Abstract
Oseltamivir carboxylate is a potent and specific inhibitor of influenza neuraminidase (NA). An influenza A/H1N1 variant selected in vitro with reduced susceptibility to oseltamivir carboxylate contains a His274Tyr mutation. To understand the mechanism by which a His274Tyr mutation gives rise to drug resistance, we studied a series of NA variant proteins containing various substitutions at position 274. Replacement of His274 with larger side chain residues (Tyr or Phe) reduced the NA sensitivity to oseltamivir carboxylate. In contrast, replacement of His274 with smaller side chain residues (Gly, Asn, Ser, and Gln) resulted in enhanced or unchanged sensitivity to oseltamivir carboxylate. Previous studies have suggested that the slow-binding inhibition of NA by oseltamivir carboxylate is a result of the reorientation of Glu276. Loss of this slow-binding inhibition in the His274Tyr and His274Phe mutant NA but not in His274Asn, His274Gly, His274Ser, or His274Gln supports the conclusion that the conformational change of Glu276 is restricted in the His274Tyr and His274Phe mutant NA upon oseltamivir carboxylate binding. Interestingly, His274Asn, as well as His274Gly, His274Ser, and His274Gln, also displayed reduced sensitivity to zanamivir and its analogue, 4-amino-Neu5Ac2en. Substitution of His274 with Tyr in influenza A/Tokyo/3/67 (H3N2) recombinant NA did not affect the susceptibility to oseltamivir carboxylate. These data indicate that the volume occupied by the amino acid side chain at position 274 can influence the sensitivities of influenza N1 NA but not of N2 NA to both oseltamivir carboxylate and zanamivir.
Influenza continues to be a significant health concern, and reemergence of pandemics is a constant threat (8, 24, 47). Until late 1999, two closely related compounds, amantadine and rimantadine, were the only antiviral drugs approved for clinical use for the prevention and treatment of influenza A virus infections. However, the clinical usefulness of these drugs is limited by the lack of activity against influenza B viruses and the rapid and frequent emergence of drug-resistant viruses that remain transmissible and pathogenic (15, 33).
The influenza neuraminidase (NA), a viral surface glycoprotein, has long been considered a valid target for antiviral therapy (36). This enzyme, which cleaves terminal sialic acid residue from glycoconjugates, is essential for virus proliferation and infectivity. The amino acid residues in the enzyme active site are highly conserved among different influenza NA subtypes (2-4), and NA inhibitors (NAI) have been shown to have antiviral activities against a broad range of influenza viruses (43-45).
Several potent and selective inhibitors, e.g., oseltamivir carboxylate (Ro64-0802, GS4071) and zanamivir (Relenza, GG167), of the influenza NA have been discovered through structure-based rational drug design (19, 21, 46). Oseltamivir carboxylate, the active metabolite of oseltamivir phosphate (Tamiflu, Ro64-0796, GS4104), is a potent and specific inhibitor of influenza A and B virus NA (26, 27, 48). Oseltamivir has been approved for the prevention and treatment of influenza virus infection in adults and for the treatment of influenza infection in children (14, 16).
Development of drug resistance is a potential concern for all antiviral agents (28, 31, 32, 38, 39). The potential for drug resistance to emerge rapidly in influenza virus has been demonstrated through previous clinical experience with amantadine and rimantadine (15). The high incidence of resistance with amantadine and rimantadine is related to their mechanism of action (13). These two closely related drugs function by inhibiting the ion channel activity of the influenza A viral membrane M2 protein. The drug-resistant mutant M2 proteins still possess normal ion channel activity. Thus, resistance to these agents can arise without compromising viral function. Hence the drug-resistant viruses are transmissible and pathogenic. In contrast, oseltamivir carboxylate binds specifically to the highly conserved active site of NA, and NA mutations conferring reduced susceptibilities to oseltamivir carboxylate would be expected to diminish the enzymatic activity (22, 23). Thus, emergence of resistance to oseltamivir carboxylate through NA mutations would be disadvantageous to the virus, and therefore such resistant viruses would be expected to be self limited in infectivity.
Influenza variants with reduced susceptibility to oseltamivir carboxylate have been generated in vitro in cell culture by applying increasing selective pressure of the drug over a prolonged period of time (40, 42). The most common mutation selected by exposure to NAI in vitro was an amino acid substitution at position 292 (Arg292Lys) in influenza A N2 NA. The Arg292Lys mutation has been selected by oseltamivir carboxylate and zanamivir, as well as BCX-1812 (1, 6, 11, 30, 42). Virus carrying the Arg292Lys mutation is significantly attenuated in infectivity and pathogenicity in mice and ferrets (5). Another resistant mutation, at conserved position 119, has been identified in vitro in the active site of influenza A N2 NA under selection pressure with zanamivir (7, 29). Virus carrying a zanamivir-selected mutation at the Glu119 position was also attenuated in the infectivity and pathogenicity in mice and ferrets. No mutation was selected by oseltamivir carboxylate at position 119 in vitro, but virus carrying the Glu119Val mutation has been identified in two patients infected with influenza A/H3N2 and treated with oseltamivir phosphate (9, 48). The virus carrying the Glu119Val mutation has dramatically reduced infectivity in vivo (17). An influenza A/Texas H1N1 variant containing a His274Tyr mutation in NA has been isolated from a challenge study (9), and it has been shown to have substantially reduced infectivity and pathogenicity in mice and ferrets (18). There has been no success generating a mutation in influenza B NA by oseltamivir carboxylate in vitro or in vivo, though zanamivir induced the development of a resistant mutation in influenza B NA in an immunocompromised child (10).
