Abstract
A comprehensive analysis of the TP53 gene and its protein status was carried out on a panel of 56 colorectal cancer cell lines. This analysis was based on a combination of denaturing HPLC mutation screening of all exons of the p53 gene, sequencing the cDNA, and assessing the function of the p53 protein by assaying the induced expression of phosphorylated p53 and p21 after exposing cells to γ-rays. In a few cases where there was no production of p53 message nor evidence of functional p53 protein, all of the p53 exons were sequenced directly. Thirteen of the 56 cell lines had functional p53, 21 lines had missense mutations (one of which made no detectable protein), 4 lines produced no p53 transcripts, and the remaining 18 lines carried truncating TP53 mutations. Thus, our results showed a relatively high frequency of TP53 mutations (76.8%) in our cell lines, with almost half of the mutations being truncating mutations. This is a rather higher frequency of such mutations than usually reported. Of the 18 cell lines with truncating mutations, 12 had detectable truncated protein based on Western blot analysis, whereas no protein was detected in the remaining 6 cell lines. Our data provide a valuable source of TP 53 mutations for further studies and raise the question of the extent to which truncating mutations may have dominant negative effects, even when no truncated protein can be detected by standard methods.
Keywords: DNA damage, p21, oncogenicity, γ-ray, tumorigenesis
The gene encoding p53 is found mutated in >50% of all types of human cancers. Its most important normal function is probably to direct cell cycle arrest at the G1 or G2 phase of the cell cycle after certain types of DNA damage and to induce apoptosis when the damage is too severe (1). The normal function of the p53 protein can be assessed by its response to DNA damage, for example, γ-irradiation. TP53 mutations in tumors are most probably primarily selected for, because they interfere with the apoptotic process. To date, >75% of the TP53 mutations reported in colorectal carcinomas (CRC) are missense mutations, which have been the focus of in vitro and in vivo studies. The in vitro studies clearly show that (i) many mutant p53 proteins can inhibit normal p53 function, (ii) the mutant proteins can acquire new abnormal functions, and (iii) different mutants vary in their oncogenicity (2). Recent in vivo studies using mice transgenic for two mutations, R270H and R172H, further support these findings (3, 4). These studies have also, however, raised further questions. What are the mechanisms that cause the variation in oncogenicity in different mutants? Could they involve interactions between p53 and its relatives, such as p63 and p73 (5)? In the two mouse studies, the p53 mutants examined were minimally present in normal tissues and became stabilized only in tumors. It was therefore suggested that a key p53 regulatory network must be altered for the mutant protein to be selected for during tumor evolution (6). Thus, the role of mutant p53 in the process of tumorigenesis may be much more complicated than previously thought, involving cell-type specificity and potential interactions with changes in other genes.
TP53 is estimated to be mutated in 40–50% of CRCs (http://www-p53.iarc.fr/index.html and http://p53.free.fr/). It is much more frequently mutated in high-grade dysplastic polyps, which are thought to mark the transition from adenoma to carcinoma, than in early adenomas. This finding implies that most TP53 mutations probably occur before metastasis (7, 8). The mechanism of how, or whether, p53 plays a role in the metastasis of CRC remains unknown. In an effort to address the above questions, we have carried out a thorough analysis of p53 status in a panel of 56 genetically well characterized CRC cell lines. The implications of the results and comparisons with published data are discussed.
Results
TP53 Mutation Detection. Primers located at least 50 bp away from the ends of each exon were designed to amplify exons 1–11 of the TP53 gene (Table 1). All amplicons had exactly the size expected, except in one case, namely exon 1 in cell line SW1222, where a smaller amplicon than expected was observed. Sequencing confirmed that the anomalous amplicon was due to a 113-bp homozygous deletion. All amplicons were subjected to denaturing HPLC (DHPLC) analysis on the WAVE machine (Transgenomic, Omaha, NE). Samples showing abnormal patterns were subsequently sequenced. However, all of the exon 3 and 4 amplicons were sequenced directly because they did not show clear-cut peaks in DHPLC analysis. Based on this analysis, mutations were found in 37 cell lines (Table 2).
Table 1. PCR amplification conditions for exons 1–11 of TP53 and their corresponding DHPLC analysis conditions.
