Abstract
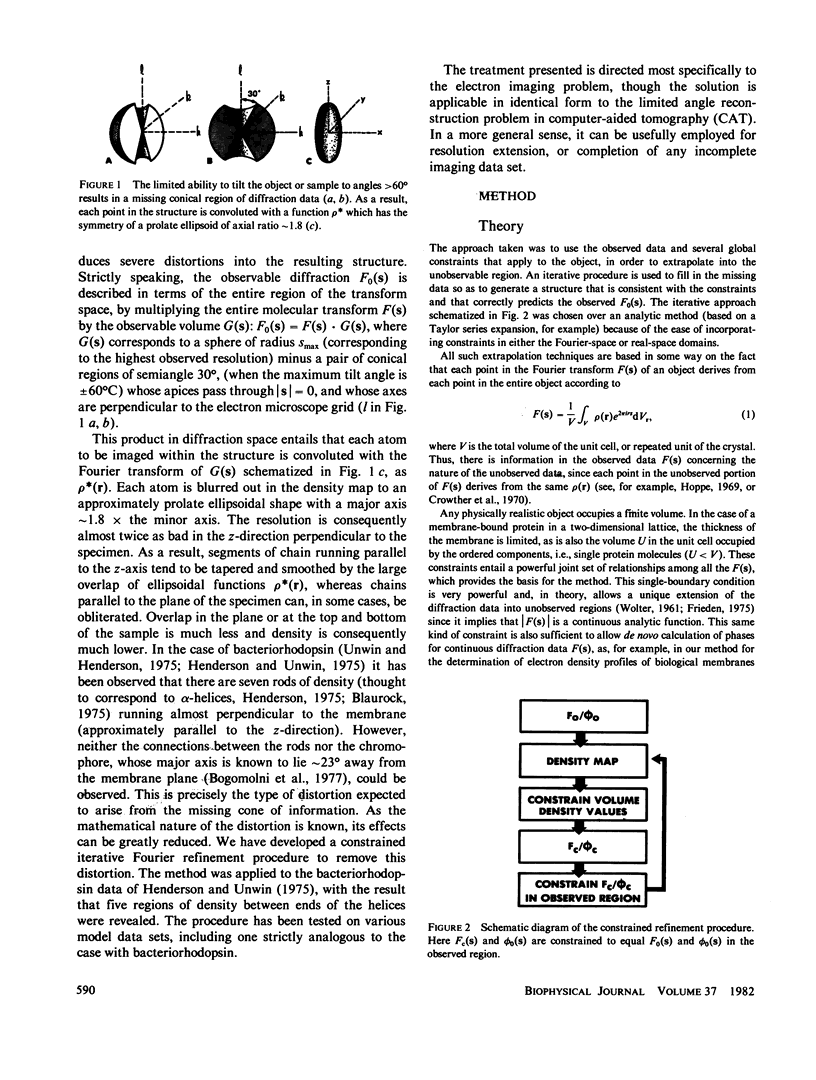
