Abstract
Depending on the degree of underlying resistance present, optimization of the pharmacokinetics of protease inhibitors may result in improved virologic suppression. Thirty-seven human immunodeficiency virus (HIV)-infected subjects who had chronic detectable viremia and who were receiving 800 mg of indinavir three times a day (TID) were switched to 400 mg of indinavir BID with 400 mg of ritonavir two times a day (BID) for 48 weeks. Full pharmacokinetic evaluations were obtained for 12 subjects before the switch and 3 weeks after the switch. Combination therapy increased the indinavir predose concentrations in plasma by 6.47-fold, increased the minimum concentration in serum by 3.41-fold, and reduced the maximum concentration in serum by 57% without significantly changing the area under the plasma concentration-time curve at 24 h. At week 3, 58% (21 of 36) of the subjects for whom postbaseline measurements were available achieved a viral load in plasma of <50 copies/ml or a reduction from the baseline load of ≥0.5 log10 copies/ml. Of these subjects, 82% (14 of 17) whose viruses had three or fewer protease inhibitor mutations and 88% (14 of 16) whose viruses had an indinavir virtual phenotypic susceptibility test of more than sixfold less than that for the baseline isolate were considered virologic responders. The indinavir virtual inhibitory quotient, which is a function of baseline indinavir phenotypic resistance (estimated by virtual phenotype) and the indinavir predose concentration in plasma achieved with indinavir-ritonavir combination therapy, was the best predictor of a viral load reduction. Sixteen subjects discontinued the study by week 48 due to adverse events, predominantly related to hyperlipidemia. Pharmacokinetic intensification of indinavir-based therapy with ritonavir reduced the viral loads in subjects but added toxicity. The virtual inhibitory quotient, which incorporates both baseline viral resistance and the level of drug exposure in plasma, was superior to either baseline resistance or drug exposure alone in predicting the virologic response.
Suboptimal drug exposure in patients receiving combination antiretroviral therapy has been increasingly recognized as an important contributor to virologic failure, particularly with drugs in the protease inhibitor (PI) class (1, 2, 5, 6, 15, 19). Subinhibitory concentrations of drugs may be a consequence of poor adherence to the drug regimen, but they may also occur due to the wide variabilities in metabolism and the levels in plasma seen with many of the PIs. Indinavir is an example of a PI that is rapidly metabolized by the cytochrome P450 system and that has reduced bioavailability when taken with food, making it necessary for patients to take the drug every 8 h in a fasting state. The pharmacokinetic profile of indinavir is characterized by large interpatient variability and large differences between peak concentrations in plasma (Cmax) and predose concentrations in plasma (Ctrough) (1, 12, 17, 19). Indinavir Ctroughs frequently fall near or below the in vitro 50% inhibitory concentration (IC50) for wild-type human immunodeficiency virus (HIV) (1, 12, 17, 19).
Ritonavir is a licensed HIV PI that is also a very potent inhibitor of cytochrome P450 3A4 (CYP3A4). Coadministration of ritonavir with other PIs increases the bioavailabilities and half-lives of the PIs by inhibiting CYP3A4-mediated metabolism (8, 13). Addition of ritonavir to indinavir-containing regimens may have several potential advantages. In particular, this combination can be administered without the food restrictions required with indinavir as a single PI-based regimen and can be administered twice a day (BID) as opposed to three times a day (TID) (11; A. Hsu, G. R. Granneman, M. Heath-Chiozzi, E. Ashbrenner, L. Manning, R. Brooks, P. Bryan, K. Erdman, and E. Sun, Program Abstr. 12th Int. AIDS Conf., abstr. 22361, 1998). When ritonavir at 400 mg BID is administered with indinavir at 400 mg BID, the pharmacokinetic parameters for indinavir are differentially altered compared to those for indinavir administered at 800 mg TID: the indinavir Cmaxs are reduced by 50%, while the indinavir Ctoughs are increased nearly fourfold and the areas under the plasma concentration-time curves (AUCs) remain unchanged (11). Furthermore, the interpatient variabilities of indinavir pharmacokinetic parameters are substantially reduced with ritonavir coadministration (11).
This study was designed to evaluate the safety, tolerability, and efficacy of pharmacokinetic intensification or boosting of indinavir-based therapy with ritonavir in subjects who were receiving indinavir therapy and in whom HIV RNA was detectable in plasma. Ctroughs of PIs have been associated with virologic suppression in several clinical studies (1, 5, 6, 9, 15), as have AUCs in other studies (1, 14). Enhancement of indinavir Ctroughs by the addition of ritonavir to the antiretroviral drug regimen could theoretically enhance virologic suppression. Furthermore, the differential alteration of indinavir pharmacokinetic parameters by ritonavir in this particular regimen could help to discriminate which of these parameters are more closely associated with in vivo PI activity. Finally, the study design also allowed assessment of the degree of indinavir resistance that could be overcome with pharmacokinetic boosting.
(This study was presented in part at the 7th Conference on Retroviruses and Opportunistic Infections, San Francisco, Calif., 30 January to 2 February 2000 [N. Shulman et al., 7th Conf. Retrovir. Opportunistic Infect., abstr. 534, 2000]; the 4th International Workshop on HIV Drug Resistance and Treatment Strategies, Sitges, Spain, 12 to 16 June 2000 [A. Zolopa et al., 4th Int. Workshop on HIV Drug Resist. Treatment Strategies, abstr. 95, 2000]; the 13th World AIDS Conference, Durban, South Africa, July 2000 [D. Havlir et al., 13th Int. AIDS Conf., abstr. WePeB4122, 2000]; the 8th Conference on Retroviruses and Opportunistic Infections, Chicago, Ill., 4 to 8 February 2001 [A. Hsu et al., 8th Conf. Retrovir. Opportunistic Infect., abstr. 337, 2001; D. Kempf et al., 8th Conf. Retrovir. Opportunistic Infect., abstr. 523, 2001]; and the 2nd International Workshop on Clinical Pharmacology of HIV Therapy, Noordwijk, The Netherlands, 2 to 4 April 2001 [A. Hsu et al., 2nd Int. Workshop Clin. Pharmacol. HIV Ther., abstr. 7.3, 2001].)
MATERIALS AND METHODS
Study population and design.
This was a single-arm, prospective, open-label study conducted at four clinical centers in the United States. HIV-infected adults who had been receiving indinavir plus two nucleoside reverse transcriptase (RT) inhibitors (NRTIs) for at least 3 months and who had detectable HIV RNA levels (between 50 and 50,000 copies/ml) at the time of screening were eligible for the study. Patients who had received ritonavir prior to this study were excluded. The institutional review board or independent ethics committee for each site approved the protocol prior to site participation in this clinical trial.