Here we report the characterization of an influenza A/WS/33 (H1N1) virus variant carrying a His274Tyr mutation in NA selected in vitro in the presence of oseltamivir carboxylate and identification of the mechanism by which mutations at His274 alter the sensitivity of influenza A virus N1 NA to oseltamivir carboxylate and zanamivir.
MATERIALS AND METHODS
Inhibitors.
The NAI, oseltamivir carboxylate ([3R,4R,5S]-4-acetamido-5-amino-3-[1-ethylpropoxy]-1-carboxylic acid), zanamivir (4-guanidino-Neu5Ac2en), and its 4-amino analogue (4-amino-Neu5Ac2en), were synthesized at Gilead Sciences. Chemical structures are shown in Fig. 1.
FIG. 1.
Chemical structures of sialic acid and influenza NAI.
Viruses and cells.
Influenza A/WS/33 (H1N1) virus, Madin-Darby canine kidney (MDCK) cells, and HeLa cells were from the American Type Culture Collection. MDCK cells and HeLa cells were grown in Eagle's minimum essential medium containing Earle's salts and Dulbecco's modified Eagle's medium, respectively, supplemented with 10% fetal bovine serum (Irvine Scientific) at 37°C in a 5% CO2-containing atmosphere.
In vitro selection of mutant virus with decreased susceptibility to oseltamivir carboxylate.
Influenza A/WS/33 (H1N1) virus was passaged in MDCK cells in 24-well tissue culture plates in the presence or absence of oseltamivir carboxylate, as previously described (42). Virus was allowed to replicate until >90% death of the cell monolayer was observed or until the infection no longer progressed. Selections were started at low concentrations of oseltamivir carboxylate, and the concentration of inhibitor was increased two- or threefold at each subsequent passage. The inhibition of virus replication by oseltamivir carboxylate was determined using a plaque reduction assay in MDCK cells. The variant clones were isolated from the virus pools through three rounds of plaque purification in the presence of oseltamivir carboxylate.
Viral RNA samples were prepared from a viral supernatant of cell culture (42). The NA and hemagglutinin genes were amplified by reverse transcriptase PCR and sequenced. The DNA sequences were analyzed with DNAStar software. Amino acid residue numbering for NA was assigned according to homology with the N2 NA sequence.
Expression of influenza NA.
A plasmid containing the full-length cDNA encoding influenza virus A/Tokyo/3/67 NA (N2) was a generous gift from Gillian Air (University of Oklahoma) (25). The NA gene of influenza virus A/WS/33 was cloned by reverse transcriptase PCR, with the following primers: 5′ GATCAAGCTT GAAAATGAAT CCAAACCAGA AAATA 3′ and 5′ GATCGAATTC CTACTTGTCA ATGGTGAACG G 3′. PCR fragments of NA genes were cloned into the pcDNA3 vector (Invitrogen) at HindIII and EcoRI sites. The pcDNA3 expression plasmids containing NA genes were transfected into HeLa cells using Lipofectamine reagents (Gibco BRL).
Site-directed mutagenesis was conducted with the QuickChange mutagenesis kit (Stratagene). All the sequences were confirmed by automated ALFexpress sequencing (Amersham Pharmacia Biotech).
NA enzymatic activity assay.
HeLa cells transfected with expression plasmids for NA were harvested by treatment with phosphate-buffered saline at pH 7.4 containing 0.02% EDTA at 72 h posttransfection. The cell pellets were then washed twice with the enzyme assay buffer containing 33 mM morpholineethanesulfonic acid (pH 6.5), 4 mM CaCl2, and 120 mM NaCl. NA enzymatic activity was measured as previously described using the fluorogenic substrate 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUNANA) (Sigma) (42). All the reactions were conducted at 37°C in white 96-well plates (DYNEX) in a total volume of 200 μl, and the fluorescence was monitored every minute for 60 min using a GEMINI spectrofluorometer (Molecular Devices) with excitation and emission wavelengths of 330 and 445 nm, respectively.
To measure the kinetic parameters of slow-binding inhibition, the reactions were initiated by adding enzyme to a prewarmed mixture of substrate and inhibitors at 37°C. The progression data were fitted to the equation below:
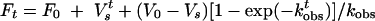 |
(1) |
where F0 and Ft represent the fluorescent signal values at time zero and time t, respectively; and where V0, Vs, and kobs represent the initial velocity, the steady-state velocity, and the first-order transition rate constant from initial to steady state, respectively (12, 20, 34, 35, 37). The association rate constant (kon) and dissociation rate constant (koff) were then determined with the equation below:
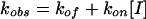 |
(2) |
RESULTS
Isolation of an A/WS/33 variant with decreased susceptibility to oseltamivir carboxylate.