| Exon | Primer sequences (5′-3′) | PCR product size, bp | PCR Tm, °C | DHPLC denaturating temperature, °C/gradient initial buffer B, % |
|---|---|---|---|---|
| 1 | cacagctctggcttgcaga | 442 | 66 | 60/58, 64/52 |
| agcgattttcccgagctga | ||||
| 2 | agctgtctcagacactggca | 317 | 64* | 65/47 |
| gagcagaaagtcagtcccatg | ||||
| 3-4 | agacctatggaaactgtgagtgga | 631 | 56* | N/A |
| gaagcctaagggtgaagagga | ||||
| 5-6 | cgctagtgggttgcagga | 550 | 64 | 61/57, 66/49 |
| cactgacaaccacccttaac | ||||
| 7 | ctgcttgccacaggtctc | 283 | 64 | 58/54, 66/46 |
| tggatgggtagtagtatggaag | ||||
| 8-9 | gttgggagtagatggagcct | 455 | 64 | 57/57, 63/50 |
| ggcattttgagtgttagactg | ||||
| 10 | ctcaggtactgtgtatatacttac | 351 | 59 | 57/55, 65/46 |
| atactacgtggaggcaagaat | ||||
| 11 | tcccgttgtcccagcctt | 476 | 58 | 56/58, 62/53 |
| taacccttaactgcaagaacat |
With the addition of 0.5 × Q (Qiagen PCR amplification kit).
Table 2. TP53 mutations identified in 43 CRC cell lines.
| Mutation type | Cell line | Location | Nucleotide substitution | Mutation effect | Mutation recorded in CRCs at IARC* | P53 protein detected |
|---|---|---|---|---|---|---|
| Point mutation | C10 | E7 codon 245† | G to A (GGC to AGC)‡ | Gly to Ser | Yes | + |
| C106 | E4 codon 125 | C to T (ACG to ATG) | Thr to Met | Yes | + | |
| C75 | E7 codon 249 | G to C (AGG to AGC) | Arg to Ser | Yes | + | |
| CaR-1 | E8 codon 272 | G to A (GTG to ATG) | Val to Met | Yes | + | |
| CC07 | E7 codon 245 | G to A (GGC to AGC) | Gly to Ser | Yes | + | |
| CCK-81 | E8 codon 278 | C to A (CCT to CAT) | Pro to His | Yes | + | |
| COLO320DM | E7 codon 248 | C to T (CGG to TGG) | Arg to Trp | Yes | + | |
| CX-1 | E8 codon 273 | G to A (CGT to CAT) | Arg to His | Yes | + | |
| DLD1 | E7 codon 241 | C to T (TCC to TTC) | Ser to Phe | Yes | + | |
| HRA19 | E8 codon 273 | G to A (CGT to CAT) | Arg to His | Yes | + | |
| HT55 | E6 codon 213 | G to T (CGA to CTA) | Arg to Leu | Yes | + | |
| LIM1863 | E7 codon 234 | T to C (TAC to CAC) | Tyr to His | Yes | + | |
| LS1034 | E7 codon 245 | G to A (GGC to AGC) | Gly to Ser | Yes | + | |
| LS123 | E5 codon 175 | G to A (CGC to CAC) | Arg to His | Yes | + | |
| NCl-H716 | E6 codon 224 | G to T (GAG to GAT) | Glu to Asp | Not, but in other cancers | - | |
| SNU-C2B | E8 codon 273 (hetero) | C to T (CGT to TGT) | Arg to Cys | Yes | + | |
| G to A (CGT to CAT) | Arg to His | Yes | + | |||
| SW1116 | E5 codon 159 | C to A (GCC to GAC) | Ala to Asp | Yes | + | |
| SW480 | E8 codon 273 | G to A (CGT to CAT) | Arg to His | Yes | + | |
| SW837 | E7 codon 248 | C to T (CGG to TGG) | Arg to Trp | Yes | + | |
| VACO10MS | E5 codon 175 | G to A (CGC to CAC) | Arg to His | Yes | + | |
| VACO5 | E8 codon 282 | C to T (CGG to TGG) | Arg to Trp | Yes | + | |
| C125-PM | E6 codon 196 | C to T (CGA to TGA) | Arg to Stop | Yes | + | |
| C80 | E4 codon 52 | C to T (CAA to TAA) | Gln to Stop | Not, but in other cancers | + | |
| C84 | E10 codon 342 | C to T (CGA to TGA) | Arg to Stop | Yes | + | |
| CACO2 | E6 codon 204 | G to T (GAG to TAG) | Glu to Stop | Yes | - | |
| CC20 | E5 codon 126 | C to G (TAC to TAG) | Tyr to Stop | Not, but in other cancers | + | |
| CoCM-1 | E6 codon 196 | C to T (CGA to TGA) | Arg to Stop | Yes | - | |
| LS411 | E5 codon 126 | C to A (TAC to TAA) | Tyr to Stop | Yes | - | |
| RCM-1 | E8 codon 306 | C to T (CGA to TGA) | Arg to Stop | Yes | + | |
| SW403 | E4 codon 51 | G to T (GAA to TAA) | Glu to Stop | Not, but in other cancers | - | |
| VACO429§ | E8 codon 306 (Hetero) | C to T (CGA to TGA) | Arg to Stop | Yes | + | |
| C70 | E8 3′ + GT to GG (1170 + 2T to G) | T to G | Frameshift | Not, but in other cancers | + | |
| PC/JW | E1 3′ + GT to 3′ + AT (223 + 1G to A) | G to A | Frameshift¶ | Not | - | |
| T84 | E6 5′ - AG to 5′ - AT (811 - G to T) | G to T | Frameshift¶ | Yes | - | |
| VACO 400 | E1 3′ + GT to 3′ + GC (223 + 2T to C) | T to C | Frameshift¶ | Not | - | |
| Insertion | COLO741 | E9 codon 321 | AA insertion | Frameshift | No | + |
| VACO4A | E7 codon 239 | A insertion | Frameshift | Yes | + | |
| COLO201§ | E4 codon 103 | T insertion | Frameshift | No | + | |
| Deletion | COLO201 | E4 codon 103 | 25 bp deletion | Frameshift | No | + |
| HCA46 | E8 codon 272 | 2 bp `TG' deletion | Frameshift | No | - | |
| HCA7 | E8 codon 300 (hetero) | 1 bp `C' deletion | Frameshift | Yes | + | |
| SW1222 | E1 114th of exon 1 | 113 bp deletion | Frameshift¶ | No | - | |
| SW1417 | E7 codon 238 | 14 bp deletion | Frameshift | Yes | + | |
| SW948 | E4 codon 117 | 1 bp `G' deletion | Frameshift | Yes | - | |
| VACO429 | E2 codon 58 (hetero) | 1 bp `T' deletion | Frameshift | No | + |
Data from the IARC TP53 mutation database.
