Abstract
Streptococcus pyogenes strains inducibly resistant (iMLS phenotype) to macrolide, lincosamide, and streptogramin B (MLS) antibiotics can be subdivided into three phenotypes: iMLS-A, iMLS-B, and iMLS-C. This study focused on inducibly erythromycin-resistant S. pyogenes strains of the iMLS-B and iMLS-C types, which are very similar and virtually indistinguishable in a number of phenotypic and genotypic features but differ clearly in their degree of resistance to MLS antibiotics (high in the iMLS-B type and low in the iMLS-C type). As expected, the iMLS-B and iMLS-C test strains had the erm(A) methylase gene; the iMLS-A and the constitutively resistant (cMLS) isolates had the erm(B) methylase gene; and a control M isolate had the mef(A) efflux gene. mre(A) and msr(A), i.e., other macrolide efflux genes described in gram-positive cocci, were not detected in any test strain. With a radiolabeled erythromycin method for determination of the intracellular accumulation of the drug in the absence or presence of an efflux pump inhibitor, active efflux of erythromycin was observed in the iMLS-B isolates as well as in the M isolate, whereas no efflux was demonstrated in the iMLS-C isolates. By the triple-disk (erythromycin plus clindamycin and josamycin) test, performed both in normal test medium and in the same medium supplemented with the efflux pump inhibitor, under the latter conditions iMLS-B and iMLS-C strains were no longer distinguishable, all exhibiting an iMLS-C phenotype. In conjugation experiments with an iMLS-B isolate as the donor and a Rifr Fusr derivative of an iMLS-C isolate as the recipient, transconjugants which shared the iMLS-B type of the donor under all respects, including the presence of an efflux pump, were obtained. These results indicate the existence of a novel, transferable efflux system, not associated with mef(A) or with other known macrolide efflux genes, that is peculiar to iMLS-B strains. Whereas the low-level resistance of iMLS-C strains to MLS antibiotics is apparently due to erm(A)-encoded methylase activity, the high-level resistance of iMLS-B strains appears to depend on the same methylase activity plus the new efflux system.
Two principal mechanisms have so far been found to be responsible for acquired erythromycin resistance in Streptococcus pyogenes: target site modification and active efflux (12, 27). Target site modification is mostly based on N6 dimethylation of an adenine residue (A2058) in the peptidyl transferase circle of 23S rRNA domain V through the action of a family of enzymes encoded by erm class genes. Methylation is thought to induce a conformational change in the 50S ribosomal subunit, leading to reduced binding of and coresistance to macrolide, lincosamide, and streptogramin B (MLS) antibiotics, whose binding sites probably overlap (12, 26). It is well established that MLS resistance can be expressed either constitutively (cMLS phenotype) or inducibly (iMLS phenotype) in streptococci (11, 12), and in S. pyogenes it is mediated by two classes of methylase genes, the conventional erm(B) (27) and the recently described erm(A), subclass erm(TR) (20), determinants, hereafter designated erm(A) according to current nomenclature (17).
iMLS strains of S. pyogenes, all showing susceptibility (or, occasionally, intermediate susceptibility) to lincosamides that turns to high-level resistance after induction, have recently been shown to be genotypically and phenotypically heterogeneous and were subdivided into three distinct types, designated iMLS-A, iMLS-B, and iMLS-C (9). From a genotypic point of view, iMLS-A as well as cMLS strains are strictly associated with the erm(B) gene, whereas iMLS-B and iMLS-C strains are strictly associated with the erm(A) gene (9). From a phenotypic point of view, iMLS-A strains are characterized by high-level resistance to 14-, 15-, and 16-membered macrolides and by reduced susceptibility to resistance to ketolides; iMLS-B strains are characterized by high-level resistance to 14- and 15-membered macrolides, susceptibility to 16-membered macrolides turning to high-level resistance after induction, and marked susceptibility to ketolides turning to intermediate susceptibility or resistance after induction; and iMLS-C strains are characterized by low-level resistance to 14- and 15-membered macrolides (with MICs increasing by two- to fourfold after induction), susceptibility to 16-membered macrolides turning to low-level resistance after induction, and marked susceptibility to ketolides also after induction (8, 9). Inducibly erythromycin-resistant S. pyogenes strains of the iMLS-A, iMLS-B, and iMLS-C phenotypes can easily be differentiated with a triple-disk (erythromycin plus clindamycin and josamycin) test (9).
Besides the posttranscriptional modification of the 23S rRNA caused by erm class methylases, mutations in 23S rRNA or ribosomal proteins leading to macrolide resistance have also been recently described in streptococci, first in laboratory-derived mutants (24) and then in clinical isolates (5, 23) of Streptococcus pneumoniae and lately also in clinical isolates of S. pyogenes (1, 13).