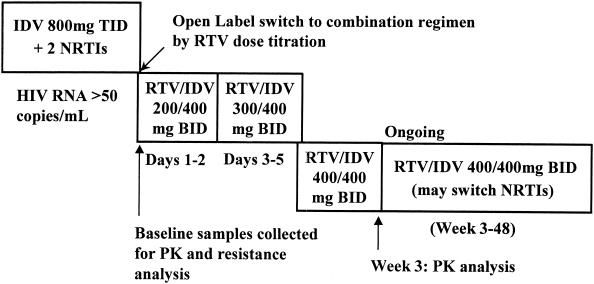
Subjects were switched from 800 mg of indinavir TID to 400 mg of indinavir BID and 400 mg of ritonavir BID. The dose of ritonavir was introduced at 200 mg BID for the first 2 days, escalated to 300 mg BID for the next 3 days, and then further escalated to 400 mg BID, as tolerated. This dose escalation schedule could have been lengthened to 10 days in the case of tolerability-related events. Reduction of the ritonavir dose to 300 mg BID was permitted for subjects who were unable to tolerate 400 mg of BID. On day 1 of the study, the food restrictions and fluid intake recommended with indinavir therapy were removed. In order to determine the isolated effect of ritonavir-mediated alteration of indinavir pharmacokinetics on virologic suppression, no other changes in the subjects' antiretroviral regimens were permitted during the first 3 weeks. After week 3, subjects were permitted to change the NRTI portion of their antiretroviral regimen at the discretion of their physicians. The study design is shown in Fig. 1.
FIG. 1.
Study design. RTV, ritonavir; IDV, indinavir; PK, pharmacokinetic.
Study evaluations.
Subjects were evaluated for the development of adverse events every 3 weeks for the first 12 weeks and then at weeks 16, 24, 36, and 48. Blood samples for the determination of plasma HIV RNA levels (using the Amplicor HIV-1 Monitor Ultrasensitive assay; sensitivity, <50 copies/ml; Roche Diagnostics, Inc., Indianapolis, Ind.) and CD4 cell counts were obtained at each scheduled visit. Blood chemistries, complete blood counts, cholesterol and triglyceride determinations, coagulation studies, and urinalyses were monitored at the baseline and at weeks 6, 12, 24, 36, and 48. All laboratory evaluations were performed at a central laboratory.
For all subjects, indinavir Ctroughs were measured at the baseline, week 3, and week 24. Intensive pharmacokinetic profiling was performed for a subset of 16 subjects at the baseline and week 3. At the baseline, these subjects received 800 mg of indinavir with water after an overnight fast and were then given a standardized breakfast 2 h later. Plasma indinavir levels were measured at time zero (predosing) and at 1, 1.5, 2, 3, 4.5, 6, and 8 h postdosing. At week 3, these same subjects received observed doses of 400 mg of indinavir and 400 mg of ritonavir at the time of a standardized breakfast. Indinavir and ritonavir levels were measured at time zero (predosing) and at 2, 4, 6, 8, 10, and 12 h postdosing. Subjects received their NRTIs at the same time that they received the indinavir and ritonavir doses. The meals and snacks given after breakfast were not standardized or restricted.
The plasma indinavir and ritonavir concentrations were measured by previously validated reverse-phase high-performance liquid chromatography procedures with UV detection at 205 nm and a modified mobile phase (11). The lower limits of quantitation for the assay are 2 ng/ml for indinavir with 0.5 ml of plasma and 6 ng/ml for ritonavir with 1 ml of plasma. The assay is linear between 2 and 400 ng/ml for indinavir and 6 and 3,500 ng/ml for ritonavir. Samples quantified above the highest standard were diluted with blank plasma and reassayed. The following pharmacokinetic parameters were evaluated: AUC, the minimum concentration in plasma (Cmin), Cmax, and Ctrough.
Baseline genotypic analysis was performed for all subjects. HIV RNA was extracted from plasma, and nested PCR amplification was used to generate a 1.3-kb fragment encompassing Pr and the first 900 nucleotides of RT (18). Direct dideoxynucleotide terminator cycle sequencing of the PCR product was performed as described previously (18). Sequencing of both strands was performed. Sequencing reactions were analyzed on an ABI 377 instrument (Perkin-Elmer Applied Biosystems, Foster City, Calif.) and manually proofread and edited. Sequences were compared with the HIV type 1 (HIV-1) clade B consensus sequence, and differences in amino acid sequences, including positions that contained a mixture of wild-type and mutant residues, were classified as mutations. RT codons 1 to 300 and Pr codons 1 to 99 were analyzed. Mutations at the following amino acids in HIV Pr were classified as PI mutations according to the Data Analysis Plan of the HIV Resistance Collaborative Working Group (3): 10, 20, 24, 30, 32, 33, 36, 46, 47, 48, 50, 54, 71, 73, 77, 82, 84, 88, and 90. Mutations observed at the following amino acids in RT were classified as major NRTI mutations: 41, 67, 69, 70, 75, 151, 184, 210, 215, and 219.
HIV phenotyping was performed with plasma obtained at the baseline by the Antivirogram method (Tibotec-Virco, Mechelen, Belgium) (10). Phenotypic data were expressed as the fold change in the IC50, which was calculated by dividing the IC50 of the drug for the recombinant virus (containing the HIV Pr and RT genes of the baseline patient plasma sample) by the IC50 of the drug for the standard wild-type recombinant virus.
Virtual phenotype (VP) values were calculated from the entire Pr and RT sequences through a proprietary method by Tibotec-Virco (B. Larder, S. Kemp, and K. Hertogs, Program Abstr. 4th Int. Workshop HIV Drug Resist. Treatment Strategies, abstr. 63, 2000). Briefly, this method involves comparison of the Pr or RT sequence to matching sequences in a large database of sequences for isolates for which both the genotype and the phenotype have been determined. The VP is the ratio of the average IC50 for the isolates with the corresponding genotypic matches in the Virco database divided by the IC50 for the wild-type virus. This method gives a reasonable estimate of the fold change in IC50 for a given genotype compared to the IC50 for the wild type. The VP may then be applied to the IC50s for wild-type virus determined under specific conditions, such as serum-adjusted IC50s, to estimate the IC50s for other viruses in the presence of human serum.