E means exon in all cases.
The underlined base in each case is that which is changed.
Also included with the Deletion category.
RNA message was found in all cases except where indicated by this keynote.
For the 19 cell lines in which no mutations were detected by DHPLC analysis, RT-PCR was carried out to amplify the full length of the p53 ORF. The complete TP53 ORF could be amplified in 11 of these cell lines. All of these ORF amplicons were sequenced directly, and mutations were identified in two further cell lines, CCK-81 and SNU-C2B. The p53 mRNA expression in the 8 cell lines that had not yielded an amplified p53 ORF was subsequently tested by using primers designed to amplify small regions of the mRNA (see Expression of Mutant and Truncated p53).
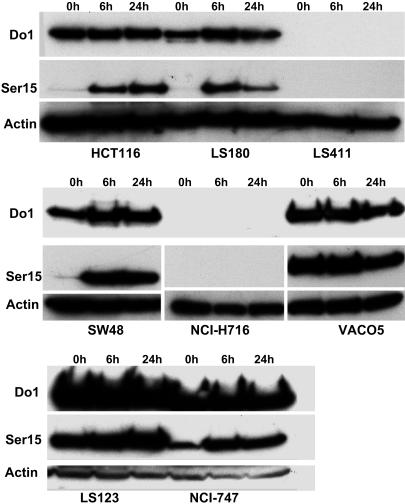
In the 17 cell lines in which no mutations had so far been identified, DNA damage experiments were carried out to ascertain whether they had functional p53 protein. Each of these cell lines in exponential growth phase was treated with 6 Gy γ-ray and harvested at 6 and 24 h after irradiation. The relative amounts of total p53 and Ser-15-phosphorylated p53 were assessed by Western blotting by using appropriate antibodies. Thirteen of these cell lines showed obviously increased expression of phosphorylated p53, and, in some cases total p53 (Fig. 1 and Table 3). Four cell lines, however, showed unexpected p53 expression patterns after treatment. Thus, in cell lines NCI-H716 and CoCM1, no total or phosphorylated p53 could be detected; in cell line RCM1, there was a smaller size p53 protein and no expression of phosphorylated p53; in cell line CC20, there was no difference between the expression of total p53 and phosphorylated p53 before or after DNA damage. To clarify the cause of the abnormal expression of p53, direct sequencing of exons 1–11 of these 4 cell lines was carried out. This sequencing revealed that each of them carried mutations in TP53 (Table 2).
Fig. 1.
P53 status in CRC cell lines after γ-radiation. Cell lines were irradiated with 6 Gy and harvested after 6 and 24 h. The corresponding cell extracts were then analyzed by Western blotting with the anti-p53 antibodies DO-1 and Ser15P as described in Materials and Methods. An increase in Ser15P-detected activity shows that P53 was stabilized in cell lines with WT p53: HCT116, LS180, SW48, and NCI-747. There was either no change in reactivity after irradiation, or lack of reactivity with either antibody in cell lines with mutant p53: LS411, NCI-H716, LS123, and VACO5. LS411 is a mutant line with truncated p53 that cannot be detected, NCI-H716 has a missense mutation whose mutant p53 protein product cannot be detected, and LS123 and VACO5 both have a mutant p53 that is stabilized without stimulation by DNA damage.