Active efflux, which reduces the intracellular antibiotic concentration to subtoxic levels, thus leading to resistance, is due to drug pumps acting through proton- or ATP-driven membrane transporters (2, 15). This is one of the major resistance mechanisms for macrolide antibiotics (30). Only recently has a macrolide efflux mechanism been described in S. pyogenes and other streptococci (22), in which it is associated with a new resistance pattern (M phenotype) characterized by resistance, among MLS antibiotics, only to 14- and 15-membered macrolides, and then usually at a low level (19, 22). M resistance is mediated in S. pyogenes by the mef(A) gene (4), which encodes a membrane protein responsible for macrolide efflux. The mef(A) determinant is also occasionally detected in strains of other phenotypes.
This study focused on inducibly erythromycin-resistant S. pyogenes strains of the iMLS-B and iMLS-C types, which are very similar and virtually indistinguishable in several phenotypic and genotypic features but clearly differ in their degree of macrolide resistance (high in the iMLS-B type and low in the iMLS-C type) (9). We show here that this difference is due to the presence in iMLS-B but not iMLS-C isolates of a new, non-mef(A)-mediated erythromycin efflux system.
MATERIALS AND METHODS
Bacterial strains and macrolide resistance phenotypes.
The 14 S. pyogenes strains used in this study, 13 erythromycin-resistant (MIC, ≥1 μg/ml) isolates and one erythromycin-susceptible isolate, are listed in Table 1 together with their susceptibilities to MLS antibiotics (in some instances also after induction by growth in 0.05 μg of erythromycin per ml) and their macrolide resistance phenotypes and genotypes (see below). All test strains were clinical isolates recovered from throat cultures from different symptomatic patients in the course of recent extensive studies in Italy (9, 16, 25; G. C. Schito, E. A. Debbia, G. Nicoletti, D. Pavesio, S. Ripa, G. Tempera, and P. E. Varaldo, Program Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., 1999, abstr. 1211, p. 154). All strains with similar phenotypic features differed in typing characteristics as determined by SmaI macrorestriction fragment pattern analysis by pulsed-field gel electrophoresis (PFGE) (16) and/or random amplified polymorphic DNA analysis (10) with primers M13 and H2 (18).
TABLE 1.
The 14 S. pyogenes test strains used, their antimicrobial susceptibilities, and their macrolide resistance phenotypes and genotypes
| Strain | MICa (μg/ml)
|
Macrolide resistance
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | CLR | AZM | JOS | JOS (ind.) | TEL | TEL (ind.) | CLI | CLI (ind.) | Phenotyeb | Genotype | |
| iB1 | >128 | >128 | >128 | 0.03 | >128 | ≤0.015 | 8 | 0.06 | >128 | iMLS-B | erm(A) |
| iB2 | >128 | >128 | >128 | 0.03 | >128 | ≤0.015 | 8 | 0.03 | >128 | iMLS-B | erm(A) |
| iB3 | >128 | >128 | >128 | 0.03 | >128 | ≤0.015 | 4 | 0.12 | >128 | iMLS-B | erm(A) |
| iB4 | >128 | >128 | >128 | 0.06 | >128 | ≤0.015 | 8 | 0.06 | >128 | iMLS-B | erm(A) |
| iC1 | 2 | 0.5 | 8 | 0.06 | 16 | ≤0.015 | 0.06 | 0.03 | >128 | iMLS-C | erm(A) |
| iC2 | 1 | 0.5 | 4 | 0.06 | 8 | ≤0.015 | 0.06 | 0.03 | >128 | iMLS-C | erm(A) |
| iC3 | 2 | 1 | 4 | 0.03 | 16 | ≤0.015 | 0.06 | 0.06 | >128 | iMLS-C | erm(A) |
| iC4 | 1 | 0.5 | 4 | 0.06 | 8 | ≤0.015 | 0.06 | 0.03 | >128 | iMLS-C | erm(A) |
| iA1 | >128 | >128 | >128 | >128 | >128 | 4 | 16 | 0.25 | >128 | iMLS-A | erm(B) |
| iA2c | >128 | >128 | >128 | >128 | >128 | 2 | 8 | 0.06 | >128 | iMLS-A | erm(B) mef(A) |
| c1 | >128 | >128 | >128 | >128 | >128 | 4 | 8 | >128 | >128 | cMLS | erm(B) |
| c2c | >128 | >128 | >128 | >128 | >128 | 2 | 4 | >128 | >128 | cMLS | erm(B) mef(A) |
| M1 | 16 | 8 | 16 | 0.06 | 0.12 | 0.25 | 0.25 | 0.03 | 0.03 | M | mef(A) |
| s1 | ≤0.015 | 0.03 | 0.03 | 0.06 | 0.06 | ≤0.015 | ≤0.015 | 0.06 | 0.06 | Susceptible | |
ERY, erythromycin; CLR, clarithromycin; AZM, azithromycin; JOS, josamycin; TEL, telithromycin; CLI, clindamycin; ind., after induction by growth in 0.05 μg of erythromycin per ml.