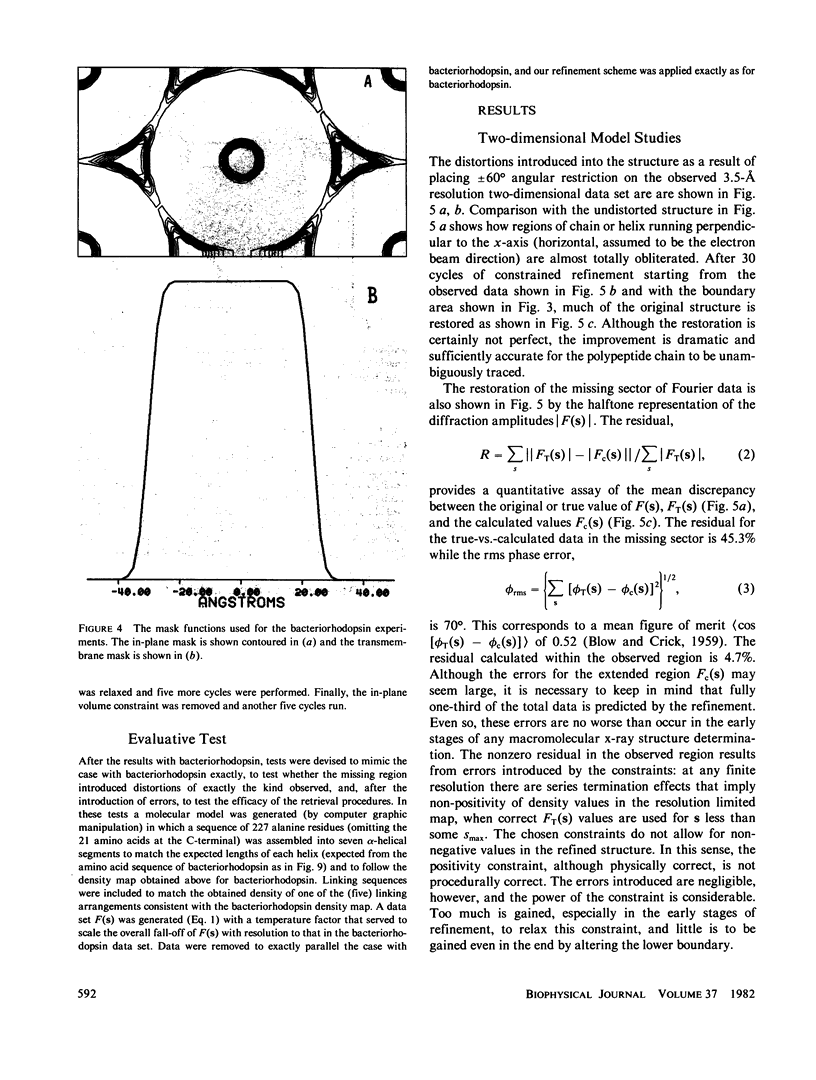
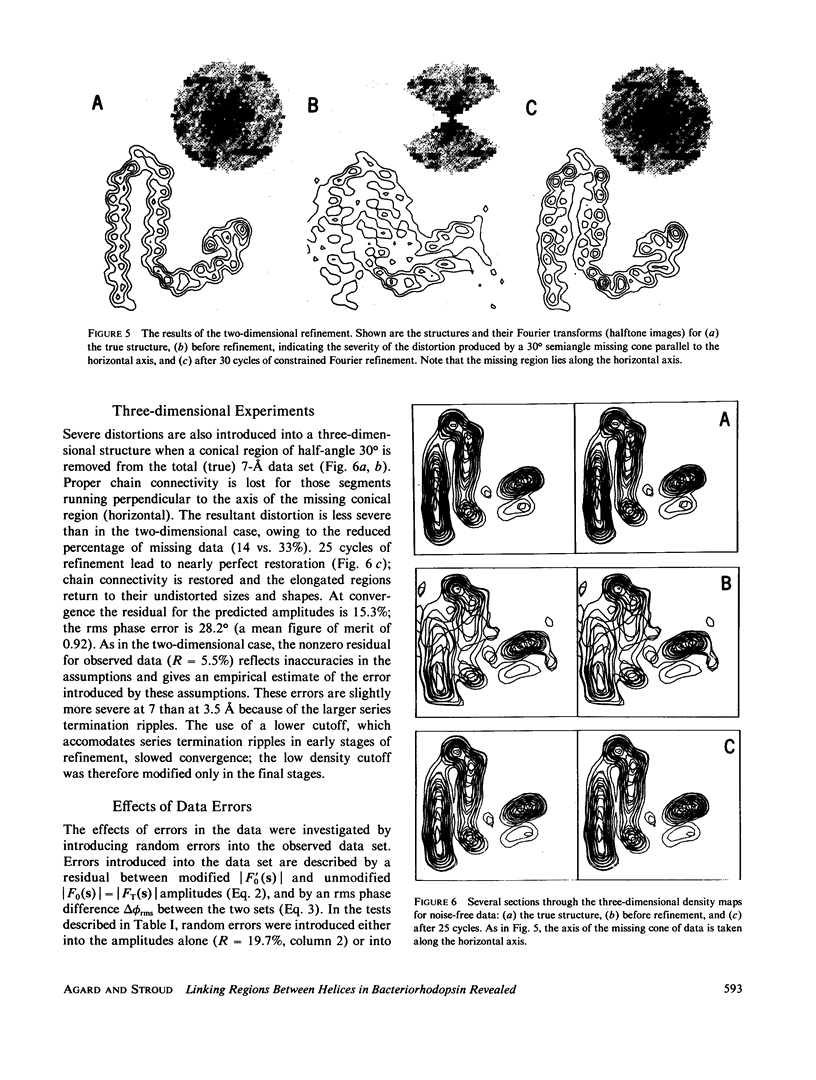
