody, and PE-conjugated streptavidin was used for detection. Fluorochrome-conjugated antibodies against CD5, CD19, and CD38 were obtained from Becton Dickinson. For the evaluation of CD200 expression on a large panel of CLL patients, PE-conjugated anti-CD200 antibody (clone OX-104, Serotec) was used.
Cell ELISA. ELISA plates were coated with ConA (10 mg/ml in 0.1 M NaHCO3, pH 8.6/0.1 mM CaCl2). Cells (105) in PBS were added per well, and the plates were centrifuged at 250 × g for 10 min. Cells were fixed by overnight incubation with 0.02% glutaraldehyde in PBS. After two PBS washes, nonspecific binding sites were blocked with 4% nonfat dry milk in PBS for 3 h, followed by a 2-h incubation with 50 μl of HA-tagged scFv antibody at room temperature. After six PBS washes, bound scFv antibodies were detected by using an anti-HA antibody (clone 12CA5) and AMDEX alkaline phosphatase (AP)-conjugated anti-mouse IgG (Amersham Pharmacia Biotech). AP-AMDEX antibody was detected with PNPP substrate and measured at 405 nm using a microplate reader.
Maturation of Dendritic Cells from Blood Monocytes. Buffy coats were obtained from the San Diego Blood Bank and primary blood lymphocytes were isolated by using Ficoll (StemCell Technologies, Vancouver). Cells were adhered for 1 h in Eagle's minimal essential medium containing 2% human serum followed by vigorous washing with PBS. Cells were cultured for 5 d in the presence of 800 units/ml human recombinant granulocyte–macrophage colony-stimulating factor/500 units/ml human recombinant IL-4 (StemCell Technologies)/100 μg/ml human recombinant IFN-γ (R & D Systems). Matured cells were harvested and irradiated at 2,000 rad by using a γ-irradiator (University of California, San Diego).
Mixed-Lymphocyte Reaction. Mixed-lymphocyte reactions were set up in 24-well plates by using 200,000 dendritic cells and 1 × 106 responder cells. Responder cells were T cell-enriched lymphocytes purified from peripheral blood by Ficoll separation. T cells were enriched by incubating the cells for 1 h in tissue culture flasks and taking the nonadherent cell fraction. Two hundred thousand CD200-transfected 293-EBNA cells or CLL cells were added to the dendritic cells in the presence or absence of 20 μg/ml anti-CD200 antibody 2–4 h before lymphocyte addition. Supernatants were collected after 48 and 68 h and analyzed for the presence of cytokines.
Cytokine Analysis. Cytokines such as IL-2, IFN-γ, IL-4, IL-10, and IL-6 found in the tissue culture supernatant were quantified by using ELISA. Matched capture and detection antibody pairs for each cytokine were obtained from R & D Systems, and a standard curve for each cytokine was produced by using recombinant human cytokine. Anticytokine capture antibody was coated on the plate in PBS at the optimum concentration. After overnight incubation, the plates were washed and blocked for 1 h with PBS containing 1% BSA and 5% sucrose. After third washes with PBS containing 0.05% Tween, supernatants were added at the indicated dilutions in PBS containing 1% BSA. Captured cytokines were detected with the appropriate biotinylated anticytokine antibody followed by the addition of alkaline phosphatase-conjugated streptavidin and SigmaS substrate. Color development was assessed with an ELISA plate reader (Molecular Devices).
Supporting Information. All methods for isolation of the CLL-AAT cell line, immunoprecipitation, mass spectrometry, cloning and expression of CD200, conversion of scFv-9E2 to rabbit-human chimeric IgG, and ZAP-70 analysis are detailed in Supporting Text, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Christopher Hinkel and Peter Calveley for antibody production and purification. We are grateful to Dr. Martha Wild for critical reviewing of the manuscript. We thank Genoptix (San Diego) for expert ZAP-70 correlation studies.
Conflict of interest statement: Communicating member R.A.L. is a member of the Scientific Advisory Board of Alexion Antibody Technologies. A.S. has declared a financial interest in a company whose potential product was studied in the present work; and J.R.M., A.K.-R., T.M., E.P.R., F.Q., and K.S.B. are employed by a company whose potential product was studied in the present work.