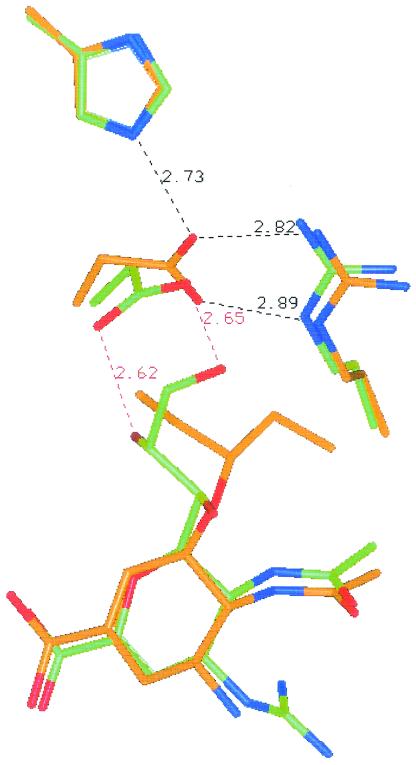
The human influenza A/WS/33 (H1N1) virus was passaged in MDCK cells in the presence of concentrations of oseltamivir carboxylate that were increased two- or threefold at each passage. Two separate selections were conducted, with different starting inhibitor concentrations of 0.6 and 90 nM oseltamivir carboxylate. When the final selection step, the 14th passage in study I (550 μM oseltamivir carboxylate) and the 11th passage in study II (400 μM oseltamivir carboxylate), respectively, was reached, the susceptibilities of the virus pools to oseltamivir carboxylate were examined in plaque reduction assays. Both virus pools displayed a dramatically decreased (4,000-fold) susceptibility to oseltamivir carboxylate in plaque reduction assays. Genotypic analysis of plaque-purified variants did not detect any mutation in the hemagglutinin gene but revealed a single mutation in the NA gene, resulting in a His→Tyr substitution at amino acid 274 (His274Tyr). This conserved His residue is located very near the conserved residue Glu276 in the enzyme active site (see Fig. 5).
FIG. 5.
Interaction of oseltamivir carboxylate (brown) and zanamivir (green) with amino acid residues located in the active site of influenza A NA (N9).
The effect of the His274Tyr NA mutation on the properties of the NA enzyme was determined by comparing the wild-type and mutant enzyme activity in an in vitro assay using the fluorogenic substrate MUNANA. As shown in Fig. 2A and Table 1, the affinity of the mutant NA for the substrate was reduced by less than twofold relative to that for the wild-type enzyme as indicated by the increase in Km. The His274Tyr mutation also affected the activity of the enzyme, causing a twofold decrease in the turnover rate as indicated by measurements of Vmax. These assays were performed with a standardized amount of detergent-solubilized virus based on measurements of total virus protein and hemagglutinating activity. Based on these data, the catalytic efficiency (Vmax/Km) of the mutant NA was approximately 30% of that of the wild-type enzyme.
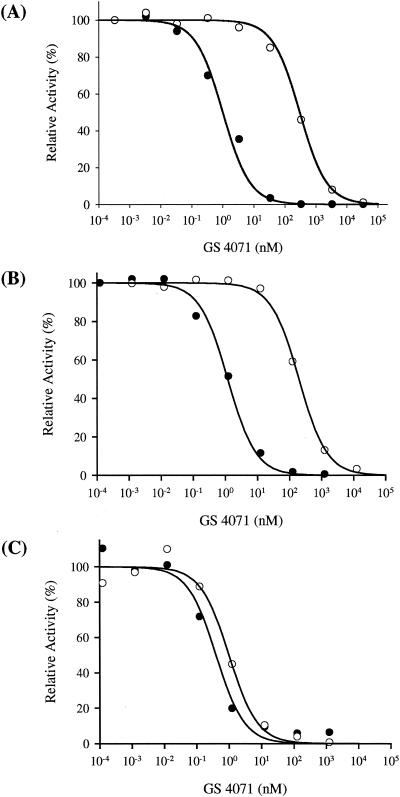
FIG. 2.
Inhibition of wild-type (solid circle) and H274Y mutant (open circle) influenza NA by oseltamivir carboxylate. (A) Native NA from A/WS/33 (H1N1) virus. (B) Recombinant A/WS/33 (H1N1) NA expressed in HeLa cells. (C) Recombinant A/Tokyo/3/67 (H3N2) NA expressed in HeLa cells.
TABLE 1.
Characterization of A/WS/33 (H1N1) virus-associated NA enzymea
| Enzyme | Substrate (MUNANA)
|
Ki (nM)
|
||
|---|---|---|---|---|
| Km (μM) | Relative Vmax | Oseltamivir carboxylate | Zanamivir | |
| Wild type (274His) | 31 | 1 | 0.3 | 0.1 |
| Mutant (274Tyr) | 52 | 0.5 | 10.5 | 0.3 |
Values were means of data from three or more independent experiments. Standard deviations were less than 6% of the mean value.
The sensitivity of the wild-type and mutant NA to inhibition by oseltamivir carboxylate and zanamivir was also determined. As shown in Table 1, the inhibition constant (Ki) for oseltamivir carboxylate was 300-fold higher for the mutant enzyme than for the wild-type enzyme. In contrast, there was only a threefold increase in the Ki for zanamivir.
Characterization of recombinant influenza NA expressed in mammalian cells.
In order to understand the biochemical mechanism of drug resistance of NA containing the His274Tyr mutation, an in vitro heterologous expression system was established to express NA in mammalian cells. The wild-type recombinant influenza A/WS/33 (H1N1) NA expressed in HeLa cells displayed substrate binding affinity similar to that of the native viral NA (Table 1; also see Table 3).
TABLE 3.
Summary of recognition of substrate (Km) and inhibitors (Ki) by wild-type and mutant influenza A/WS/33 (H1N1) NAa
| Type of recombinant NAb (A/WS/33 H1N1) | Km (μM) of MUNANA |
Ki (nM)
|
||
|---|---|---|---|---|
| Oseltamivir carboxylate | Zanamivir | 4-Amino-Neu5Ac2en | ||
| Wild type (His274) | 31 | 0.3 | 0.1 | 50 |
| His274Gly | 100 | 0.2 | 5.7 | 1,140 |
| His274Ser | 110 | 0.1 | 14.9 | 890 |
| His274Asn | 133 | 0.1 | 6.0 | 1,250 |
| His274Gln | 125 | 0.5 | 3.8 | 940 |
| His274Phe | 57 | 86 | 0.4 | 130 |
| His274Tyr | 55 | 105 | 0.3 | 150 |
Values were means of data from three or more independent experiments. Standard deviations were less than 10% of the mean value.