The inhibitory quotient (IQ) characterizes the relationship between drug exposure and drug susceptibility (7). For each sample for which baseline VP and week 3 Ctroughs were available, virtual IQs (vIQs) for indinavir and ritonavir were calculated by the following formula: Ctrough at week 3/(VP × serum-adjusted IC50 for wild-type virus).The serum-adjusted IC50s of indinavir and ritonavir were calculated as the mean IC50 of each drug for three wild-type laboratory strains (strains pNL4-3, HXB2, and HIV-1IIIB) as determined in side-by-side experiments in the presence of 50% human serum and 10% fetal calf serum. The serum-adjusted IC50s of indinavir and ritonavir obtained by this method are 0.053 and 0.96 μg/ml, respectively (16).
Statistical analysis.
All statistical analyses were performed with the SAS system (release 6.12; SAS Institute Inc., Cary, N.C.). All statistical tests were two tailed and were performed at the 0.05 level of statistical significance unless specified otherwise. Changes in the indinavir pharmacokinetic results from the baseline (indinavir at 800 mg TID) to week 3 (indinavir at 400 mg BID and ritonavir at 400 mg BID) were evaluated by a paired t test. In addition, the association between phenotype and VP (for indinavir and ritonavir), the association between the number of PI mutations and the indinavir VP, the association between Ctrough and VP (for indinavir and ritonavir), and the association between the indinavir vIQ and the ritonavir vIQ were evaluated by using the Spearman rank correlation coefficient (r).
Virologic response was defined as achieving a viral load below the limit of quantitation (i.e., <50 copies/ml) or achieving a viral load reduction from that at the baseline of at least 0.5 log10 copies/ml. Response was analyzed by using the dropouts-as-censored method, in which data for subjects who discontinued treatment as responders are censored, while those who discontinued treatment as nonresponders are counted as nonresponders for the duration of the study. Virologic response was stratified by the number of baseline PI mutations (≤3, ≥4), the number of baseline RT mutations (≤3, ≥4), the indinavir VP (<6-fold, >6-fold), the indinavir Ctrough at week 3 (<0.75 μg/ml, >0.75 μg/ml), and the indinavir vIQ (<2, >2). Between-stratum comparisons were performed by Fisher's exact test.
Logistic regression was used to evaluate the virologic response as a function of the baseline viral load, baseline genotypic and phenotypic resistance parameters, and week 3 indinavir pharmacokinetic parameters. In particular, stepwise logistic regression was used to evaluate the association between virologic response and potential predictors of virologic response including baseline viral load, number of PI mutations, number of RT mutations, indinavir VP, indinavir Ctrough at week 3, fold change in indinavir Ctrough between the baseline and week 3, and vIQ (for indinavir or ritonavir). The stepwise logistic regression was performed by using entry and exit significance levels of 0.10. In addition, all independent (predictor) variables with the exception of the number of PI mutations and the number of RT mutations were transformed (log10) prior to performance of the stepwise logistic regression.
RESULTS
Subject enrollment.
A total of 51 subjects were screened for participation in this study after informed consent was obtained, and 37 subjects were subsequently enrolled in the study. Of the 14 subjects who were not enrolled, 9 did not meet HIV RNA level requirements, 2 had exclusionary laboratory values, it was anticipated that 1 would be noncompliant, 1 withdrew consent, and 1 was excluded for other reasons.
Study population.
Thirty-seven male subjects were included in the study. The median age of the subjects was 42 years. Twenty-five subjects were white, five were black, six were Hispanic, and one was Asian. Prior antiretroviral treatment, including indinavir exposure, was extensive. In particular, subjects had previously received a median of 34.6 months (range, 5.8 to 43.8 months) of indinavir treatment and a median of 71.1 months (range, 19.2 to 239.1 months) of antiretroviral therapy. The subjects had a median baseline CD4 cell count of 325 cells/μl (range, 28 to 1,372 cells/μl) and a median HIV RNA level of 3.3 log10 copies/ml (range, 1.7 to 4.8 log10 copies/ml). The majority of subjects (81%; 30 of 37) had baseline plasma HIV RNA levels of <10,000 copies/ml, with 24% (9 of 37) having baseline HIV RNA levels between 50 and 400 copies/ml. Baseline characteristics are summarized in Table 1.
TABLE 1.
Baseline characteristics and resistance
| Characteristic | Result (n = 37) |
|---|---|
| Age (yr) | |
| Median | 42 |
| Range | 26-60 |
| No. of men | 37 |
| Race (no. of subjects) | |
| White | 25 |
| Black | 5 |
| Hispanic | 6 |
| Asian | 1 |
| HIV RNA level (log10 copies/ml) | |
| Median | 3.3 |
| Range | 1.7-4.8 |
| No. of subjects with HIV RNA levels (log10 copies/ml) | |
| <2.6 | 9 |
| 2.6-3.0 | 21 |
| >3.0 | 7 |
| CD4 count (no. of cells/μl) | |
| Median | 325 |
| Range | 28-1,372 |
| Duration of indinavir experience (mo) | |
| Median | 34.6 |
| Range | 5.8-43.8 |
| Duration of antiretroviral experience (mo) | |
| Median | 71.1 |
| Range | 19.2-239.1 |
| No. of NRTI mutationsa (no.) | |
| Median | 3 |
| Range | 0-6 |
| No. of PI mutationsb (no.) | |
| Median | 3 |
| Range | 0-7 |
| Indinavir VP (fold change) | |
| Median | 4.4 |
| Range | 0.7-24.3 |
NRTI mutations: amino acids 41, 67, 69, 70, 75, 151, 184, 210, 215, and 219.
PI mutations: amino acids 10, 20, 24, 30, 32, 33, 36, 46, 47, 48, 50, 54, 71, 73, 77, 82, 84, 88, and 90.