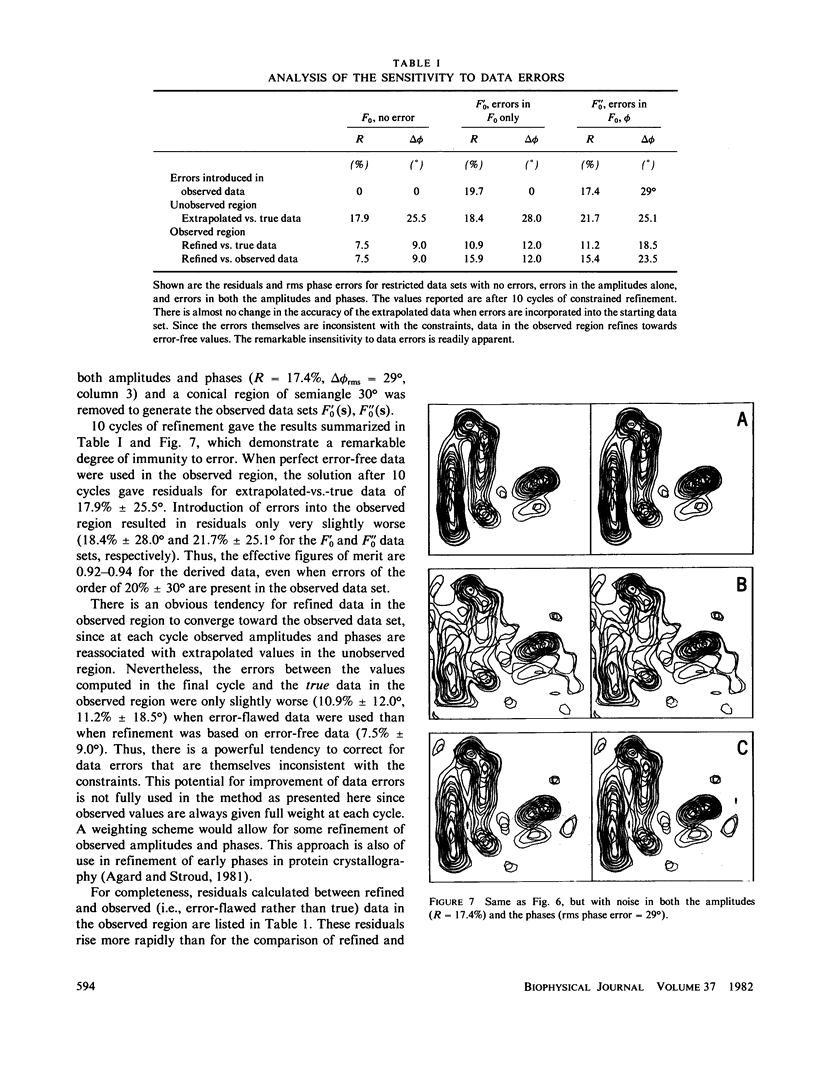