Table 3. Presence of total p53, phosphorylated p53 (Ser15P), and p21 in cell lines with WT TP53 before and after γ-irradiation.
| Cell line | Total p53 before γ-irradiation | Total p53 increased after γ-irradiation | Ser15P p53 increased after γ-irradiation | P21 increased after γ-irradiation |
|---|---|---|---|---|
| C32 | + | No | Yes | Yes |
| C99 | + | No | Yes | Yes |
| COLO678 | + | No | Yes | Yes |
| Gp2D | + | Yes | Yes | Yes |
| HCT116 | + | Yes | Yes | Yes |
| LOVO | + | Yes | Yes | Yes |
| LS180 | + | No | Yes | Yes |
| LS174T | + | No | Yes | Yes |
| LS513 | + | Yes | Yes | Yes |
| NCl-747 | + | No | Yes | No* |
| RKO | + | Yes | Yes | Yes |
| SKCO-1 | + | No | Yes | Yes |
| SW48 | + | Yes | Yes | Yes |
The data are based on western blot analysis using appropriate antibodies, as described in Materials and Methods.
This cell line produced no detectable p21.
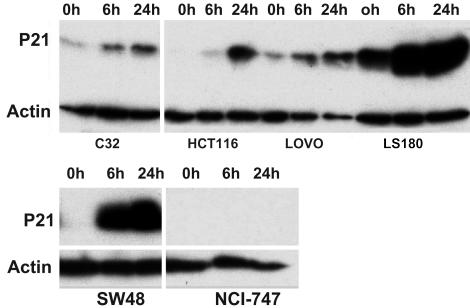
To further confirm that the phosphorylated p53 in the 13 cell lines from the above study could indeed have an effect on its expected target genes, the expression of p21 protein was analyzed in those cell lines. Twelve of the 13 cell lines showed the expected increase in p21 protein, whereas one cell line, NCI-747, did not show any expression of p21 either before or after DNA damage (Fig. 2 and Table 2). This finding may reflect mutation or deletion of the p21 gene in this cell line.
Fig. 2.
P21 expression detected by p21 antibody in CRC cell lines after DNA damage. Cells from each line were irradiated with a dose of 6 Gy γ-ray and harvested after 6 and 24 h. Cell extracts were then analyzed by Western blotting with an anti-p21 antibody as described in Materials and Methods. P21 expression was increased after radiation in cell lines HCT116, LS180, SW48, C32, and LOVO, but was not detected at all in NCI-747. None of these lines carried TP53 mutations.
Our results confirm that only one of the cell lines with mutant p53, HCA7, also has WT p53 (Table 3). This result is in agreement with our loss of heterozygosity (LOH) data for this region of chromosome 17p (data can be provided on request). The two cell lines SNU-C2B and VACO429 were each apparently heterozygous for two different mutations. Full-length ORFs from these two lines were amplified, cloned, and sequenced. The results revealed that the different mutations in these two cell lines were indeed located on different alleles (Table 2).
In summary, of the 56 cell lines analyzed, 40 have one mutation and 3 have two mutations. The resulting 46 mutations include 22 missense mutations, 10 nonsense mutations, and 14 frameshift mutations that are due to changes at a splice site or insertions or deletions in the coding region (Table 2).
Expression of Mutant and Truncated p53. The missense mutant p53 product in cell line NCI-H716 could not be detected at the protein level by Western blot analysis (Fig. 1). This finding is in contrast to previous reports that missense mutations produce stable mutant p53 protein (8–10). To ascertain whether the rest of the cell lines with missense mutations in TP53 contain detectable mutant p53 protein, Western blot analysis for p53 was carried out by using the DO-1 antibody. The results showed that, of the 21 cell lines with mutant p53 caused by missense mutations, only one line, NCI-H716, did not show any expression of p53 protein.
Expression of p53 protein was also investigated by Western blotting, as described above, by using antibody DO-1, in the 22 cell lines with nonsense or frameshift mutations in TP53, which were expected to cause truncated ORFs (Table 2). Eleven cell lines had detectable expression of p53 by using DO-1, 9 of which showed a single truncated p53 protein product, and 2 cell lines, HCA7 and C70, each expressed two truncated products (Fig. 3). These double products are presumably due to the presence of WT p53 in HCA7 and an alternative splice variant due to the splice site mutation in C70. Freshly made Western blots from the remaining 11 cell lines, in which no protein had been detected by antibody DO-1, were hybridized with a different N-terminal antibody to p53, PAb 1801. A single band with the expected truncated protein size was detected in an additional cell line C80. This protein product was not detectable by DO-1, perhaps because the relevant epitope was obscured by the conformation of the mutant protein. All of the cell lines with truncation mutations were also analyzed with another p53 antibody, PAb1802, with specificity for a determinant near the C terminus of p53. As expected, they were all negative for this C-terminal antibody (data not shown). No protein product was detected consistently in the remaining 10 cell lines with truncation or frameshift mutations in p53.
Fig. 3.