In ascending order of amino acid side chain volume.
The recombinant wild-type influenza A/WS/33 (H1N1) NA displayed susceptibility to oseltamivir carboxylate similar to that of the native viral NA (Fig. 2A and B). Replacement of His274 with Tyr in A/WS/33 (H1N1) recombinant NA resulted in a 200-fold reduced susceptibility to oseltamivir carboxylate, which is comparable to the reduced sensitivity observed with the mutant viral enzyme (Table 2). However, replacement of His274 by Tyr in A/Tokyo/3/67 (H3N2) recombinant NA did not affect the susceptibility of N2 NA to oseltamivir carboxylate (Fig. 2C and Table 2). These data suggest that the involvement of His274 in the binding of oseltamivir carboxylate to N2 NA is different from that in N1 NA.
TABLE 2.
Fifty percent inhibitory concentrations for the inhibition of NA by oseltamivir carboxylatea
| Virus | IC50 (nM)b | |
|---|---|---|
| Native viral NA (A/WS/33, H1N1) | ||
| Wild type | 1.0 | |
| His274Tyr | 300 | |
| Recombinant NA (A/WS/33, H1N1) | ||
| Wild type | 1.1 | |
| His274Tyr | 200 | |
| Recombinant NA (A/Tokyo/3/67, H3N2) | ||
| Wild type | 0.4 | |
| His274Tyr | 1.0 |
Values were means of data from three or more independent experiments. Standard deviations were less than 8% of the mean value.
IC50, 50% inhibitory concentration.
Effect of side chain volume on Km and Ki.
To understand the mechanism of reduced susceptibility to oseltamivir carboxylate due to the His274Tyr mutation in N1 NA, we systematically replaced His274 with amino acids containing either a basic side chain (Arg), an acidic side chain (Asp), uncharged polar side chains (Asn, Gln, Ser, and Thr), or uncharged nonpolar side chains (Ala, Val, Gly, and Phe). NA variants containing the His274Ala, His274Val, His274Thr, His274Arg, and His274Asp mutations did not display NA activity, possibly due to reduced protein expression, disruption of protein conformation, or posttranslational processing as a result of the mutations.
As shown in Table 3, the binding affinity for the substrate (Km) to the NA active site is dependent on the side chain volume of amino acid at position 274, with smaller side chains (Gly, Ser, Asn, and Gln) displaying Km values approximately twofold higher than do larger side chains (Tyr and Phe) (41). The wild-type NA with His at position 274 displays the highest substrate binding (lowest Km) (Fig. 3A).
FIG. 3.
Influenza NA recognition of substrate (MUNANA) (A) and inhibitors oseltamivir carboxylate (B), zanamivir (C), and 4-amino-Neu5Ac2en (D) is affected by the side chain volume of amino acid at position 274 in A/WS/33 (H1N1).
In contrast to the effect seen on binding affinity of the substrate, the opposite effect of a mutation at position 274 is seen for binding of oseltamivir carboxylate to NA (Table 3). As shown in Fig. 3B, replacement of His274 with residues bearing side chains with larger volumes (Tyr or Phe) resulted in 200- to 300-fold-reduced sensitivity to oseltamivir carboxylate, whereas replacement of His274 with residues bearing side chains with smaller volumes (Gly, Asn, Ser, or Gln) resulted in either unchanged or slightly increased sensitivity to oseltamivir carboxylate.
Slow-binding inhibition of oseltamivir carboxylate.
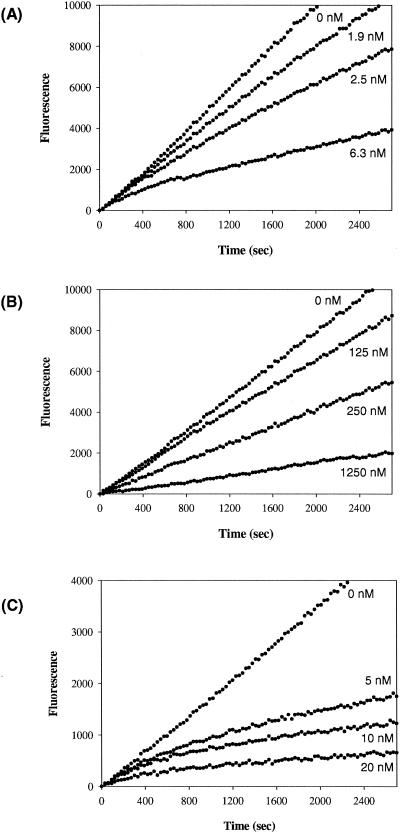
Previous studies have shown that oseltamivir carboxylate is a slow-binding inhibitor of both influenza A N2 NA and influenza B NA (12, 20, 37, 42). As shown in Fig. 4A, oseltamivir carboxylate is a slow-binding inhibitor of influenza A N1 NA. The apparent kon and koff values for the wild-type A/WS/33 NA were 5.8 × 105 M−1 s−1 and 5.7 × 10−4 s−1 (Table 4,) respectively, which are comparable with those reported previously for A/Tokyo/3/67 and B/Memphis/3/89 NA (12, 20, 37).
FIG. 4.
Evaluation of the slow-binding inhibition kinetics of wild-type (inhibitor concentrations of 0, 1.9, 2.5, and 6.3 nM) (A), H274Y mutant (inhibitor concentrations of 0, 125, 250, and 1,250 nM) (B), and H274N mutant (inhibitor concentrations of 0, 5, 10, and 20 nM) (C) recombinant NA of A/WS/33 (H1N1) by oseltamivir carboxylate.