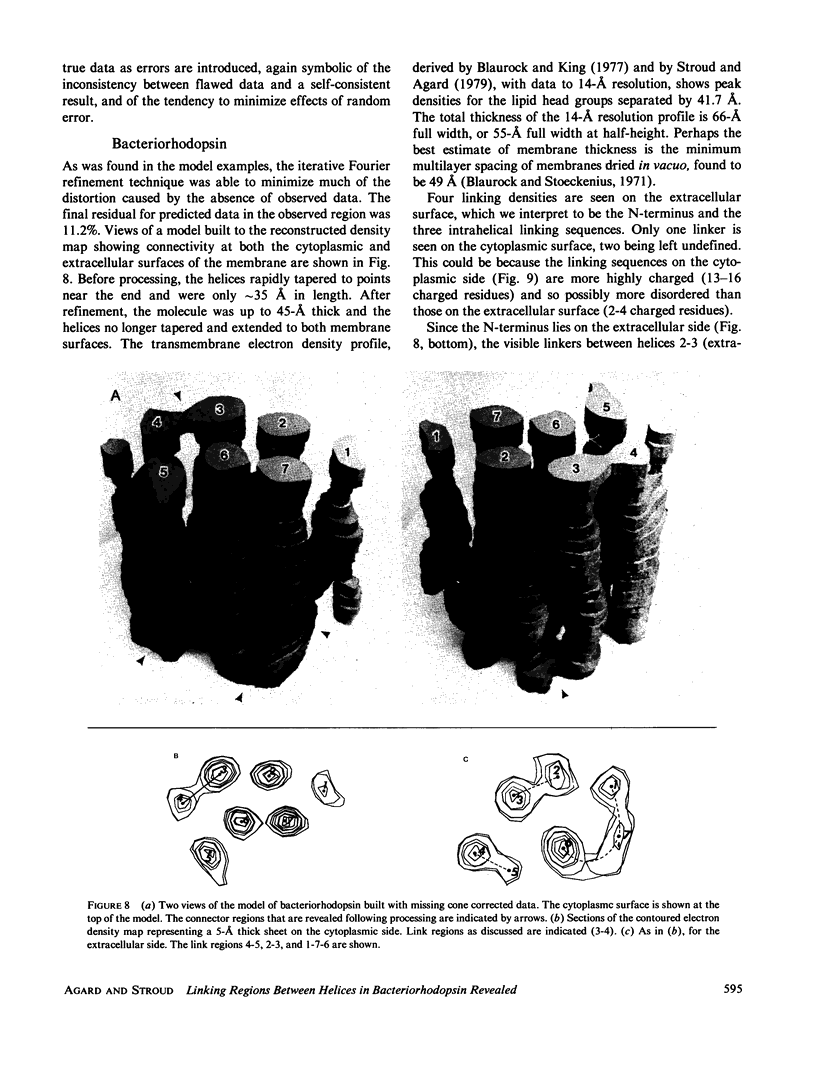