Abbreviations: CLL, chronic lymphocyte leukemia; B-CLL, B cell CLL; PBMC, peripheral blood mononuclear cell; scFv, single-chain variable region fragment; HA, hemagglutinin.
References
- 1.Jemal, A., Thomas, A., Murray, T. & Thun, M. (2002) CA Cancer J. Clin. 52, 23–47. [DOI] [PubMed] [Google Scholar]
- 2.Jewell, A. P. (2002) Br. J. Biomed. Sci. 59, 235–238. [DOI] [PubMed] [Google Scholar]
- 3.Kay, N. E., Hamblin, T. J., Jelinek, D. F., Dewald, G. W., Byrd, J. C., Farag, S., Lucas, M. & Lin, T. (2002) Hematology, 193–213. [DOI] [PubMed]
- 4.Stevenson, F. K. & Caligaris-Cappio, F. (2004) Blood 103, 4389–4395. [DOI] [PubMed] [Google Scholar]
- 5.Shanafelt, T. D., Geyer, S. M. & Kay, N. E. (2004) Blood 103, 1202–1210. [DOI] [PubMed] [Google Scholar]
- 6.Byrd, J. C., Stilgenbauer, S. & Flinn, I. W. (2004) Hematology, 163–183. [DOI] [PubMed]
- 7.Robak, T. (2004) Leuk. Lymphoma 45, 205–219. [DOI] [PubMed] [Google Scholar]
- 8.Gorczynski, R. M., Cattral, M. S., Chen, Z., Hu, J., Lei, J., Min, W. P., Yu, G. & Ni, J. (1999) J. Immunol. 163, 1654–1660. [PubMed] [Google Scholar]
- 9.Hoek, R. M., Ruuls, S. R., Murphy, C. A., Wright, G. J., Goddard, R., Zurawski, S. M., Blom, B., Homola, M. E., Streit, W. J., Brown, M. H., et al. (2000) Science 290, 1768–1771. [DOI] [PubMed] [Google Scholar]
- 10.Tendler, C. L., Burton, J. D., Jaffe, J., Danielpour, D., Charley, M., McCoy, J. P., Pittelkow, M. R. & Waldmann, T. A. (1994) Cancer Res. 54, 4430–4435. [PubMed] [Google Scholar]
- 11.Takeuchi, T., Ueki, T., Sasaki, Y., Kajiwara, T., Li, B., Moriyama, N. & Kawabe, K. (1997) Cancer Immunol. Immunother. 43, 375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filella, X., Alcover, J., Zarco, M. A., Beardo, P., Molina, R. & Ballesta, A. M. (2000) Prostate 44, 271–274. [DOI] [PubMed] [Google Scholar]
- 13.Lauerova, L., Dusek, L., Simickova, M., Kocak, I., Vagundova, M., Zaloudik, J. & Kovarik, J. (2002) Neoplasma 49, 159–166. [PubMed] [Google Scholar]
- 14.Tabata, T., Hazama, S., Yoshino, S. & Oka, M. (1999) Am. J. Surg. 177, 203–208. [DOI] [PubMed] [Google Scholar]
- 15.Gorczynski, R. M., Chen, Z., Kai, Y., Wong, S. & Lee, L. (2004) Transplantation 77, 1138–1144. [DOI] [PubMed] [Google Scholar]
- 16.Fallarino, F., Asselin-Paturel, C., Vacca, C., Bianchi, R., Gizzi, S., Fioretti, M. C., Trinchieri, G., Grohmann, U. & Puccetti, P. (2004) J. Immunol. 173, 3748–3754. [DOI] [PubMed] [Google Scholar]
- 17.Gorczynski, R. M., Lee, L. & Boudakov, I. (2005) Transplantation 79, 488–491. [PubMed] [Google Scholar]
- 18.Wright, G. J., Jones, M., Puklavec, M. J., Brown, M. H. & Barclay, A. N. (2001) Immunology 102, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorczynski, R. M. (2001) Eur. J. Immunol. 31, 2331–2337. [DOI] [PubMed] [Google Scholar]
- 20.Kupsch, J. M., Tidman, N. H., Kang, N. V., Truman, H., Hamilton, S., Patel, N., Newton Bishop, J. A., Leigh, I. M. & Crowe, J. S. (1999) Clin. Cancer Res. 5, 925–931. [PubMed] [Google Scholar]