TABLE 4.
Summary of kinetic parameters for the interaction of oseltamivir carboxylate with different variants of influenza N1 NAb
| Mutation | kon (M−1 s−1) | koff (s−1) | Ki (nM)a |
|---|---|---|---|
| Wild type (His274) | 5.8 × 105 | 5.7 × 10−4 | 1.0 (0.3) |
| His274Asn | 1.7 × 106 | 1.8 × 10−4 | 0.1 (0.1) |
| His274Gly | 1.8 × 106 | 3.7 × 10−4 | 0.2 (0.2) |
| His274Ser | 1.1 × 106 | 5.8 × 10−4 | 0.5 (0.1) |
| His274Gln | 1.7 × 106 | 5.7 × 10−4 | 0.3 (0.5) |
The values in parentheses were obtained from experiments designed for direct measurement of Ki.
Values were means of data from three or more independent experiments. Standard deviations were less than 7% of the mean value.
Therefore, the effect of mutations of His274Tyr, His274Gly, His274Ser, His274Asn, and His274Gln on the slow-binding inhibition behavior of oseltamivir carboxylate was investigated. As shown in Fig. 4B, His274Tyr mutant NA did not display slow-binding inhibition by oseltamivir carboxylate, lacking the time-dependent change in the rate of substrate hydrolysis in reactions containing oseltamivir carboxylate. All other four substitutions with smaller side chain volumes at position 274 (Gly, Ser, Asn, and Gln) displayed slow-binding inhibition by oseltamivir carboxylate. For example, the kinetics of the slow-binding inhibition of His274Asn mutant NA by oseltamivir carboxylate is shown in Fig. 4C.
DISCUSSION
A mutation in influenza A virus containing N1 NA, His274Tyr, has been identified in our in vitro selection studies with oseltamivir carboxylate. In the two independent in vitro selection studies, influenza A/WS/33 (H1N1) virus developed a His274Tyr mutation in the NA gene. The mutant NA enzyme exhibited 300-fold-reduced sensitivity to oseltamivir carboxylate. However, we were not able to select the His274Tyr mutation in influenza A N2 NA despite our extensive in vitro drug selection studies.
The establishment of an in vitro expression system for NA provided us a means to explore the molecular mechanisms of drug resistance in N1 NA containing the His274Tyr mutation and to assess whether the susceptibility of N2 NA to oseltamivir carboxylate would be affected by the His274Tyr mutation. Our data indicated that the replacement of His274 with Tyr in influenza A/WS/33 (H1N1) recombinant NA resulted in a 200-fold-reduced susceptibility to oseltamivir carboxylate, whereas the same replacement (His274 with Tyr) in influenza A/Tokyo/3/67 (H3N2) recombinant NA did not affect the susceptibility to oseltamivir carboxylate. These data explain our in vitro selection observation that the influenza A variant containing His274Tyr mutation in NA could only be selected in influenza A viruses with N1 NA but not with N2 NA.
These in vitro observations of a difference between N1 and N2 NA in terms of resistance profile are consistent with the observations from in vivo studies that resistance mutations to NAI in influenza NA are subtype specific and that the resistance profile from in vitro analysis is highly predictive of that of viruses derived from clinical studies (38, 39).
Our data indicated that the catalytic activity (Vmax/Km) of the His274Tyr mutant NA was only 30% of that of the wild-type enzyme, suggesting that the infectivity and replicative capacity of the mutant virus containing the His274Tyr substitution in NA may be compromised in vivo. Not surprisingly, a purified clinical isolate (influenza A/Texas H1N1) containing the His274Tyr mutation in NA derived from a challenge study (9) has been shown to have substantially reduced infectivity and pathogenicity in mice and ferrets (18).
The observations from systematic substitutions of His274 with amino acid residues containing different side chain properties (charge, polarity, and volume) led us to hypothesize that introducing amino acid residues with different side chain volumes at position 274 would shift the equilibrium between the two orientation states of the side chain of Glu276, as illustrated in Fig. 5. Introducing a bulky side chain residue at position 274 would block the reorientation of Glu276 required for the high-affinity binding of oseltamivir carboxylate to NA, resulting in reduced sensitivity to oseltamivir carboxylate. Conversely, introducing a smaller side chain residue at position 274 would create the conformation of NA favoring the reorientation of Glu276 even in the absence of oseltamivir carboxylate, which would result in an unfavorable binding pocket for substrate MUNANA due to the hydrophobicity of the pocket. If this hypothesis stands, we would expect to observe an opposite effect of side chain volume on sensitivity to zanamivir and 4-amino-Neu5Ac2en, since both of these compounds possess glycerol-like moiety that is the same as for the natural substrate, sialic acid (Fig. 1). As predicted, replacement of His274 with residues bearing side chains with smaller volumes (Gly, Asn, Ser, and Gln) resulted in substantially reduced sensitivity to zanamivir (Fig. 3C) and 4-amino-Neu5Ac2en (Fig. 3D), whereas replacement of His274 with residues bearing side chains with larger volumes (Tyr or Phe) resulted in slightly decreased sensitivity to zanamivir and 4-amino-Neu5Ac2en.
As shown in Fig. 5, a hydrogen bond is formed between His274 and reoriented Glu276 in the cocrystal structure of oseltamivir carboxylate and N9 NA. However, our mutagenesis studies suggested that the binding affinity of oseltamivir carboxylate to N1 NA is determined by the side chain volume of amino acid residue at position 274, not by the capability of hydrogen-bonding formation. This observation suggests that analysis of resistance to NAI in N9 NA might not be used to predict NAI resistance in other NA subtypes.