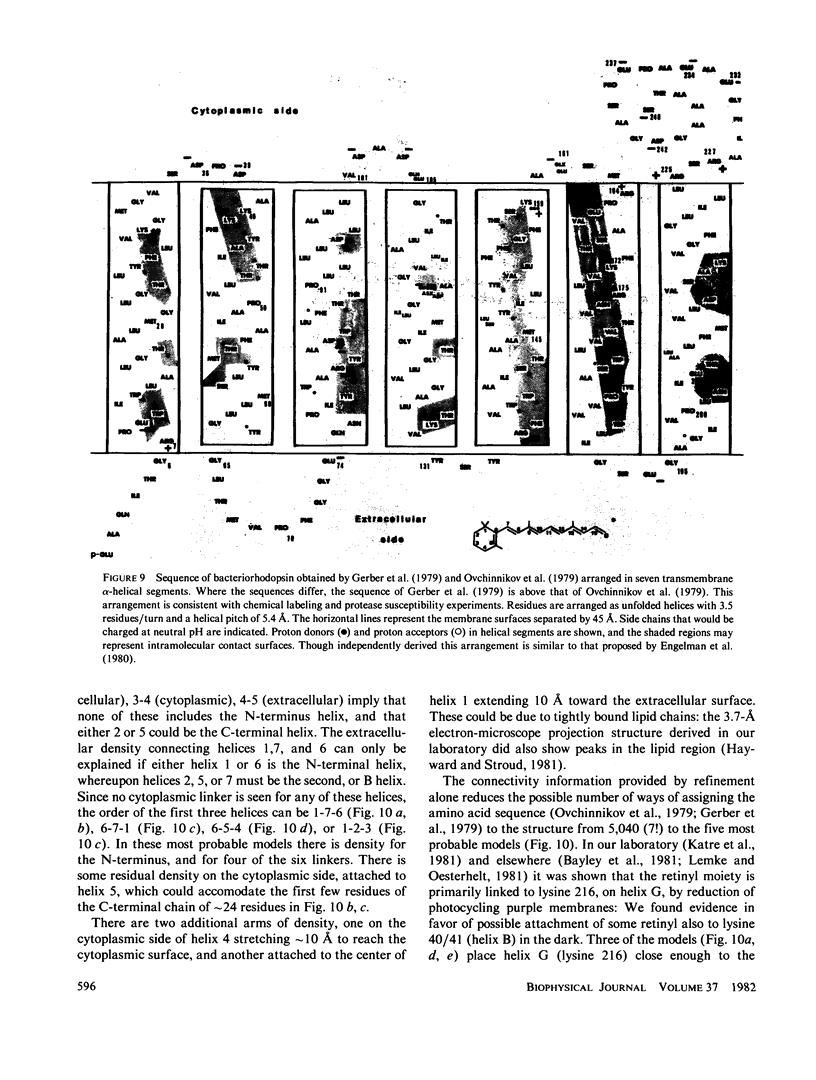
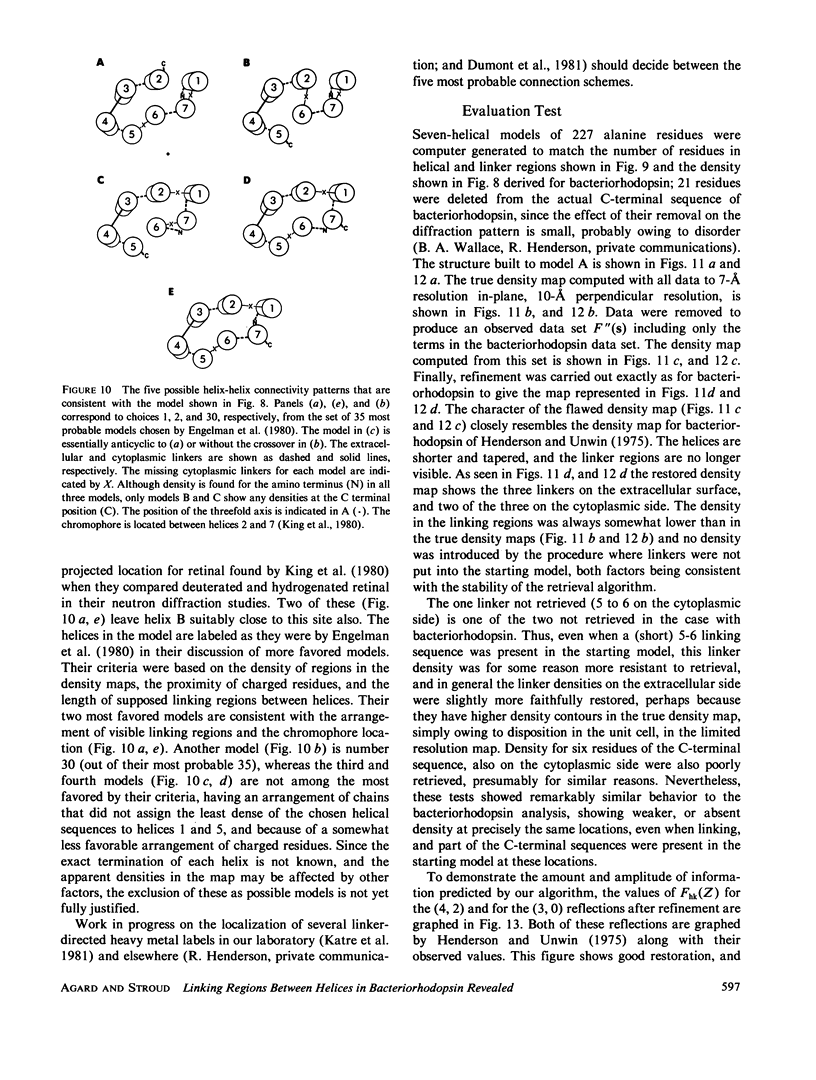