Detection of truncated p53 in CRC cell lines with anti-p53 antibody DO-1. Cell extracts were analyzed by Western blotting by using DO-1 as described in Materials and Methods. Samples from 1–10 are: C70, C84, COLO678, COLO741, Gp2d, HCA7, LS513, SW948, VACO4A, and VACO429. Cell line LS513 (position 7) is a positive control from a line with normal TP53. C70 (position 1) and HCA7 (position 6) each showed two different products and SW948 (position 8) lacked evidence of any product, whereas the remaining 6 lines each showed a single product.
For all of the 11 cell lines in which no mutant or truncated p53 could be detected by Western blot analysis, RT-PCR was performed by using primers designed to amplify small regions of the TP53 mRNA from exons 1–11 (Table 4, which is published as supporting information on the PNAS web site) to ascertain whether there was any, even relatively low, or partial expression of the transcripts. Based on the results of this RT-PCR analysis, 7 of these cell lines expressed TP53 mRNA, whereas 4 cell lines, namely PC/JW, T84, VACO400, and SW1222, did not show evidence of any expression corresponding to any of the small overlapping regions. This result was consistent with the prediction that cell lines PC/JW, VACO400, and SW1222 would have no transcripts due to mutations in exon1. For T84, the absence of any observed transcript could be due to the mutation at the splice site leading to the production of a novel unstable transcript.
Discussion
An exhaustive study of a panel of 56 CRC cell lines for the presence of TP53 mutations and the consequent expression of p53 mRNA and protein was carried out. This study was based both on DNA and RNA analysis, as well as functional tests. DHPLC screening detected 40 mutations, three of which were in cell lines that each carried two mutations. Further sequencing of cDNA or genomic DNA, from all of the lines where no mutation had been detected by DHPLC, revealed 6 additional mutations, giving a total of 46 mutations detected in 43 of the 56 cell lines. This is a detection rate of 86.96% by DHPLC, which is comparable with that previously reported (11). DNA damage studies confirmed the expected function for p53 in the 13 cell lines with a normal functional TP53 sequence. Only in cell lines is it possible to do such a thorough analysis, especially using functional assays.
We have compared our data with those in the International Agency for Research on Cancer (IARC) database (http://www-p53.iarc.fr/index.html) (12). This is one of the two most regularly updated databases for TP53 mutations, the other one being the Universal Mutation Database-p53 database at http://p53.free.fr/ (13). The comparison of the distributions of the positions of single base pair substitutions in our cell line data with that in the IARC database shows them to be quite similar. In particular, it is clear that a high proportion of the missense mutations are in or near the p53 DNA-binding region that is between codons 102 and 292 (14). We have also found very similar positions with a high frequency of mutational events to those found in colon carcinomas. Thus, identical mutations at codons 245 and 273 were found in three different cell lines, identical mutations at codons 248 and 175 in two different lines, whereas nonsense mutations at codon 126 were also found in two different lines. We found only one example of a cell line with both a WT and mutant form of TP53, which is in agreement with essentially all previous reports on data from carcinomas.
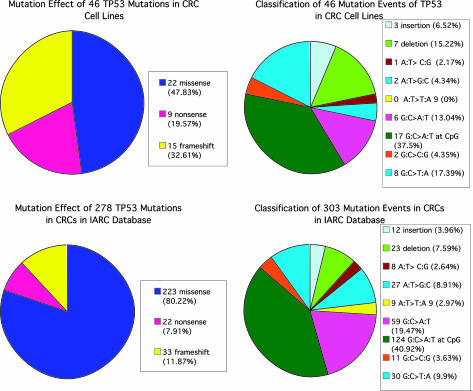
The main differences between our data and those in the database are as follows. (i) The proportion of the cell lines with p53 mutations, 76.8% (43/56), is much higher than the average of 50% usually reported for colon carcinomas (http://www-p53.iarc.fr/index.html). (ii) The proportion of missense mutations among these is significantly lower, at 47.83% (22/46) as compared with 80.22% (223/278) in the IARC database (P = 0.0018) (Fig. 4 Left). A more detailed comparison of the types of mutations we have found in the cell lines and those reported in the IARC database is illustrated in Fig. 4 Right. This comparison shows quite similar proportions for the different types of point mutations. In particular, the proportion of the point mutations that are transitions at CpG sites is 37.5%, which is comparable with that in the IARC database. The figures indicate the expected relatively high mutation rate at CpG positions, based on the arguments of Sved and Bird (15), and as also found for the APC gene in CRCs (16).
Fig. 4.
Comparisons of the distributions of types of TP53 mutations in cell lines and in the IARC database. (Left) Overall TP53 mutation effects in 43 CRC cell lines (Upper Left) compared with that in colon carcinomas in the IARC database updated in July 2005 (Lower Left). (Right) Detailed distribution of TP53 mutational events observed in 43 CRC cell lines (Upper Right) compared with that in CRCs in the IARC database updated in July 2005 (Lower Right).