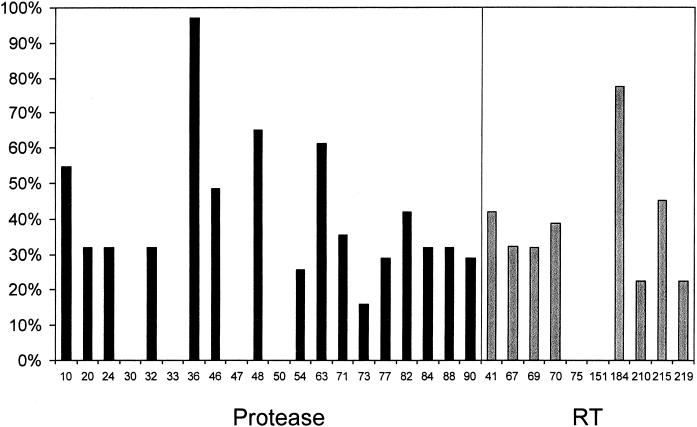
Baseline viral genotype results were available for 31 of the 37 subjects. The viral load of one sample was too low, and five samples were not available for genotyping. The HIV isolates contained a median of three mutations in the PI sequence (range 0 to 7). Isolates from 8 of the 31 subjects (26%) had no PI mutations, while those from 14 subjects (45%) had four or more PI mutations. Isolates from 15 subjects (48%) had a mutation at M46 (I/L), and those from 13 subjects (42%) had a mutation at V82 (A/T/F) (Fig. 2); both of these mutations commonly develop while patients are receiving indinavir therapy. Isolates also contained a median of three NRTI-associated mutations (range, zero to six mutations), including 14 (45%) that displayed the T215Y/F mutation, which reduces susceptibility to zidovudine, stavudine, and, possibly, other NRTIs. Twenty-four isolates (77%) contained the M184V/I mutation, which confers high-level resistance to lamivudine and lower degrees of resistance to abacavir and didanosine. In addition, 22 isolates (71%) had two or more major mutations in the RT sequence (Fig. 2).
FIG. 2.
Baseline prevalence of mutations.
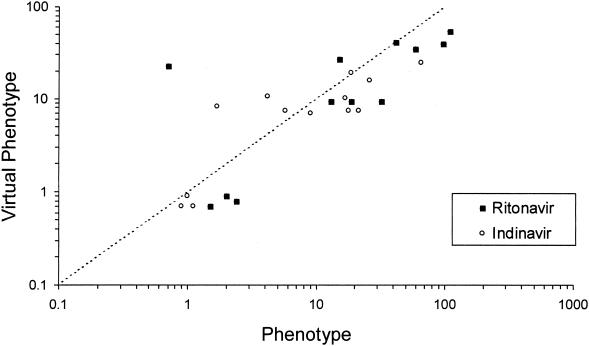
Phenotype values for indinavir and ritonavir were available for a limited number of baseline isolates (n = 13 and 12, respectively) due to the low baseline viral loads (<1,000 copies/ml) of many subjects enrolled in this study. As shown in Fig. 3, phenotype and VP appeared to be strongly correlated for both indinavir (r = 0.74) and ritonavir (r = 0.78). Therefore, the baseline VP, which was available for isolates from 31 subjects, was used for the subsequent analyses. The median indinavir VP derived from the baseline genotypes was a 4.4-fold change (range, 0.7- to 24.3-fold change) in susceptibility.
FIG. 3.
Relationship between phenotype and VP.
Pharmacokinetics analysis.
The effect of coadministration of ritonavir with indinavir was evaluated in the 12 subjects in the pharmacokinetic substudy for whom matching plasma samples obtained before the switch and after the switch from 800 mg of indinavir TID to 400 mg of indinavir BID and 400 mg of ritonavir BID for 3 weeks were available (Table 2). Coadministration of ritonavir resulted in a 57% decrease in the indinavir Cmax and a 3.41-fold increase in the indinavir Cmin (P ≤ 0.001 for both comparisons). The AUC at 24 h (AUC24) for indinavir did not change significantly, decreasing by 18% (P = 0.061). These pharmacokinetic changes in HIV-infected subjects were similar to those previously observed in healthy volunteers (11).
TABLE 2.
Indinavir pharmacokinetics at baseline (indinavir TID) and week 3 (indinavir and ritonavir BID)a
| Regimen | Cmin (μg/ml) | Cmax (μg/ml) | AUC24 (μg · h/ml) |
|---|---|---|---|
| Indinavir TID (n = 12) | 0.157 ± 0.156 | 9.41 ± 3.27 | 62.3 ± 21.2 |
| Indinavir and ritonavir BID (n = 12) | 0.536 ± 0.317 (341)b | 4.01 ± 1.04 (−57)c | 50.8 ± 14.7 (−18)d |
Values are means ± standard deviations. The values in parentheses are percent change.
P = 0.001.
P < 0.001.
P = 0.061.
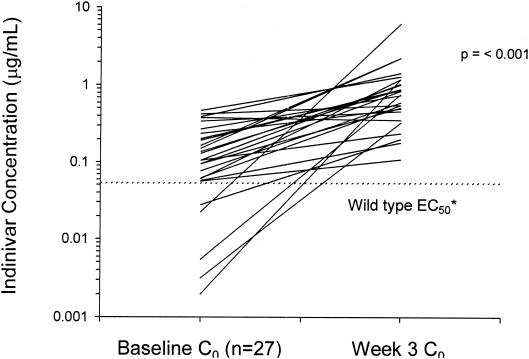
The indinavir Ctroughs before and after coadministration of ritonavir were compared for 27 of 29 subjects for whom plasma samples were available at both the baseline and week 3. Two subjects were determined to have had the sample for indinavir Ctrough determination collected close to the time of Cmax for the last indinavir-ritonavir dose, so the data for these subjects were excluded from the analysis. The Ctrough increased 6.47-fold (P = 0.001). As demonstrated in Fig. 4, the Ctrough increased in all but one subject.
FIG. 4.
Indinavir Ctroughs following the change from indinavir at 800 mg TID to ritonavir at 400 mg BID and indinavir at 400 BID. ∗, the 50% effective concentration (EC50) is from the Crixivan package insert (Merck & Co., Inc.); C0, predose concentration.
vIQs.
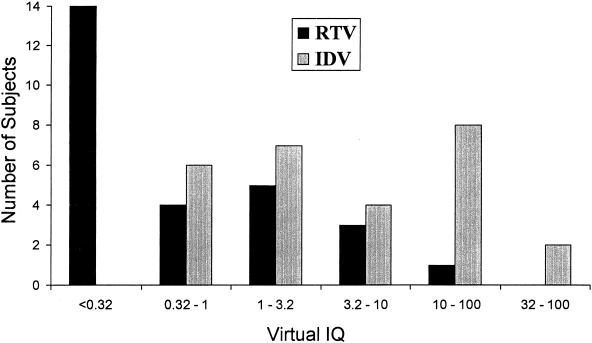
vIQs were calculated for the 27 subjects described above for whom samples were available at week 3 for indinavir Ctrough determinations. The Ctroughs of indinavir and ritonavir were correlated (r = 0.73), as were the VPs (r = 0.95). Consequently, the calculated vIQs for indinavir and ritonavir were also highly correlated (r = 0.89). Because of differences in the serum-adjusted IC50s between the two PIs, the median indinavir vIQ was more than 20-fold higher than the median ritonavir vIQ (5.25 versus 0.24). For 67% (18 of 27) of these subjects, the ritonavir vIQ was <1, while for 22% (6 of 27) of these subjects, the indinavir vIQ was <1 (Fig. 5).