According to Giovanetti et al. (9).
Isolates iA2 and c2 were chosen, among the strains of their phenotypes (iMLS-A and cMLS, respectively), because of the presence of an amplification product in preliminary PCR experiments targeting the mef(A) gene.
Determination of macrolide resistance phenotype.
As described previously (9), the macrolide resistance phenotype was determined on the basis of the pattern of susceptibility to MLS antibiotics and confirmed on the basis of the triple-disk (erythromycin plus clindamycin and josamycin) test, set up to facilitate the laboratory discrimination of the three types (iMLS-A, iMLS-B, and iMLS-C) of inducibly erythromycin-resistant strains.
Antibiotics.
Erythromycin and clindamycin were purchased from Sigma Chemical Co. (St. Louis, Mo.). The other antibiotics were obtained as follows: clarithromycin from Abbott Laboratories (Abbott Park, Ill.), azithromycin from Pfizer Inc. (New York, N.Y.), josamycin from ICN Biomedicals (Costa Mesa, Calif.), and telithromycin from Aventis Pharma (Lainate, Italy).
Susceptibility tests.
Antibiotic MICs were determined by broth microdilution (14). Mueller-Hinton II broth (BBL Microbiology Systems, Cockeysville, Md.) supplemented with 3% lysed horse blood was used as the test medium, and the inoculum was 5 × 105 CFU/ml. Streptococcus pneumoniae ATCC 49619 was used for quality control.
Detection of erythromycin resistance genes.
Erythromycin resistance genes erm(B) and erm(A) were detected by PCR with the oligonucleotide primer pair described by Sutcliffe et al. (21) and the primers designated III8 and III10 by Seppälä et al. (20), respectively. The erythromycin resistance gene mef(A) was primarily detected by PCR with the primer pair described by Sutcliffe et al. (21). For confirmation, amplicons obtained with this primer pair were sequenced. Nucleic acid sequencing was carried out on a 373A automated DNA sequencer (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.) with a Taq fluorescent dideoxy terminator cycle sequencing kit (Perkin-Elmer). Primers for the elucidation of the sequence were those designed to detect mef(A) by Sutcliffe et al. (21), referred to above. Suitable restriction endonucleases (Roche Molecular Biochemicals, Mannheim, Germany) were used for DNA digestion. Another primer pair designed to detect mef(A) by Clancy et al. (4) was used in other experiments. Detection by PCR of the mre(A) and msr(A) genes was performed with the primer pairs designed by Clancy et al. (3) and Wondrak et al. (29), respectively.
Macrolide efflux studies.
The existence of a macrolide efflux mechanism was evaluated in exponentially growing cells of S. pyogenes by comparing the uptake of N-methyl-[14C]erythromycin (DuPont NEN, Boston, Mass.) in the absence or presence (25 μM) of carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma), an energy uncoupler acting as an efflux pump inhibitor. To ensure full induction of the efflux determinant, the cultures were grown in the presence of unlabeled erythromycin prior to the addition of radiolabeled erythromycin, according to the procedure described by Sutcliffe et al. (22).
Conjugation experiments.
Conjugative transfer was performed on membrane filters (28) with both the recipient and the donor grown to an optical density of 0.4 ± 0.05 units at 540 nm and then mixed at a donor/recipient ratio of 1:5. The filter, placed on a warmed plate of Columbia agar (Difco Laboratories, Detroit, Mich.) supplemented with 5% sheep blood cells, was incubated at 37°C for 18 h, and the cells recovered were resuspended in 1 ml of sterile saline. This suspension was plated onto Columbia blood agar supplemented with 10 μg of rifampin per ml, 10 μg of fusidic acid per ml, and 64 μg of erythromycin per ml (1 μg of erythromycin per ml was used when an erythromycin-susceptible isolate was used as a recipient). The inoculated plates were incubated at 37°C for 48 to 72 h and then examined for the presence of transconjugants. The frequencies of transfer were expressed as the number of transconjugants per recipient CFU after mating.
Macrorestriction analysis.
SmaI macrorestriction fragment patterns were analyzed by PFGE; macrorestriction and PFGE were performed, and the relevant patterns were analyzed and compared, as recently described elsewhere (16).