Among the 46 mutations identified in our cell lines, 13 have not been identified in CRCs before, although 5 have been reported in other types of cancers (Table 2). This result reflects a wider range of types of mutations we have found in the cell lines than is reported in the IARC database. Overall, 74.42% (32/43) of p53 mutant cell lines made p53 protein product detectable by Western blots using two different antibodies. However, only 4 of the 43 lines made no TP53 transcripts, raising the question as to whether protein product was actually made by the remaining 7 cell lines that was not detectable by the Western blot analysis. This finding could be (i) because both the p53 antibody epitopes (DO-1 and PAb1801) were obscured by the mutations in these cell lines or (ii) because the turnover rate of these protein products was too rapid for detection, or, and perhaps the least likely (iii) because the protein product was somehow sequestered in a compartment of the cell not allowing efficient extraction. Another distinct possibility is that other antibodies might detect protein products associated with alternative p53 transcripts in the 7 lines that make at least some fragments of p53 message. This suggestion is based on recent evidence for the existence of p53 isoforms even in normal tissues (17). The data presented here are entirely consistent with a dominant negative effect for most p53 mutations, as has often been suggested. If only lack of p53 activity were selected for, then a much higher proportion of mutations with small deletions and insertions and complete lack of function would be expected. Thus, as emphasized before, TP53 is not really a tumor suppressor gene, although loss of the normally functioning version of the gene is almost universal after the first partially dominant mutation has been selected. Further investigation is needed to establish whether the truncated products do indeed have dominant negative effects.
There are three main possible reasons why the distribution of p53 mutations found in this study of cell lines is somewhat different from that in the public database. The first, and most obvious, is that our exhaustive approach to finding p53 mutations in the cell lines has detected mutations that might well be missed in analysis of fresh tumor material. This possibility is accentuated by the fact that primary tumor material is often contaminated with normal tissue, which makes mutation detection less efficient. There is also a bias in screening for mutations based on expectations from previously reported results. For example, exon 1 of TP53 has been overlooked in nearly all of the mutation detection studies reported so far (14). Our data emphasize the need to sequence every exon of a candidate gene to be sure of detecting all, or at least the great majority, of the mutations that may be present in a tumor. Such sequencing is demanding, especially when dealing with primary tumor material. A second possibility as to why our distribution of TP53 mutations is different is that cell lines may grow out preferentially from primary tumors containing a particular range of p53 mutations. It is well known that only ≈10–15% of colon carcinomas generally give rise to cell lines so that there is certainly plenty of opportunity for such selection to occur. The third, and we believe by far the least likely, possibility is that some of the mutations have been selected for in tissue culture. This possibility is made unlikely by the consistency of genetic data obtained from cell lines from the same tumor source, but which have been cultured independently for long periods of time.
Our comprehensive study of p53 mutations in a panel of 56 cell lines has revealed some unexpected mutations and has widened the scope of studies needed to understand the role of p53 mutations in the evolution of colorectal, and possibly most other, carcinomas.
Materials and Methods
Cell Lines and Cell Culture. Fifty-six different CRC cell lines were studied. The references for these cell lines can be obtained from the authors. A summary of the tissue culture conditions for the cell lines is as follows: cell lines CACO2, CC07, CC20, CCK-81, COLO201, COLO320DM, CX-1, DLD1, Gp2d, HCA7, HCT116, HT29, HT55, LIM1863, LOVO, LS180, LS1034, LS123, LS174T, LS411, PC/JW, RCM-1, SKCO-1, SW1116, SW403, SW48, SW480, SW837, SW948, and T84 were cultured in DMEM [provided by Cancer Research UK (CRUK)] in 10% CO2; cell lines COLO678, COLO741, LS513, NCI-H716, NCI-747, RKO, SNU-C2B, SW1222, and SW1417 were cultured in RPMI medium 1640 (provided by CRUK) in 5% CO2; and cell lines C10, C106, C125-PM, C32, C70, C75, C80, C84, C99, CaR-1, CoCM-1, HCA46, HRA19, VACO400, VACO10MS, VACO429, VACO4A, and VACO5 were cultured in Iscove's modified Dulbecco's medium (Invitrogen) in 10% CO2. All of the media were used with 10% FBS (Autogen Bioclear, Wiltshire, U.K.) and 6 mM l-glutamine (CRUK). All cultures were mycoplasma free and maintained in a humidified atmosphere with controlled CO2 content as indicated above.