FIG. 5.
vIQ range. RTV, ritonavir; IDV, indinavir.
HIV RNA response.
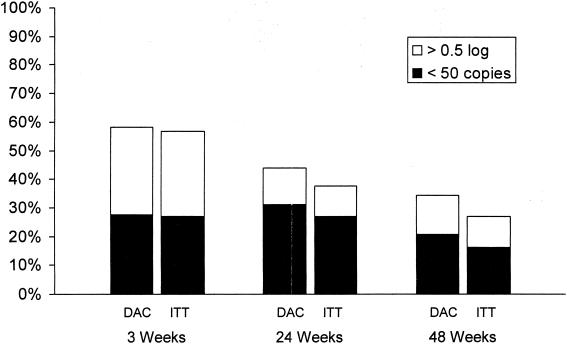
Virologic response was defined as achievement of a viral load below the limit of quantitation (i.e., <50 copies/ml) or achievement of a reduction in the viral load from that at the baseline of at least 0.5 log10 copies/ml. Virologic response was analyzed by using the dropouts-as-censored method (see Materials and Methods). At week 3, prior to any change in the NRTI portion of the antiretroviral regimen, 58% (21 of 36) of subjects achieved a virologic response, including 28% (10 of 36) with viral loads <50 copies/ml. Five subjects subsequently altered their NRTIs after the visit at week 3: four due to persistent viremia and one due to an adverse event (lipodystrophy). At week 24, 44% (14 of 32) of subjects were considered virologic responders, including 31% (10 of 32) with <50 copies/ml. At week 48, 34% (10 of 29) of subjects were considered virologic responders, with 21% (6 of 29) achieving viral loads <50 copies/ml. By using an intent-to-treat analysis, in which dropouts were considered virologic nonresponders, the corresponding virologic response rates at weeks 3, 24, and 48 were lower (Fig. 6).
FIG. 6.
Overall response rates obtained by the dropouts-as-censored (DAC) and intent-to-treat (ITT) methods
Predictors of virologic response.
Baseline virologic and pharmacokinetic parameters were analyzed by using the dropouts-as-censored method for their ability to predict virologic response (Table 3). At week 3, 82% (14 of 17) of subjects whose baseline isolates contained three or fewer PI mutations were considered virologic responders, whereas only 36% (5 of 14) of subjects whose baseline isolates contained four or more PI mutations were considered virologic responders (P = 0.012). This difference was maintained through week 24 (71% [10 of 14 subjects] versus 21% [3 of 14 subjects]; P = 0.021) but was no longer statistically significant at week 48 (55% [6 of 11 subjects] versus 21% [3 of 14 subjects]; P = 0.115) (Table 3). Of the seven subjects whose baseline isolates had six or more Pr mutations, six (86%) were nonresponders at weeks 3, 24, and 48.
TABLE 3.
Predictors of response
| Predictor | % of subjects
|
||
|---|---|---|---|
| Wk 3 | Wk 24 | Wk 48 | |
| No. of baseline PI mutations | |||
| 0-3 | 82 | 71 | 55 |
| ≥4 | 36 (0.012)a | 21 (0.021) | 21 (0.115) |
| No. of baseline NRTI mutations | |||
| 0-3 | 72 | 67 | 58 |
| ≥4 | 46 (0.262) | 23 (0.030) | 15 (0.041) |
| Change in baseline indinavir VP | |||
| Less than sixfold | 88 | 77 | 60 |
| More than sixfold | 33 (0.003) | 20 (0.007) | 20 (0.087) |
| Wk 3 indinavir Ctrough | |||
| >0.75 μg/ml | 79 | 77 | 70 |
| <0.75 μg/ml | 40 (0.060) | 14 (0.002) | 7 (0.002) |
| Indinavir vIQ | |||
| >2 | 89 | 75 | 62 |
| <2 | 11 (<0.001) | 0 (<0.001) | 0 (0.006) |
P values are in parentheses.
The virologic response at week 3 was not associated with the number of baseline NRTI mutations; however, 67% (10 of 15) of subjects whose isolates had three or fewer baseline NRTI mutations were considered responders at week 24, where only 23% (3 of 13) of those whose isolates had four or more NRTI mutations were considered responders at week 24 (P = 0.030). This difference was sustained at week 48 (58% [7 of 12 subjects] versus 15% [2 of 13 subjects]; P = 0.041) (Table 3). Of the five subjects who switched their NRTIs after week 3, only one subject experienced an altered virologic response at week 24 (i.e., the subject changed from a nonresponder to a responder). This subject subsequently discontinued the study therapy prior to week 48, and data for the subject were censored from the analysis performed at week 48 by the dropouts-as-censored method. In the event that this subject is considered a virologic nonresponder due to the change in antiretroviral therapy, the difference between subjects whose isolates had zero to three baseline NRTI mutations and those whose isolates had four or more baseline NRTI mutations was no longer statistically significant at week 24 (60% [9 of 15 subjects] versus 23% [3 of 13 subjects]; P = 0.067) or week 48 (54% [7 of 13 subjects] versus 15% [2 of 13 subjects]; P = 0.097).
The baseline indinavir VP was found to be associated with virologic responses at week 3 and week 24. We found that 88% (14 of 16) of subjects with baseline indinavir VPs less than sixfold were virologic responders at week 3, whereas 33% (5 of 15) with baseline indinavir VPs more than sixfold were virologic responders at week 3 (P = 0.003). Similarly, 77% (10 of 13) of subjects with baseline indinavir VPs less than sixfold were virologic responders at week 24, whereas 20% (3 of 15) with baseline indinavir VPs more than sixfold were virologic responders at week 24 (P = 0.007). However, the difference between subjects with baseline indinavir VPs less than sixfold and those with indinavir VPs more than sixfold at week 48 (60% [6 of 10 subjects] versus 20% [3 of 15 subjects]) was no longer statistically significant (P = 0.087). No subject with baseline indinavir VPs ≥12-fold responded to intensification of indinavir therapy with ritonavir.
Higher indinavir Ctroughs at week 3 were found to be associated with virologic responses at weeks 24 and 48. In particular, 77% (10 of 13) of subjects with an indinavir Ctrough >0.75 μg/ml were considered virologic responders at week 24, whereas 14% (2 of 14) of subjects with indinavir Ctroughs <0.75 μg/ml were considered virologic responders at week 24 (P = 0.002). Similarly, 70% (7 of 10) of subjects with indinavir Ctroughs >0.75 μg/ml were considered virologic responders at week 48, whereas 7% (1 of 14) with indinavir Ctroughs <0.75 μg/ml were considered virologic responders at week 48 (P = 0.002).