According to the above results, one would expect that another mutation at position 274, His274Phe, could arise from the in vitro selection studies with oseltamivir carboxylate. We reasoned that the failure to isolate the His274Phe mutation might be due to the fact that switching from His to Tyr requires only one nucleotide change in a viral codon, whereas switching from His to Phe requires two nucleotide changes in a codon.
Previous studies have shown that oseltamivir carboxylate is a slow-binding inhibitor of both influenza A N2 NA and influenza B NA. Our data indicated that oseltamivir carboxylate is also a slow-binding inhibitor to influenza A N1 NA. It has been shown previously that an Arg292Lys mutation of influenza A N2 NA reduced the sensitivity of the mutant NA to oseltamivir carboxylate and also abolished the slow-binding inhibition of the mutant NA by oseltamivir carboxylate (42). It has been postulated that the reduced sensitivity to oseltamivir carboxylate and the loss of the slow-binding inhibition by oseltamivir carboxylate of the Arg292Lys mutant NA might be due to the lack of conformational change of Glu276 upon oseltamivir carboxylate binding. This might be a result of the interaction between the side chain of the Lys292 residue and the side chain of the Glu276 residue in the Arg292Lys mutant NA being stronger than in the wild-type NA.
As stated earlier, the side chain volume of amino acid residue at position 274 affects the sensitivity of NA to oseltamivir carboxylate and this might be due to the conformational change of the Glu276 residue upon oseltamivir carboxylate binding. Therefore we hypothesized that mutations at position 274 would also have an effect on the kinetics of inhibition by oseltamivir carboxylate. Our data indicated that the His274Tyr mutation abolished the slow-binding inhibition of NA by oseltamivir carboxylate. Conversely, all the mutations with smaller side chain volumes at positions 274, His274Gly, His274Ser, His274Asn, and His274Gln displayed slow-binding inhibition by oseltamivir carboxylate. These observations are consistent with the hypothesis that the conformational change of Glu276 upon binding of oseltamivir carboxylate to NA is required for the slow-binding inhibition of NA by oseltamivir carboxylate.
Both the wild-type and the His274Tyr mutant NA of influenza A/Tokyo/3/67 (H3N2) virus displayed slow-binding inhibition by oseltamivir carboxylate (data not shown), suggesting that the conformational change of Glu276 upon oseltamivir carboxylate binding might still occur in the His274Tyr mutant N2 NA. These data further suggest that the microenvironment surrounding the active site in N2 NA might be different from that in N1 NA.
The apparent kon and koff for the wild-type and mutant NA of influenza A/WS/33 suggest that the conformational change of Glu276 required for the slow-binding inhibition by oseltamivir carboxylate is dependent on the side chain volume of amino acid 274. Blocking the reorientation of Glu276 due to a steric hindrance introduced by specific substitutions at position 274 is a possible mechanism leading to a decreased susceptibility to oseltamivir carboxylate.
Oseltamivir carboxylate and zanamivir represent a new class of anti-influenza therapeutic agents, which specifically inhibit the essential NA function through binding to the enzyme active site. The amino acid residues that line the active site are highly conserved among all NA of different influenza strains; therefore, mutations in the active site are expected to alter the substrate binding affinity. It would be expected that mutations in the active site that display reduced sensitivity to oseltamivir carboxylate would also have decreased affinity for substrate binding. Extensive studies on the identification and characterization of drug-resistant NA mutations have been conducted, which indicate that resistance development is a rare event, since the loss in NA function that accompanies active-site mutation is costly in terms of viral fitness. Therefore, resistance to the NAI is unlikely to be of clinical significance.
Acknowledgments
We gratefully acknowledge Norbert Bischofberger, Choung Kim, and Xiaowu Chen of Gilead Sciences and Noel Roberts, Bradford Graves, and Brad Sherborne of Roche Discovery, Welwyn, United Kingdom, for their insightful discussions during the course of this study. We thank Tomas Cihlar, Craig Gibbs, Mick Hitchcock, and Bill Lee of Gilead Sciences for helpful suggestions and critical review of the manuscript.