Preparation of DNA, RNA, and Protein from Cell Lines. DNA and RNA were extracted from cells by using the DNeasy Tissue Kit (Qiagen, Crawley, U.K.) and the RNeasy Mini Kit (Qiagen), respectively, following the manufacturer's protocols. For protein extraction, cells were rinsed with PBS and lysed in RIPA buffer [150 mM NaCl/1% (wt/vol) Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS/50 mM Tris·Cl, pH 7.5] supplemented with a protease inhibitor mixture (Complete Mini tablets, Roche Diagnostics). For routine quantitation of proteins, the bicinchoninic acid procedure was used, according to the manufacturer's protocol (Pierce).
PCR Amplification of Exons 1–11 of TP53 for Mutation Detection. Primers were designed to amplify exons 1–11 of TP53 (Table 1). The standard PCR was in a volume of 50 μl containing 50 ng template DNA, 10 pmol of each primer, 0.25 mM each dNTP, 2.5 mM MgCl2, 5 units of AmpliTaq Gold (Applied Biosystems), 1× Taq Gold buffer (Applied Biosystems), and the appropriate volume of H2O. For exons that could not be amplified by using the above condition, 0.5 × Q solution (Qiagen) was used following the manufacturer's protocol (Table 1). “Touchdown” PCR programs (18) were then used to amplify the exons: 1 cycle of denaturation (12 min at 95°C), 15 cycles of denaturation (95°C for 30 s), annealing temperature (Tm; +7.5°C for 30 s, with –0.5°C per cycle), and extension (72°C for 30 s), followed by 20 cycles of denaturation (95°C for 30 s), annealing (Tm, 30 s), and extension (72°C for 30 s), and a final extension cycle of 72°C for 10 min.
Mutation Detection Using DHPLC. Each PCR amplicon amplified as above was mixed with an equal amount of the corresponding PCR amplicon from a known normal sample. The mixed PCR amplicons were denatured at 95°C for 4 min, followed by 1-min cycles of decreasing temperature at a rate of 1.6°C per cycle down to 15°C. After this procedure, the amplicon mixtures were analyzed by DHPLC on a WAVE Machine (Transgenomic). A 2% per minute increase in buffer B (as provided by the manufacturer) was used for all of the fragments. The denaturing temperatures for each exon and the corresponding initiating buffer B percentages are indicated in Table 1.
RT-PCR. cDNA from each cell line was synthesized from 2 μg of denatured RNA (65°C for 5 min) by incubation at 37°C for 60 min with final quantities or concentrations of 1 μM of oligo(dT), 4 units of Omniscript reverse transcriptase (Qiagen), 10 units of RNase inhibitor (Ambion, Huntingdon, U.K.), and 0.5 mM each dNTP. Primers used for p53 RT-PCR are summarized in Table 5, which is published as supporting information on the PNAS web site. Primers used for β actin RT-PCR were: forward, 5′-ACACCTTCTACAATGAGC-3′ and reverse, 5′-ACGTCACACTTCATGATG-3′. All RT-PCRs were carried out in a 25-μl PCR containing 0.2 μM each primer, 2.5 units of AmpliTaq Gold, 1.5 mM MgCl2, 200 μM each dNTP, and 200 μg (for p53 amplification) or 100 μg (for β-actin amplification) of cDNA. The cycling conditions used for these PCRs were as follows: 95°C for 10 min, 30 (for p53) or 25 (for β-actin) cycles of 95°C for 30 s, the appropriate annealing temperature for p53 as shown in Table 4 and 56°C (β-actin) for 45 s, and 72°C for 60 s, with a final extension step of 72°C for 10 min.
Sequencing. PCR amplicons showing abnormal DHPLC patterns were sequenced directly by using the appropriate PCR primers and Big Dye Sequencing kit (Applied Biosystems) on an ABI 377 (Applied Biosystems) sequencer. Before the sequencing reaction was performed, 20 μl of the PCR was treated with 4 units of Exonuclease I (New England Biolabs) and 1 unit of Shrimp Alkaline Phosphatase (SAP, Amersham Pharmacia) at 37°C for 1.5 h and 80°C for 20 min. Approximately 100 ng of treated amplicon was used for each sequencing reaction. RT-PCR amplicons were sequenced in a similar manner but with two additional internal pairs of primers: forward 5′-ACAGCCAAGTCTGTGACTTG-3′, reverse 5′-GTGATGATGGTGAGGATGG-3′; forward 5′-AGGTTGGCTCTGACTGTACCA-3′ and reverse 5′-TCCTTCCACTCGGATAAGATGC-3′. In two cell lines where two mutations were identified, the corresponding p53 ORF amplicons were purified with QIAquick kit (Qiagen) and then subcloned into TOPO TA vector (Invitrogen). At least 10 clones from each cloning were sequenced by using vector primers and internal primers (Table 4). Sequencing data were analyzed by using the sequencher program (Gene Codes, Ann Arbor, MI).