RESULTS
Erythromycin resistance genes.
The presence of the erythromycin resistance genes erm(B), erm(A), and mef(A) in the erythromycin-resistant test strains was primarily investigated by PCR. The erm(B) gene was detected in the two strains of the cMLS phenotype and in the two inducibly resistant strains of the iMLS-A phenotype. The erm(A) gene was detected in all inducibly resistant strains of the iMLS-B and iMLS-C phenotypes.
The situation was initially less clear for the mef(A) gene. With the primer pair described by Sutcliffe et al. (21), an amplicon of the expected size was detected in the strain of the M phenotype, in one strain (c2) of the cMLS phenotype, in one strain (iA2) of the iMLS-A phenotype, in all four isolates of the iMLS-B phenotype, and in three (iC1 to iC3) of the four isolates of the iMLS-C phenotype. It is worth noting that the putative mef(A) amplicon was fainter in the iMLS-B and iMLS-C isolates than in the others. By sequencing, the known 348-bp sequence of the mef(A) gene (4) was confirmed in the M isolate (M1) as well as in the cMLS (c2) and iMLS-A (iA2) isolates, whereas a completely different and shorter sequence [325 bp, i.e., not shorter enough to be clearly distinguished from the true mef(A) sequence in PCR assays] with no significant homology to any known GenBank sequence was observed in the iMLS-B and iMLS-C isolates. With another primer pair designed to detect mef(A) by Clancy et al. (4), only the true mef(A) gene was detected, the one present in strains M1 (M phenotype), c2 (cMLS phenotype), and iA2 (iMLS-A phenotype), whereas no amplification was observed with iMLS-B and iMLS-C isolates. The overall results of the distribution of erythromycin resistance genes among the S. pyogenes test strains are summarized in Table 1.
On the basis of the relevant sequences, the restriction endonuclease PvuII had one digestion site in the 348-bp mef(A) gene, whereas HindIII, XbaI, and NheI had one digestion site each in the 325-bp sequence yielded by nonspecific amplification. Besides the set of test strains, all iMLS-B and iMLS-C isolates available in our laboratory and showing the faint, false mef(A) amplicon in addition to erm(A) proved to be susceptible to digestion with HindIII, XbaI, and NheI but not with PvuII, whereas the opposite (i.e., susceptibility to digestion with PvuII but not with HindIII, XbaI, or NheI) was observed with the M isolate as well as the cMLS and iMLS-A isolates positive for mef(A) in addition to erm(B) (data not shown). In following studies, the faint, false mef(A) amplicon was avoided by carefully controlling the PCR conditions (21, 22), especially the annealing temperature, which must never drop below 52 to 54°C.
mre(A) and msr(A), other macrolide efflux genes described in gram-positive cocci, were not detected by PCR in any test strain.
Active efflux experiments.
Active efflux of erythromycin was investigated in the four iMLS-B and the four iMLS-C test strains. An erythromycin-susceptible isolate (s1) and an M isolate (M1) were used as negative and positive controls, respectively. By the radiolabeled erythromycin method, an active efflux was detected in all iMLS-B isolates besides the M isolate, whereas no efflux was demonstrated in the iMLS-C isolates besides the susceptible isolate.
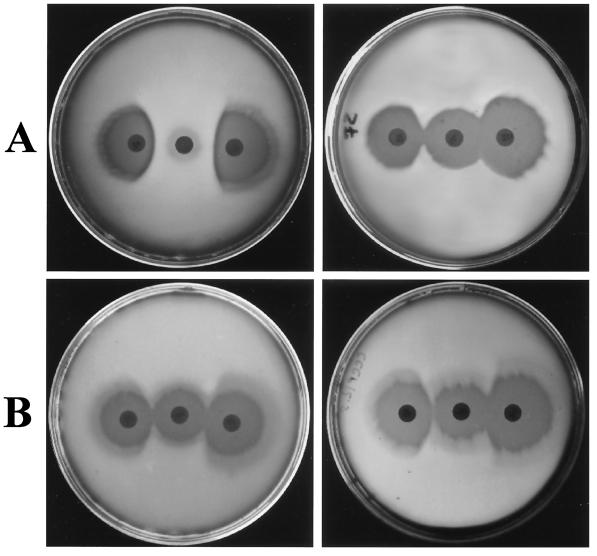
The iMLS-B and iMLS-C test strains were subjected to the triple-disk assay—set up to facilitate the laboratory discrimination of the three types (iMLS-A, iMLS-B, and iMLS-C) of inducibly erythromycin-resistant strains (9)—both in normal test medium and in the same medium supplemented with the CCCP efflux pump inhibitor (12.5 μM). Under the latter conditions, iMLS-B and iMLS-C strains were no longer distinguishable, and all exhibited an iMLS-C phenotype (Fig. 1). iMLS-A and cMLS test strains iA1 and c1, respectively, were tested in similar experiments as efflux-negative controls.