REFERENCES
- 1.Babu, Y. S., P. Chand, S. Bantia, P. Kotian, A. Dehghani, Y. El-Kattan, T. H. Lin, T. L. Hutchison, A. J. Elliott, C. D. Parker, S. L. Ananth, L. L. Horn, G. W. Laver, and J. A. Montgomery. 2000. BCX-1812 (RWJ-270201): discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based drug design. J. Med. Chem. 43:3482-3486. [DOI] [PubMed] [Google Scholar]
- 2.Bossart, P. J., Y. S. Babu, W. J. Cook, G. M. Air, and W. G. Laver. 1988. Crystallization and preliminary X-ray analyses of two neuraminidases from influenza B virus strains B/Hong Kong/8/73 and B/Lee/40. J. Biol. Chem. 263:6421-6423. [PubMed] [Google Scholar]
- 3.Bossart-Whitaker, P., M. Carson, Y. S. Babu, C. D. Smith, W. G. Laver, and G. M. Air. 1993. Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid. J. Mol. Biol. 232:1069-1083. [DOI] [PubMed] [Google Scholar]
- 4.Burmeister, W. P., R. W. Ruigrok, and S. Cusack. 1992. The 2.2 A resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J. 11:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr, J., J. Ives, L. Kelly, R. Lambkin, J. Oxford, D. Mendel, L. Tai, and N. Roberts. 2002. Influenza virus carrying neuraminidase with reduced sensitivity to oseltamivir carboxylate has altered properties in vitro and is compromised for infectivity and replicative ability in vivo. Antivir. Res. 54:79-88. [DOI] [PubMed] [Google Scholar]
- 6.Goto, H., R. C. Bethell, and Y. Kawaoka. 1997. Mutations affecting the sensitivity of the influenza virus neuraminidase to 4-guanidino-2,4-dideoxy-2,3-dehydro-N-acetylneuraminic acid. Virology 238:265-272. [DOI] [PubMed] [Google Scholar]
- 7.Gubareva, L. V., R. Bethell, G. J. Hart, K. G. Murti, C. R. Penn, and R. G. Webster. 1996. Characterization of mutants of influenza A virus selected with the neuraminidase inhibitor 4-guanidino-Neu5Ac2en. J. Virol. 70:1818-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gubareva, L. V., L. Kaiser, and F. G. Hayden. 2000. Influenza virus neuraminidase inhibitors. Lancet 355:827-835. [DOI] [PubMed] [Google Scholar]
- 9.Gubareva, L. V., L. Kaiser, M. N. Matrosovich, Y. Soo-Hoo, and F. G. Hayden. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J. Infect. Dis. 183:523-531. [DOI] [PubMed] [Google Scholar]
- 10.Gubareva, L. V., M. N. Matrosovich, M. K. Brenner, R. C. Bethell, and R. G. Webster. 1998. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J. Infect. Dis. 178:1257-1262. [DOI] [PubMed] [Google Scholar]
- 11.Gubareva, L. V., M. J. Robinson, R. C. Bethell, and R. G. Webster. 1997. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J. Virol. 71:3385-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart, G. J., and R. C. Bethell. 1995. 2,3-didehydro-2,4-dideoxy-4-guanidino-N-acetyl-D-neuraminic acid (4-guanidino-Neu5Ac2en) is a slow-binding inhibitor of sialidase from both influenza A virus and influenza B virus. Biochem. Mol. Biol. Int. 36:695-703. [PubMed] [Google Scholar]
- 13.Hay, A. J., A. J. Wolstenholme, J. J. Skehel, and M. H. Smith. 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 4:3021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayden, F. G., R. L. Atmar, M. Schilling, C. Johnson, D. Poretz, D. Paar, L. Huson, P. Ward, and R. G. Mills. 1999. Use of the selective oral neuraminidase inhibitor oseltamivir to prevent influenza. N. Engl. J. Med. 341:1336-1343. [DOI] [PubMed] [Google Scholar]
- 15.Hayden, F. G., and R. B. Couch. 1992. Clinical and epidemiological importance of influenza A viruses resistant to amantadine and rimantadine. Rev. Med. Virol. 2:89-96. [Google Scholar]
- 16.Hayden, F. G., L. Jennings, R. Robson, G. Schiff, H. Jackson, B. Rana, G. McClelland, D. Ipe, N. Roberts, and P. Ward. 2000. Oral oseltamivir in human experimental influenza B infection. Antivir. Ther. 5:205-213. [PubMed] [Google Scholar]
- 17.Ives, J., J. Carr, N. A. Roberts, C. Y. Tai, D. B. Mendel, L. Kelly, R. Lambkin, and J. Oxford. 2000. An oseltamivir treatment-selected influenza A/Wuhan/359/95 virus with a E119V mutation in the neuraminidase gene has reduced infectivity. J. Clin. Virol. 18:251-269. [Google Scholar]
- 18.Ives, J. A., J. A. Carr, D. B. Mendel, C. Y. Tai, R. Lambkin, L. Kelly, J. S. Oxford, F. G. Hayden, and N. A. Roberts. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antivir. Res. 55:307-317. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, H. C., N. Roberts, Z. M. Wang, and R. Belshe. 2000. Management of influenza: use of new antivirals and resistance in perspective. Clin. Drug Investig. 20:447-454. [Google Scholar]
- 20.Kati, W. M., A. S. Saldivar, F. Mohamadi, H. L. Sham, W. G. Laver, and W. E. Kohlbrenner. 1998. GS4071 is a slow-binding inhibitor of influenza neuraminidase from both A and B strains. Biochem. Biophys. Res. Commun. 244:408-413. [DOI] [PubMed] [Google Scholar]
- 21.Kim, C. U., X. Chen, and D. B. Mendel. 1999. Neuraminidase inhibitors as anti-influenza virus agents. Antivir. Chem. Chemother. 10:141-154. [DOI] [PubMed] [Google Scholar]
- 22.Kim, C. U., W. Lew, M. A. Williams, H. Liu, L. Zhang, S. Swaminathan, N. Bischofberger, M. S. Chen, D. B. Mendel, C. Y. Tai, W. G. Laver, and R. C. Stevens. 1997. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J. Am. Chem. Soc. 119:681-690. [DOI] [PubMed] [Google Scholar]