Western Blotting. Protein samples were denatured, separated by SDS/PAGE gel, and transferred onto Hybond-P nitrocellulose membrane (Amersham Pharmacia) by using the MiniPROTEAN 3 apparatus (Bio-Rad). Membranes were rinsed and blocked with 0.05% Tween 20 (Sigma) and 1% skimmed milk in PBS for 1 h with 3 changes of this blocking buffer every 20 min at room temperature. Membranes were then incubated with primary antibodies in blocking buffer after conditions described in Table 5. After washing membranes in blocking buffer for 1 h with a change of the buffer every 15 min at room temperature, they were incubated for 1 h with appropriate secondary antibody (HRP Rabbit Anti-Mouse Immunoglobulin for monoclonal primary antibody, and HRP Goat Anti-Rabbit Immunoglobulin for polyclonal primary antibody, DAKO) in blocking buffer for 1 h with mild shaking, and then washed in the same fashion as that after incubation with the primary antibody. Membranes were visualized by using ECL plus Western blotting detection system (Amersham Pharmacia). Anti-actin monoclonal antibody (Sigma) was hybridized to membranes to serve as a measure of loading control.
Supplementary Material
Acknowledgments
We thank David Bicknell, Bruce Winney, and Jenny Wilding in our group for helpful discussions. We also thank David Lane for permission to use the P53 antibodies DO-1, PAb 1801, and PAb 1802, and the CRUK cell culture service for providing these antibodies. Y.L. is a research fellow sponsored by GlaxoSmithKline.
Conflict of interest statement: No conflicts declared.
Abbreviations: CRC, colorectal carcinoma; DHPLC, denaturing HPLC.
References
- 1.El-Deiry, W. S. (1998) Semin. Cancer Biol. 8, 345–357. [DOI] [PubMed] [Google Scholar]
- 2.Sigal, A. & Rotter, V. (2000) Cancer Res. 60, 6788–6793. [PubMed] [Google Scholar]
- 3.Lang, G. A., Iwakuma, T., Suh, Y. A., Liu, G., Rao, V. A., Parant, J. M., Valentin-Vega, Y. A., Terzian, T., Caldwell, L. C., Strong, L. C., et al. (2004) Cell 119, 861–872. [DOI] [PubMed] [Google Scholar]
- 4.Olive, K. P., Tuveson, D. A., Ruhe, Z. C., Yin, B., Willis, N. A., Bronson, R. T., Crowley, D. & Jacks, T. (2004) Cell 119, 847–860. [DOI] [PubMed] [Google Scholar]
- 5.Melino, G., Lu, X., Gasco, M., Crook, T. & Knight, R. A. (2003) Trends Biochem. Sci. 28, 663–670. [DOI] [PubMed] [Google Scholar]
- 6.Van Dyke, T. (2005) Nature 434, 287–288. [DOI] [PubMed] [Google Scholar]
- 7.Fearon, E. R. & Vogelstein, B. (1990) Cell 61, 759–767. [DOI] [PubMed] [Google Scholar]
- 8.Rodrigues, N. R., Rowan, A., Smith, M. E., Kerr, I. B., Bodmer, W. F., Gannon, J. V. & Lane, D. P. (1990) Proc. Natl. Acad. Sci. USA 87, 7555–7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartek, J., Bartkova, J., Vojtesek, B., Staskova, Z., Rejthar, A., Kovarik, J. & Lane, D. P. (1990) Int. J. Cancer 46, 839–844. [DOI] [PubMed] [Google Scholar]
- 10.Iggo, R., Gatter, K., Bartek, J., Lane, D. & Harris, A. L. (1990) Lancet 335, 675–679. [DOI] [PubMed] [Google Scholar]
- 11.Xiao, W. & Oefner, P. J. (2001) Hum. Mutat. 17, 439–474. [DOI] [PubMed] [Google Scholar]
- 12.Olivier, M., Eeles, R., Hollstein, M., Khan, M. A., Harris, C. C. & Hainaut, P. (2002) Hum. Mutat. 19, 607–614. [DOI] [PubMed] [Google Scholar]
- 13.Soussi, T., Dehouche, K. & Beroud, C. (2000) Hum. Mutat. 15, 105–113. [DOI] [PubMed] [Google Scholar]
- 14.Soussi, T. & Beroud, C. (2001) Nat. Rev. Cancer 1, 233–240. [DOI] [PubMed] [Google Scholar]
- 15.Sved, J. & Bird, A. (1990) Proc. Natl. Acad. Sci. USA 87, 4692–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodmer, W. (1999) Cytogenet. Cell Genet. 86, 99–104. [DOI] [PubMed] [Google Scholar]
- 17.Bourdon, J. C., Fernandes, K., Murray-Zmijewski, F., Liu, G., Diot, A., Xirodimas, D. P., Saville, M. K. & Lane, D. P. (2005) Genes Dev. 19, 2122–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Don, R. H., Cox, P. T., Wainwright, B. J., Baker, K. & Mattick, J. S. (1991) Nucleic Acids Res. 19, 4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.