FIG. 1.
Triple-disk test performed with the iMLS-B strain iB1 (A) and the iMLS-C strain iC1 (B). In each plate, the erythromycin disk (30 μg) is at the center, with the clindamycin disk (10 μg) on the right and the josamycin disk (30 μg) on the left. Normal test medium was used with both strains in the plates on the left side, whereas the same medium supplemented with CCCP (12.5 μM) was used in the plates on the right side. While the difference between the iMLS-B and the iMLS-C phenotypes is clearly apparent in normal test medium, no difference can be appreciated in the test medium supplemented with the efflux pump inhibitor, both strains exhibiting an iMLS-C phenotype under the latter condition.
Conjugative transfer experiments and characterization of transconjugants.
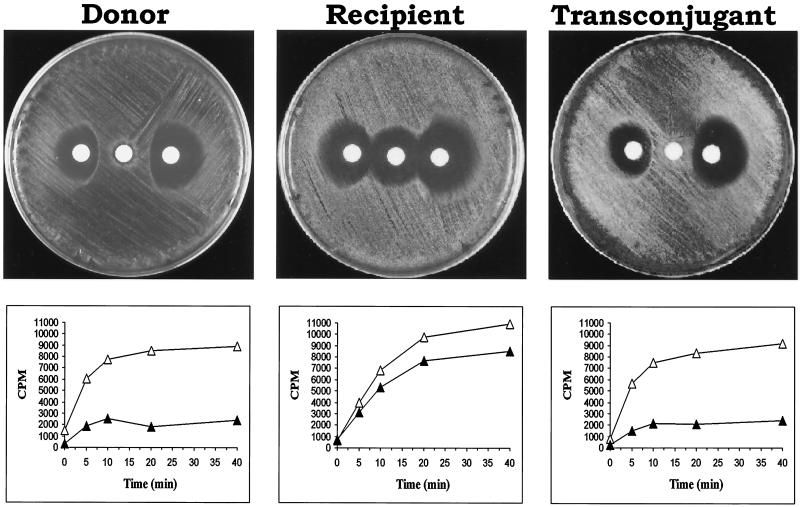
An iMLS-B isolate (iB1) was used as the donor and a fusidic acid- and rifampin-resistant derivative of an iMLS-C isolate (iC4fr) was used as the recipient in mating experiments. Five transconjugants were obtained (transfer frequency, 5.5 × 10−9), which proved to be identical in all characteristics tested. As shown in Table 2, from a genotypic point of view, the transconjugants had, of course, the same erm(A) genotype as the recipient and the donor. From a phenotypic point of view, they exhibited the iMLS-B phenotype, as demonstrated by their pattern of susceptibility to MLS antibiotics, which overlapped that of the donor strain and was confirmed by the triple-disk test (Fig. 2). These phenotypic features were associated with an erythromycin efflux mechanism that the transconjugant had apparently received from the donor, as demonstrated by the radiolabeled erythromycin method (Fig. 2). In SmaI macrorestriction analysis experiments, the PFGE profile of the tested transconjugant differed from that of the recipient for the disappearance of a band of ca. 270 kb, which was replaced by a larger one (ca. 290 kb) (Fig. 3).
TABLE 2.
Comparison of some characteristics of the erythromycin-resistant S. pyogenes strains used as the donor and recipient in transfer experiments with those of the transconjugants obtained
| Characteristica | Donor (strain iB1) | Recipient (strain iC4fr) | Transconjugants (iB1 × iC4fr) |
|---|---|---|---|
| Erythromycin resistance | |||
| Genotype | erm(A) | erm (A) | erm(A) |
| Phenotype | iMLS-B | iMLS-C | iMLS-B |
| Active efflux present | Yes | No | Yes |
| MIC (μg/ml) | |||
| Erythromycin | >128 | 1 | >128 |
| Clarithromycin | >128 | 0.5 | >128 |
| Azithromycin | >128 | 4 | >128 |
| Josamycin | 0.06 | 0.06 | 0.06 |
| Josamycin (ind.) | >128 | 8 | >128 |
| Telithromycin | ≤0.015 | ≤0.015 | ≤0.015 |
| Telithromycin (ind.) | 8 | 0.03 | 8 |
| Clindamycin | 0.06 | ≤0.015 | 0.06 |
| Clindamycin (ind.) | >128 | >128 | >128 |
ind., induced by growth in erythromycin (0.05 μg/ml).