- 21.Lee, K. J., Mao, S., Sun, C., Gao, C., Blixt, O., Arrues, S., Hom, L. G., Kaufmann, G. F., Hoffman, T. Z., Coyle, A. R., et al. (2002) J. Am. Chem. Soc. 124, 12439–12446. [DOI] [PubMed] [Google Scholar]
- 22.Liu, B., Conrad, F., Cooperberg, M. R., Kirpotin, D. B. & Marks, J. D. (2004) Cancer Res. 64, 704–710. [DOI] [PubMed] [Google Scholar]
- 23.Gorczynski, R. M., Chen, Z., Yu, K. & Hu, J. (2001) Clin. Immunol. 101, 328–334. [DOI] [PubMed] [Google Scholar]
- 24.Cameron, C. M., Barrett, J. W., Liu, L., Lucas, A. R. & McFadden, G. (2005) J. Virol. 79, 6052–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorczynski, R. M., Chen, Z., Hu, J., Kai, Y. & Lei, J. (2001) Clin. Exp. Immunol. 126, 220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Podhorecka, M., Dmoszynska, A., Rolinski, J. & Wasik, E. (2002) Leuk. Res. 26, 657–660. [DOI] [PubMed] [Google Scholar]
- 27.Kay, N. E., Han, L., Bone, N. & Williams, G. (2001) Br. J. Haematol. 112, 760–767. [DOI] [PubMed] [Google Scholar]
- 28.Fayad, L., Keating, M. J., Reuben, J. M., O'Brien, S., Lee, B. N., Lerner, S. & Kurzrock, R. (2001) Blood 97, 256–263. [DOI] [PubMed] [Google Scholar]
- 29.Trojan, A., Schultze, J. L., Witzens, M., Vonderheide, R. H., Ladetto, M., Donovan, J. W. & Gribben, J. G. (2000) Nat. Med. 6, 667–672. [DOI] [PubMed] [Google Scholar]
- 30.Harig, S., Witzens, M., Krackhardt, A. M., Trojan, A., Barrett, P., Broderick, R., Zauls, A. J. & Gribben, J. G. (2001) Blood 98, 2999–3005. [DOI] [PubMed] [Google Scholar]
- 31.Buhmann, R., Nolte, A., Westhaus, D., Emmerich, B. & Hallek, M. (1999) Blood 93, 1992–2002. [PubMed] [Google Scholar]
- 32.Jahrsdorfer, B., Wooldridge, J. E., Blackwell, S. E., Taylor, C. M., Link, B. K. & Weiner, G. J. (2005) Leukemia 19, 759–766. [DOI] [PubMed] [Google Scholar]
- 33.Orsini, E., Guarini, A., Chiaretti, S., Mauro, F. R. & Foa, R. (2003) Cancer Res. 63, 4497–4506. [PubMed] [Google Scholar]
- 34.Oka, H., Shiraishi, Y., Sasaki, H., Yoshinaga, K., Emori, Y. & Takei, M. (2003) Biol. Pharm. Bull. 26, 1336–1341. [DOI] [PubMed] [Google Scholar]
- 35.Matar, P., Rozados, V. R., Gervasoni, S. I. & Scharovsky, G. O. (2002) Cancer Immunol. Immunother. 50, 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reuben, J. M., Lee, B. N., Johnson, H., Fritsche, H., Kantarjian, H. M. & Talpaz, M. (2000) Clin. Cancer Res. 6, 1671–1677. [PubMed] [Google Scholar]
- 37.Curiel, T. J., Coukos, G., Zou, L., Alvarez, X., Cheng, P., Mottram, P., Evdemon-Hogan, M., Conejo-Garcia, J. R., Zhang, L., Burow, M., et al. (2004) Nat. Med. 10, 942–949. [DOI] [PubMed] [Google Scholar]
- 38.Beyer, M., Kochanek, M., Darabi, K., Popov, A., Jensen, M., Endl, E., Knolle, P. A., Thomas, R. K., von Bergwelt-Baildon, M., Debey, S., et al. (2005) Blood 106, 2018–2025. [DOI] [PubMed] [Google Scholar]
- 39.Barbas, C. F. (2001) Phage Display: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 40.Siegel, D. L., Chang, T. Y., Russell, S. L. & Bunya, V. Y. (1997) J. Immunol. Methods 206, 73–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.