The indinavir vIQ, which is a function of both the week 3 indinavir Ctrough and the baseline indinavir VP, appears to be the best predictor (of the parameters evaluated) of virologic response in this study. At week 3, 89% (16 of 18) of subjects with an indinavir vIQ >2 were considered virologic responders, whereas 11% (1 of 9) of subjects with an indinavir vIQ <2 were considered virologic responders (P < 0.001). Similar results were observed at week 24 (75% [12 of 16 subjects] versus 0% [0 of 9 subjects]; P < 0.001) and week 48 (62% [8 of 13 subjects] versus 0% [0 of 9 subjects]; P = 0.006).
As the ritonavir vIQ was highly correlated with the indinavir vIQ (rho = 0.89), it was not possible to differentiate between the association of the virologic response with the indinavir vIQ and the virologic response with the ritonavir vIQ. A stepwise logistic regression analysis was conducted at weeks 3, 24, and 48, with virologic response as the dependent variable and the following parameters as potential predictor (independent) variables: baseline viral load (log10 transformed), indinavir vIQ (log10 transformed), indinavir VP, the number of PI mutations, the number of NNRTI mutations, the indinavir Ctrough at week 3 (log10 transformed), and the fold change in Ctrough between the baseline and week 3 (log10 transformed), all as continuous variables. Of the independent variables considered for inclusion in the model, only indinavir vIQ entered the model at week 3 (intercept = −2.154; P = 0.012) and week 24 (intercept = −2.925; P = 0.009) (Table 4). At week 48, none of the independent variables was found to be associated with a virologic response (P > 0.10); however, this result may have been due to the small sample size at week 48. In a separate analysis, the ritonavir vIQ (log10 transformed) was substituted for the indinavir vIQ (log10 transformed). At week 3, the indinavir VP was the only significant predictor of viral response (intercept = 2.168; P = 0.015); however, the ritonavir vIQ was the only significant predictor of the viral response at week 24 (intercept = −1.675; P = 0.020). None of the independent variables was found to be associated with a virologic response at week 48 (P > 0.10).
TABLE 4.
Multiple stepwise logistic regression analysis of virologic response
| Stepwise analysis | Study visit | Parameter | Estimate | Standard error | P value | Odds ratio
|
|
|---|---|---|---|---|---|---|---|
| Estimate | 95% CIa | ||||||
| Excluding ritonavir vIQ | Wk 3 | Intercept | 0.614 | 0.609 | 0.313 | ||
| Indinavir vIQ | −2.154 | 0.858 | 0.012 | 0.116 | 0.022, 0.623 | ||
| Wk 24 | Intercept | 1.862 | 0.871 | 0.033 | |||
| Indinavir vIQ | −2.925 | 1.111 | 0.009 | 0.054 | 0.006, 0.473 | ||
| Excluding indinavir vIQ | Wk 3 | Intercept | −1.675 | 0.744 | 0.024 | ||
| Indinavir VP | 2.168 | 0.888 | 0.015 | 8.738 | 1.532, 49.84 | ||
| Wk 24 | Intercept | −0.767 | 0.630 | 0.223 | |||
| Ritonavir vIQ | −1.675 | 0.718 | 0.020 | 0.187 | 0.046, 0.765 | ||
CI, confidence interval.
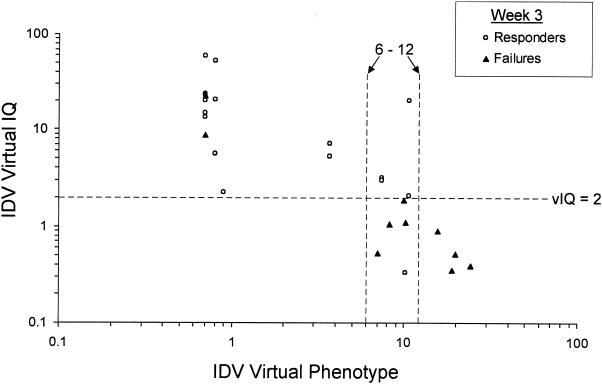
In order to determine where differences in indinavir Ctrough generated by pharmacokinetic boosting might increase the vIQ enough to affect the virologic response to indinavir-ritonavir combination therapy, the baseline indinavir VP was compared to the indinavir vIQ for each subject (Fig. 7). For all subjects for whom the indinavir VP was <6-fold, the vIQ was >2, and for all subjects for whom the VP was >12-fold, the vIQ was <2. The VP range of 6- to 12-fold resulted in the widest variabilities in vIQs, reflecting the variabilities in indinavir Ctroughs. In this 6- to 12-fold change range, 80% (four of five) of subjects for whom the vIQ was <2 were considered virologic nonresponders, while 100% (three of three) of subjects for whom the vIQ was >2 were considered virologic responders. For this group of subjects for whom there was a 6- to 12-fold reduction in susceptibility to indinavir, the vIQ resulting from ritonavir boosting is able to discriminate responders from nonresponders, whereas the VP alone cannot.
FIG. 7.
Comparison of indinavir (IDV) VP and indinavir vIQ.
Safety and tolerability.
Over the duration of the study period 34 (92%) subjects experienced at least one adverse event of moderate severity thought to be related to the study treatment. The most common treatment-related adverse events were diarrhea (46%; 17 of 37 subjects), abdominal pain (19%; 7 of 37 subjects), and weight loss (16%; 6 of 37 subjects) (Table 5). There were two episodes of nephrolithiasis (kidney stones) during the study; however, one of these episodes occurred when a subject erroneously took 800 mg of indinavir in addition to the ritonavir introduced during the first week of indinavir and ritonavir treatment. The most common laboratory abnormalities included grade 3 elevations in triglyceride (51%; 19 of 37 subjects) and cholesterol (41%; 15 of 37 subjects) levels, defined as >750 and >300 mg/dl, respectively.
TABLE 5.