FIG. 2.
Comparison of some characteristics of the iMLS-B isolate used as the donor, the iMLS-C isolate used as the recipient, and a transconjugant from a mating experiment. Above, triple-disk test (erythromycin disk at the center and josamycin and clindamycin disks on the left and right side, respectively) showing the iMLS-B phenotype of the donor, the iMLS-C phenotype of the recipient, and the iMLS-B phenotype of the transconjugant. Below, uptake of [14C]erythromycin in the absence (▴) or presence (▵) of 25 μM CCCP. The accumulation of the radiolabeled antibiotic in the presence of CCCP, denoting the presence of an active drug efflux pump, is evident in the donor and the transconjugant but not in the recipient.
FIG. 3.
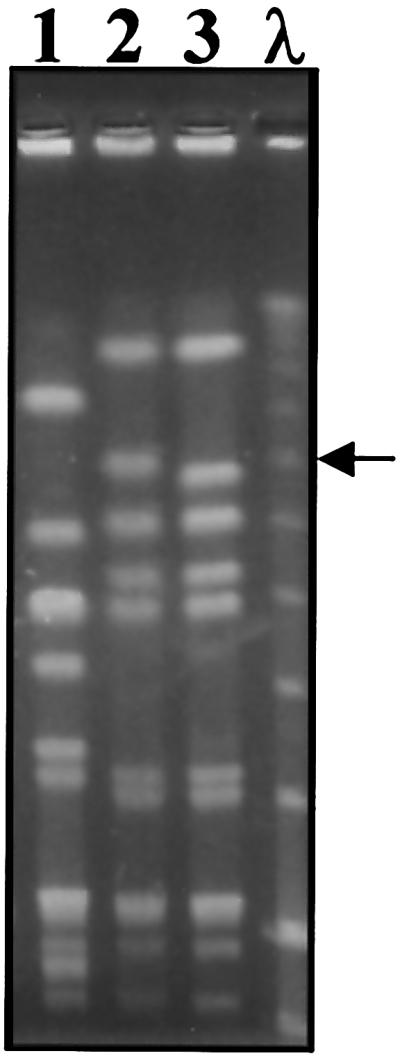
PFGE patterns of SmaI-digested genomic DNA of the S. pyogenes strains involved in the mating between an iMLS-B isolate (iB1) used as the donor and a fusidic acid- and rifampin-resistant derivative of an iMLS-C isolate (iC4fr) used as the recipient. Lane 1, donor strain iB1; lane 2, a transconjugant selected for erythromycin (64 μg/ml), fusidic acid (10 μg/ml), and rifampin (10 μg/ml) resistance; lane 3, recipient strain iC4fr. Lambda DNA concatemers (lane λ) were used as molecular size markers. The arrow indicates a ca. 290-kb band of the transconjugant that replaced a ca. 270-kb band of the recipient.
Transfer experiments with an iMLS-B isolate as the donor and a fusidic acid- and rifampin-resistant derivative of an erythromycin-susceptible isolate as the recipient were also performed, selecting for erythromycin (1 μg/ml), fusidic acid (10 μg/ml), and rifampin (10 μg/ml) resistance. However, only transconjugants that had received both the erm(A) determinant and the new efflux pump, thus displaying the iMLS-B phenotype, were obtained (7), whereas transconjugants having the new drug pump alone were not found.
DISCUSSION
A common trait of all the inducibly macrolide-resistant (iMLS) strains of S. pyogenes is susceptibility (or, occasionally, intermediate susceptibility) to lincosamides that turns to high-level resistance after induction. Among these strains, the first type (iMLS-A) is clearly separate both phenotypically (high-level resistance without induction not only to the 14- and 15- but also to the 16-membered macrolides and reduced susceptibility to ketolides) and genotypically [presence of the erm(B) gene as well as in constitutively resistant strains] (9). In contrast, the other two types of inducibly macrolide-resistant strains (iMLS-B and iMLS-C), although easily distinguishable phenotypically by the high-level resistance to MLS antibiotics expressed by iMLS-B isolates compared with the low-level resistance expressed by iMLS-C isolates, are substantially identical in all other respects. In the first place, both iMLS-B and iMLS-C isolates are genotypically characterized by their association with another, unique methylase gene, erm(A). In mating experiments, erm(A) could be successfully transferred from both iMLS-B and iMLS-C donors to macrolide-susceptible recipients of S. pyogenes, Enterococcus faecalis, and Listeria innocua (7). Moreover, in recent studies in Italy, where macrolide resistance in S. pyogenes is widespread (25), both iMLS-B and iMLS-C isolates were generally resistant to tetracycline (9) and particularly susceptible to ketolides (8); they both had the ability to invade eukaryotic cells with high efficiency (6); they both usually belonged to a unique PFGE class by SmaI macrorestriction analysis (16); and both even usually showed a faint, nonspecific amplification in PCR experiments with a particular primer pair designed to detect the mef(A) gene, as shown in the present study.