- 23.Kim, C. U., W. Lew, M. A. Williams, H. Wu, L. Zhang, X. Chen, P. A. Escarpe, D. B. Mendel, W. G. Laver, and R. C. Stevens. 1998. Structure-activity relationship studies of novel carbocyclic influenza neuraminidase inhibitors. J. Med. Chem. 41:2451-2460. [DOI] [PubMed] [Google Scholar]
- 24.Laver, W. G., N. Bischofberger, and R. G. Webster. 1999. Disarming flu viruses. Sci. Am. 280:78-87. [DOI] [PubMed] [Google Scholar]
- 25.Lentz, M. R., R. G. Webster, and G. M. Air. 1987. Site-directed mutation of the active site of influenza neuraminidase and implications for the catalytic mechanism. Biochemistry 26:5351-5358. [DOI] [PubMed] [Google Scholar]
- 26.Lew, W., X. Chen, and C. U. Kim. 2000. Discovery and development of GS 4104 (oseltamivir): an orally active influenza neuraminidase inhibitor. Curr. Med. Chem. 7:663-672. [DOI] [PubMed] [Google Scholar]
- 27.Li, W., P. A. Escarpe, E. J. Eisenberg, K. C. Cundy, C. Sweet, K. J. Jakeman, J. Merson, W. Lew, M. Williams, L. Zhang, C. U. Kim, N. Bischofberger, M. S. Chen, and D. B. Mendel. 1998. Identification of GS 4104 as an orally bioavailable prodrug of the influenza virus neuraminidase inhibitor GS 4071. Antimicrob. Agents Chemother. 42:647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKimm-Breschkin, J. L. 2000. Resistance of influenza viruses to neuraminidase inhibitors—a review. Antivir. Res. 47:1-17. [DOI] [PubMed] [Google Scholar]
- 29.McKimm-Breschkin, J. L., M. McDonald, T. J. Blick, and P. M. Colman. 1996. Mutation in the influenza virus neuraminidase gene resulting in decreased sensitivity to the neuraminidase inhibitor 4-guanidino-Neu5Ac2en leads to instability of the enzyme. Virology 225:240-242. [DOI] [PubMed] [Google Scholar]
- 30.McKimm-Breschkin, J. L., A. Sahasrabudhe, T. J. Blick, M. McDonald, P. M. Colman, G. J. Hart, R. C. Bethell, and J. N. Varghese. 1998. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases [sic] sensitivity to Neu5Ac2en-derived inhibitors. J. Virol. 72:2456-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendel, D. B., and N. A. Roberts. 1998. In-vitro and in-vivo efficacy of influenza neuraminidase inhibitors. Curr. Opin. Infect. Dis. 11:727-732. [DOI] [PubMed] [Google Scholar]
- 32.Mendel, D. B., and R. W. Sidwell. 1998. Influenza virus resistance to neuraminidase inhibitors. Drug Resist. Updates 1:184-189. [DOI] [PubMed] [Google Scholar]
- 33.Monto, A. S., and N. H. Arden. 1992. Implications of viral resistance to amantadine in control of influenza A. Clin. Infect. Dis. 15:362-369. [DOI] [PubMed] [Google Scholar]
- 34.Morrison, J. F. 1982. The slow-binding and slow, tight-binding inhibition of enzyme-catalysed reactions. Trends Biochem. Sci. 7:102-105. [Google Scholar]
- 35.Morrison, J. F., and C. T. Walsh. 1988. The behavior and significance of slow-binding enzyme inhibitors. Adv. Enzymol. Relat. Areas Mol. Biol. 61:201-301. [DOI] [PubMed] [Google Scholar]
- 36.Oxford, J. S., and R. Lambkin. 1998. Targeting influenza virus neuraminidase—a new strategy for antiviral therapy. Drug Discov. Today 3:448-456. [Google Scholar]
- 37.Pegg, M. S., and M. von Itzstein. 1994. Slow-binding inhibition of sialidase from influenza virus. Biochem. Mol. Biol. Int. 32:851-858. [PubMed] [Google Scholar]
- 38.Roberts, N. A. 2001. Anti-influenza drugs and neuraminidase inhibitors. Prog. Drug Res. 56:195-237. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, N. A. 2001. Treatment of influenza with neuraminidase inhibitors: virological implications. Philos. Trans. R. Soc. Lond. Ser. B 356:1895-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sidwell, R. W., and J. H. Huffman. 2000. In vitro and in vivo assay systems for study of influenza virus inhibitors. Antivir. Res. 48:1-16. [DOI] [PubMed] [Google Scholar]
- 41.Singh, J., and J. M. Thornton. 1992. Atlas of protein side-chain interactions, vol. 1. IRL Press, Oxford, United Kingdom.
- 42.Tai, C. Y., P. A. Escarpe, R. W. Sidwell, M. A. Williams, W. Lew, H. Wu, C. U. Kim, and D. B. Mendel. 1998. Characterization of human influenza virus variants selected in vitro in the presence of the neuraminidase inhibitor GS 4071. Antimicrob. Agents Chemother. 42:3234-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varghese, J. N., and P. M. Colman. 1991. Three-dimensional structure of the neuraminidase of influenza virus A/Tokyo/3/67 at 2.2 A resolution. J. Mol. Biol. 221:473-486. [DOI] [PubMed] [Google Scholar]
- 44.Varghese, J. N., V. C. Epa, and P. M. Colman. 1995. Three-dimensional structure of the complex of 4-guanidino-Neu5Ac2en and influenza virus neuraminidase. Protein Sci. 4:1081-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varghese, J. N., J. L. McKimm-Breschkin, J. B. Caldwell, A. A. Kortt, and P. M. Colman. 1992. The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor. Proteins 14:327-332. [DOI] [PubMed] [Google Scholar]
- 46.von Itzstein, M., J. C. Dyason, S. W. Oliver, H. F. White, W. Y. Wu, G. B. Kok, and M. S. Pegg. 1996. A study of the active site of influenza virus sialidase: an approach to the rational design of novel anti-influenza drugs. J. Med. Chem. 39:388-391. [DOI] [PubMed] [Google Scholar]
- 47.Webster, R. G. 1998. Influenza: an emerging disease. Emerg. Infect. Dis. 4:436-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitley, R. J., F. G. Hayden, K. S. Reisinger, N. Young, R. Dutkowski, D. Ipe, R. G. Mills, and P. Ward. 2001. Oral oseltamivir treatment of influenza in children. Pediatr. Infect. Dis. J. 20:127-133. [DOI] [PubMed] [Google Scholar]