Adverse events and laboratory abnormalitiesa
| Adverse event or laboratory abnormality | No. of subjects |
|---|---|
| Adverse events | |
| Body as a whole | |
| Abdominal pain | 7 |
| Asthenia | 3 |
| Headache | 2 |
| Cardiovascular system, vasodilatation | 2 |
| Digestive system | |
| Anorexia | 2 |
| Diarrhea | 17 |
| Nausea | 2 |
| Metabolic and nutritional disorders | |
| Lipodystrophy | 3 |
| Weight loss | 6 |
| Nervous system, somnolence | 2 |
| Urogenital system, kidney calculus | 2 |
| Laboratory abnormalities | |
| Hematology leukocyte count (<2.5 × 109/liter) | 2 |
| Chemistry | |
| Triglyceride levels >750 mg/dl | 19 |
| Cholesterol levels >300 mg/dl | 15 |
Adverse events refer to events of at least moderate severity with a probable or possible relationship to ritonavir or indinavir, experienced by two or more subjects. Laboratory measurements refer to those that met protocol-defined extreme criteria and that were experienced by two or more subjects. Laboratory results were obtained without regard to fasting conditions.
Nineteen of the 37 subjects enrolled in the study prematurely discontinued the study therapy. Sixteen discontinuations were due to adverse events or HIV-related events, one was due to noncompliance, and two were for personal reasons. The majority of the adverse events leading to the discontinuation of study therapy were related to elevated lipid levels or gastrointestinal symptoms.
DISCUSSION
The first key finding of this study is that reduced susceptibility to a PI can, in part, be overcome by boosting the levels of the drug in plasma. In particular, boosting of indinavir Ctroughs with ritonavir appeared to be the pharmacokinetic parameter most responsible for the improved virologic responses seen in the indinavir-experienced subjects evaluated in the present study. This finding has important implications for the many HIV-infected individuals who have developed some degree of drug resistance. Although boosting of PI regimens has become a standard of care, there are few other studies that have so clearly delineated the impact of pharmacokinetic intensification on the virologic responses in patients with some degree of drug resistance. Enhanced viral suppression was demonstrated in more than half of the subjects in the study, and one-third maintained improved responses over 48 weeks. The virologic response was dependent on both the degree of baseline resistance and the Ctroughs of indinavir that were achieved with combination therapy. Thus, significantly higher response rates were observed in subjects with lower baseline levels of indinavir (those with VPs less than sixfold, those whose viruses had three or fewer PI mutations, and/or those in whom higher indinavir Ctroughs were achieved through boosting with ritonavir (>0.75 μg/ml).
The second key finding of this study is the demonstration of the predictive value of the vIQ above and beyond that provided by resistance testing. The vIQ, a ratio that takes into consideration both the baseline resistance and the indinavir trough concentrations achieved with combination therapy, was the best predictor of a virologic response. A previous study with Pr inhibitor-experienced subjects found that IQ is better at predicting the virologic response to lopinavir- and ritonavir-based therapy than pharmacokinetic measurements alone are (D. J. Kempf, A. Hsu, J. Isaacson, P. Jiang, S. Brun, C. Renz, G. R. Granneman, and E. Sun, Program Abstr. 2nd Int. Workshop Clin. Pharmacol. HIV Ther., abstr. 7.3, 2001). In the present study, the vIQ was used due to the number of baseline samples that had HIV RNA levels too low to be analyzed for isolate phenotype; the isolates could, however, be analyzed for their genotypes. VPs were highly correlated to actual phenotypes both in the subjects whose isolates' phenotypes were analyzed in this study and in a cross-sectional study of a large database (Larder et al., Program Abstr. 4th Int. Workshop HIV Drug Resist. Treatment Strategies, abstr. 63, 2000); therefore, vIQ is likely to be a reasonable proxy for IQ.
As seen in a study with healthy volunteers (11), administration of indinavir at 400 mg BID with ritonavir at 400 mg BID enhanced the indinavir Ctrough and reduced the indinavir Cmax without altering the AUC. While previous studies have shown the cross-sectional association of the virologic response with a higher Ctrough (1, 5, 6, 9, 15), the discrimination of Ctrough, Cmax, and AUC as pharmacodynamic predictors for PI activity is generally not feasible due to the correlation between these parameters. This is the first longitudinal study to demonstrate that the Ctrough is associated with the in vivo antiviral activities of PIs, independent of Cmax and AUC.
One potential limitation to this study is that pharmacologically active doses of ritonavir were administered, which may have contributed to the virologic suppression. However, the majority of samples analyzed had ritonavir vIQs of <1 (Fig. 5). Furthermore, the ritonavir and indinavir vIQs were highly correlated, and the median indinavir vIQ was >20-fold higher than that of ritonavir. On the basis of the results of this pharmacodynamic analysis, it is likely that the increased potency observed in this study can be largely, if not exclusively, attributed to indinavir. This is supported by the results of the stepwise regression analysis, in which the indinavir vIQ was a stronger predictor of response than the ritonavir vIQ. The association of the week 24 response obtained with the ritonavir vIQ when the indinavir vIQ was excluded from the model is likely to represent a surrogate for the stronger association with indinavir vIQ.
Pharmacokinetic enhancement in this study came at a cost of substantial toxicity. More than half of the subjects discontinued therapy due to adverse events related to ritonavir or increased indinavir exposure, or both. Kidney stones were rare at these doses, supporting the hypothesis that nephrolithiasis and the nephropathy of indinavir are related to the Cmax (4). Identification of the optimal doses of indinavir and ritonavir required to maximize antiretroviral activity yet minimize toxicity when the drugs are used in combination requires further exploration.
In the clinical setting, measurement of Ctroughs of drugs, in addition to performance of resistance testing, may be useful for maximizing patient responses to PI-based therapies. Future studies, including studies with other PIs, are needed to confirm these findings.
Acknowledgments
The efforts of the following persons in support of this study are gratefully acknowledged: Jose Montoya and Michael Harbor (Stanford University); Kathy Nuffer (University of California, San Diego); Michael Allison, Suzanne Dunphy, Michelle Parish, Charles Raines, and Andrea Weiss (Johns Hopkins University); Skip Barnes, Phil Keiser, and Azadeh Mozaffari (University of Texas Southwestern Medical Center); Abigail Kelly, Karen Nowatkowski, Denise Robisch, and Nancy Underwood (Quintiles); and Anthony Japour, Jerri Swerdlow, Janet Lamm, Joan Ryan, Lourdes Manning, and Guang Yang (Abbott Laboratories).
This study was supported in part by NIH grant M01 RR 00070. Funding for this study was obtained from Abbott Laboratories (study M98-985). A.H., C.R., S.B., P.J., R.R. D.J.K. and E.S. are employees of Abbott Laboratories. Other than study support, the remaining authors (N.S., A.Z., D.H., J.G., and E.R.) received no other financial support from the sponsor.