In this report, we show that an erythromycin efflux system is present in iMLS-B but not iMLS-C isolates. This efflux system appears to be distinct from the well-known efflux system associated with mef(A), a gene which is typical of S. pyogenes strains of the M phenotype and is also occasionally detected in strains whose erythromycin resistance is primarily mediated by the erm(B)-encoded methylase (i.e., phenotypes cMLS and iMLS-A). Indeed, none of the iMLS-B isolates tested carried mef(A), but their possession of an efflux system was clearly demonstrated in the experiments with an efflux pump inhibitor (CCCP). It was also excluded that this new efflux mechanism could be associated with other macrolide efflux genes previously described in gram-positive cocci, such as mre(A) and msr(A).
In the transfer experiments from an iMLS-B donor to an iMLS-C recipient, all transconjugants showed the iMLS-B type of the donor in all respects, suggesting that the new efflux pump, once transferred to the iMLS-C recipient, transformed it into an iMLS-B strain. On the other hand, the larger PFGE band (ca. 290 kb) of the transconjugant that replaced a ca. 270-kb band of the recipient is consistent with the insertion of new DNA, probably associated with the new efflux pump, into an existing restriction fragment. In triple-disk assays aimed at discriminating the three phenotypes (iMLS-A, iMLS-B, and iMLS-C) of inducibly erythromycin-resistant strains, iMLS-B strains behaved in all respects like the iMLS-C strains when the test medium was supplemented with the CCCP efflux pump inhibitor.
In other words, these findings suggest that genuine macrolide resistance due to the erm(A)-encoded methylase is typically expressed in S. pyogenes by iMLS-C strains. Thus, the level of resistance produced by erm(A) alone appears to be low (as is the case in iMLS-C strains), while the expression of other erm genes is generally sufficient for high-level resistance, as is the case for erm(B) in cMLS and iMLS-A strains of S. pyogenes. The high-level resistance typically expressed by iMLS-B strains is due to the erm(A)-encoded methylase plus a novel drug pump, different from those already described in gram-positive cocci. So far, neither clinical nor laboratory strains bearing this new drug pump as the sole mechanism of macrolide resistance are available. However, if the pattern of macrolide susceptibility of iMLS-B strains is compared with that of iMLS-C strains, it might be inferred that this new efflux mechanism should be particularly able to actively pump out 14- and 15-membered macrolides without induction and 16-membered macrolides and, to a much lower extent, ketolides only after induction. Further studies to better characterize this new efflux system and, above all, to find the gene responsible and gain insight into its genetic control and expression are warranted.
In conclusion, our understanding of macrolide resistance mechanisms in streptococci, particularly S. pyogenes, is evolving substantially and rapidly. Until just few years ago, the sole known mechanism was posttrancriptional methylation of the 23S rRNA due to erm(B) class N-methyltransferases (26). Subsequently, an active macrolide efflux mechanism (22) encoded by a new gene called mef(A) (4) was described, and shortly afterwards erm(A), originally called erm(TR), a new erm(A) class methylase gene, was discovered (20). Most recently, target mutations (i.e., in 23S rRNA or ribosomal proteins) were reported, first in S. pneumoniae strains (5, 23, 24) and lately also in clinical isolates of S. pyogenes (1, 13). It is worth noting that in one of the last two studies, the mechanism of resistance could not be elucidated in two of seven isolates with relatively low levels of erythromycin resistance (MICs, 8 and 32 μg/ml) (1).
We now report a novel efflux system, not associated with any known efflux genes, which contributes to macrolide resistance in those erm(A)-carrying S. pyogenes isolates previously characterized as phenotype iMLS-B and renders them different from iMLS-C strains (9). Thus, erythromycin resistance in S. pyogenes and other streptococci appears to be far more complex and less able to be framed into simple schemes than was often believed.
Acknowledgments
This work was supported in part by a grant from the Italian Ministry of Education, University and Research.
REFERENCES
- 1.Bingen, E., R. Leclercq, F. Fitoussi, N. Brahimi, B. Malbruny, D. Deforche, and R. Cohen. 2002. Emergence of group A streptococcus strains with different mechanisms of macrolide resistance. Antimicrob. Agents Chemother. 46:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borges-Walmsley, M. I., and A. R. Walmsley. 2001. The structure and functions of drug pumps. Trends Microbiol. 9:71-79. [DOI] [PubMed] [Google Scholar]
- 3.Clancy, J., F. Dib-Hajj, J. W. Petitpas, and W. Yuan. 1997. Cloning and characterization of a novel macrolide efflux gene, mreA, from Streptococcus agalactiae. Antimicrob. Agents Chemother. 41:2719-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 5.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Facinelli, B., C. Spinaci, G. Magi, E. Giovanetti, and P. E. Varaldo. 2001. Association between erythromycin resistance and ability to enter human respiratory cells in group A streptococci. Lancet 358:30-33. [DOI] [PubMed] [Google Scholar]
- 7.Giovanetti, E., G. Magi, A. Brenciani, C. Spinaci, R. Lupidi, B. Facinelli, and P. E. Varaldo. 2002. Conjugative transfer of the erm(A) gene from erythromycin-resistant Streptococcus pyogenes to macrolide-susceptible S. pyogenes, Enterococcus faecalis, and Listeria innocua. J. Antimicrob. Chemother. 50:249-252. [DOI] [PubMed] [Google Scholar]
- 8.Giovanetti, E., M. P. Montanari, F. Marchetti, and P. E. Varaldo. 2000. In vitro activity of ketolides telithromycin and HMR 3004 against Italian isolates of Streptococcus pyogenes and Streptococcus pneumoniae with different erythromycin susceptibility. J. Antimicrob. Chemother. 46:905-908. [DOI] [PubMed] [Google Scholar]
- 9.Giovanetti, E., M. P. Montanari, M. Mingoia, and P. E. Varaldo. 1999. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruteke, P., A. van Belkum, L. M. Schouls, W. D. H. Hendriks, F. A. G. Reubsaet. J. Dokter, H. Boxma, and H. A. Verbrugh. 1996. Outbreak of group A streptococci in a burn center: use of pheno- and genotypic procedures for strain tracking. J. Clin. Microbiol. 34:114-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyder, S. L., and M. M. Streitfeld. 1973. Inducible and constitutive resistance to macrolide antibiotics and lincomycin in clinically isolated strains of Streptococcus pyogenes. Antimicrob. Agents Chemother. 4:327-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 13.Malbruny, B., K. Nagai, M. Coquemont, B. Bozdogan, A. T. Andrasevic, H. Hupkova, R. Leclercq, and P. C. Appelbaum. 2002. Resistance to macrolides in clinical isolates of Streptococcus pyogenes due to ribosomal mutations. J. Antimicrob. Chemother. 49:935-939. [DOI] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 15.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ripa, S., C. Zampaloni, L. A. Vitali, E. Giovanetti, M. P. Montanari, M. Prenna, and P. E. Varaldo. 2001. SmaI macrorestriction analysis of Italian isolates of erythromycin-resistant Streptococcus pyogenes and correlations with macrolide-resistance phenotypes. Microb. Drug Resist. 7:65-71. [DOI] [PubMed] [Google Scholar]
- 17.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppälä. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seppälä, H., Q. He, M. Österblad, and P. Huovinen. 1994. Typing of group A streptococci by random amplified polymorphic DNA analysis. J. Clin. Microbiol. 32:1945-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seppälä, H., A. Nissinen, Q. Yu, and P. Huovinen. 1993. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J. Antimicrob. Chemother. 32:885-891. [DOI] [PubMed] [Google Scholar]
- 20.Seppälä, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petipas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and L4 ribosomal protein account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varaldo, P. E., E. A. Debbia, G. Nicoletti, D. Pavesio, S. Ripa, G. C. Schito, G. Tempera, and the Artemis-Italy Study Group. 1999. Nationwide survey in Italy of treatment of Streptococcus pyogenes pharyngitis in children: influence of macrolide resistance on clinical and microbiological outcomes. Clin. Infect. Dis. 29:869-873. [DOI] [PubMed] [Google Scholar]
- 26.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisblum, B. 2000. Resistance to the macrolide-lincosamide-streptogramin antibiotics, p. 694-710. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 28.Willetts, N. 1988. Conjugation, p. 49-77. In J. Grinsted and P. M. Bennett (ed.), Methods in microbiology. Academic Press, London, United Kingdom
- 29.Wondrack, L., M. Massa, B. V. Yang, and J. Sutcliffe. 1996. Clinical strain of Staphylococcus aureus inactivates and causes efflux of macrolides. Antimicrob. Agents Chemother. 40:992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong, P., and V. D. Shortridge. 2000. The role of efflux in macrolide resistance. Drug Resist. Updates 3:325-329. [DOI] [PubMed] [Google Scholar]