REFERENCES
- 1.Acosta, E. P., K. Henry, L. Baken, L. M. Page, and C. Fletcher. 1999. Indinavir concentrations and antiviral effect. Pharmacotherapy 19:708-712. [DOI] [PubMed] [Google Scholar]
- 2.Burger, D. M., R. M. W. Hoetelmans, P. W. H. Hugen, J. W. Mulder, P. L. Meenhorst, P. P. Koopmans, K. Brinkman, M. Keuter, W. Dolmans, and Y. A. Hekster. 1998. Low plasma concentrations of indinavir are related to virological treatment failure in HIV-1-infected patients on indinavir-containing triple therapy. Antivir. Ther. 3:215-220. [PubMed] [Google Scholar]
- 3.DeGruttola, V., L. Dix, R. D'Aquila, D. Holder, A. Phillips, M. Ait-Khaled, J. Baxter, P. Clevenbergh, S. Hammer, R. Harrigan, D. Katzenstein, R. Lanier, M. Miller, M. Para, S. Yerly, A. Zolopa, J. Murray, A. Patrick, V. Miller, S. Castillo, L. Pedneault, and J. Mellors. 2000. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antivir. Ther. 5:41-48. [DOI] [PubMed] [Google Scholar]
- 4.Dieleman, J. P., I. C. Guyssens, M. E. van der Ende, S. de Marie, and D. M. Burger.1999. Urological complaints in relation to indinavir plasma concentrations in HIV-infected patients. AIDS 13:473-478. [DOI] [PubMed] [Google Scholar]
- 5.Dumon, C., C. Solas, I. Thuret, H. Chambost, B. Lacarelle, G. Michel, and A. Durand. 2000. Relationship between efficacy, tolerance, and plasma drug concentration of ritonavir in children with advanced HIV infection. Ther. Drug Monit. 22:402-408. [DOI] [PubMed] [Google Scholar]
- 6.Durant, J., P. Clevenbergh, R. Garraffo, P. Halfon, S. Icard, P. Del Giudice, N. Montagne, J. M. Schapiro, and P. Dellamonica. 2000. Importance of protease inhibitor plasma levels in HIV-infected patients treated with genotypic-guided therapy: pharmacological data from the Viradapt Study. AIDS 14:1333-1339. [DOI] [PubMed] [Google Scholar]
- 7.Ellner, P. D., and H. C. Neu1981. The inhibitory quotient. A method for interpreting minimum inhibitory concentration data. JAMA 246:1575-1578. [DOI] [PubMed] [Google Scholar]
- 8.Flexner, C. 2000. Dual protease inhibitor therapy in HIV-infected patients: pharmacologic rationale and clinical benefits. Annu. Rev. Pharmacol. Toxicol. 40:649-674. [DOI] [PubMed] [Google Scholar]
- 9.Haas, D. W., E. Arathoon, M. A. Thompson, R. de Jesus Pedro, J. E. Gallant, D. E. Uip, J. Currier, L. M. Noriega, D. S. Lewi, P. Uribe, L. Benetucci, P. Cahn, D. Paar, A. C. White, Jr., A. C. Collier, C. H. Ramirez-Ronda, C. Harvey, M. O. Chung, D. Mehrotra, J. Chodakewitz, and B. Y. Nguyen. 2000. Comparative studies of two-times-daily versus three-times-daily indinavir in combination with zidovudine and lamivudine. AIDS 14:1973-1978. [DOI] [PubMed] [Google Scholar]
- 10.Hertogs, K., M. P. de Bethune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels.1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu, A., G. R. Granneman, G. Cao, L. Carothers, A. Japour, T. El-Shourbagy, S. Dennis, J. Berg, K. Erdman, J. M. Leonard, and E. Sun. 1998. Pharmacokinetic interaction between ritonavir and indinavir in healthy volunteers. Antimicrob. Agents Chemother. 42:2784-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakuda, T. N., L. M. Page, P. L. Anderson, K. Henry, T. W. Schacker, F. S. Rhame, E. P. Acosta, R. C. Brundage, and C. V. Fletcher. 2001. Pharmacological basis for concentration-controlled therapy with zidovudine, lamivudine, and indinavir. Antimicrob. Agents Chemother. 45:236-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kempf, D. J., K. C. Marsh, G. Kumar, A. D. Rodrigues, J. F. Denissen, E. McDonald, M. J. Kukulka, A. Hsu, G. R. Granneman, B. A. Baroldi, E. Sun, D. Pizzuti, J. J. Plattner, D. W. Norbeck, and J. M. Leonard. 1997. Pharmacokinetic enhancement of inhibitors of the human immunodeficiency virus protease by coadministration with ritonavir. Antimicrob. Agents Chemother. 41:654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lalezari, J. 1998. Selecting the optimum dose for a new soft gelatin capsule formulation of saquinavir. NV15107 Study Group. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:195-197. [DOI] [PubMed] [Google Scholar]
- 15.Lorenzi, P., S. Yerly, K. Abderrakim, M. Fathi, O. T. Rutschmann, J. von Overbeck, D. Leduc, L. Perrin, B. Hirschel, et al. 1997. Toxicity, efficacy, plasma drug concentrations and protease mutations in patients with advanced HIV infection treated with ritonavir plus saquinavir. AIDS 11:F95-F99. [DOI] [PubMed] [Google Scholar]
- 16.Molla, A., S. Vasavanonda, G. Kumar, H. L. Sham, M. Johnson, B. Grabowski, J. F. Denissen, W. Kohlbrenner, J. J. Plattner, J. M. Leonard, D. W. Norbeck, and D. J. Kemp. 1998. Human serum attenuates the activity of protease inhibitors toward wild-type and mutant human immunodeficiency virus. Virology 250:255-262. [DOI] [PubMed] [Google Scholar]
- 17.Slain, D., A. Pakyz, D. S. Israel, S. Monroe, and R. E. Polk. 2000. Variability in activity of hepatic CYP3A4 in patients infected with HIV. Pharmacotherapy 20:898-907. [DOI] [PubMed] [Google Scholar]
- 18.Winters, M. A., J. M. Schapiro, J. Lawrence, and T. C. Merigan. 1998. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long term saquinavir treatment. J. Virol. 72:5303-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou, X. J., D. V. Havlir, D. D. Richman, E. P. Acosta, M. Hirsch, A. C. Collier, P. Tebas, and J. P. Sommadossi. 2000. Plasma population pharmacokinetics and penetration into cerebrospinal fluid of indinavir in combination with zidovudine and lamivudine in HIV-1-infected patients. AIDS 14:2869-2876. [DOI] [PubMed] [Google Scholar]