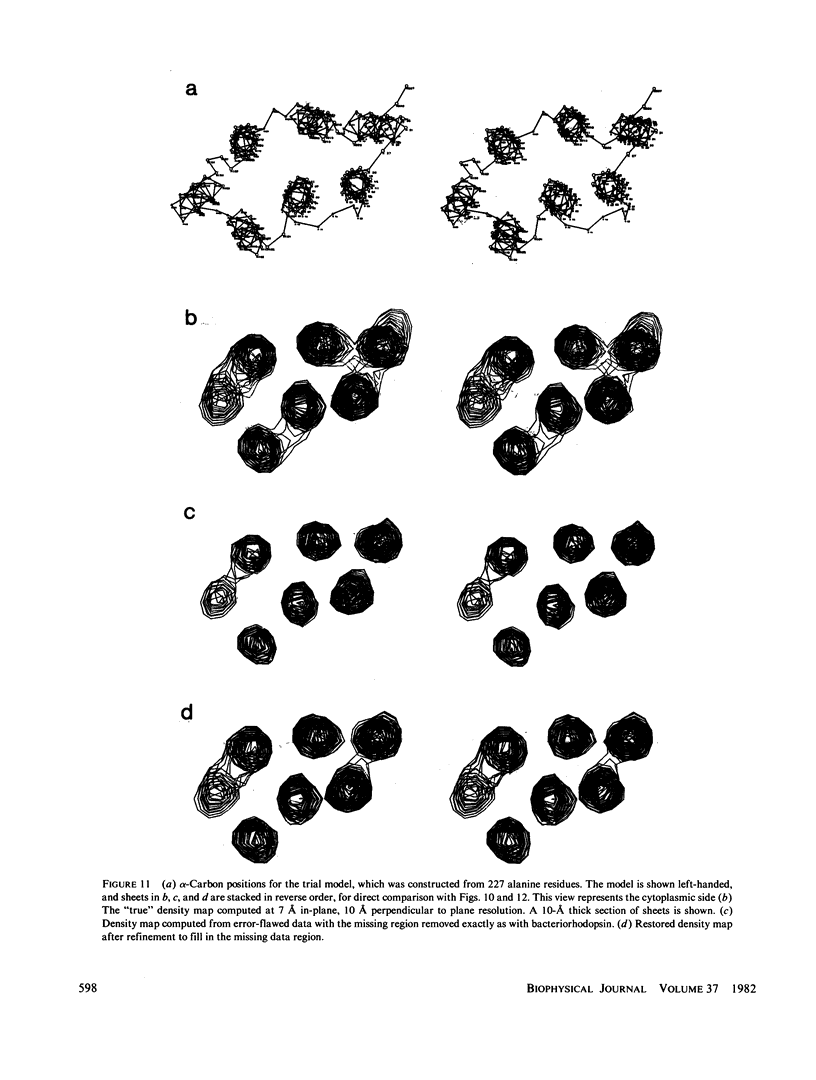
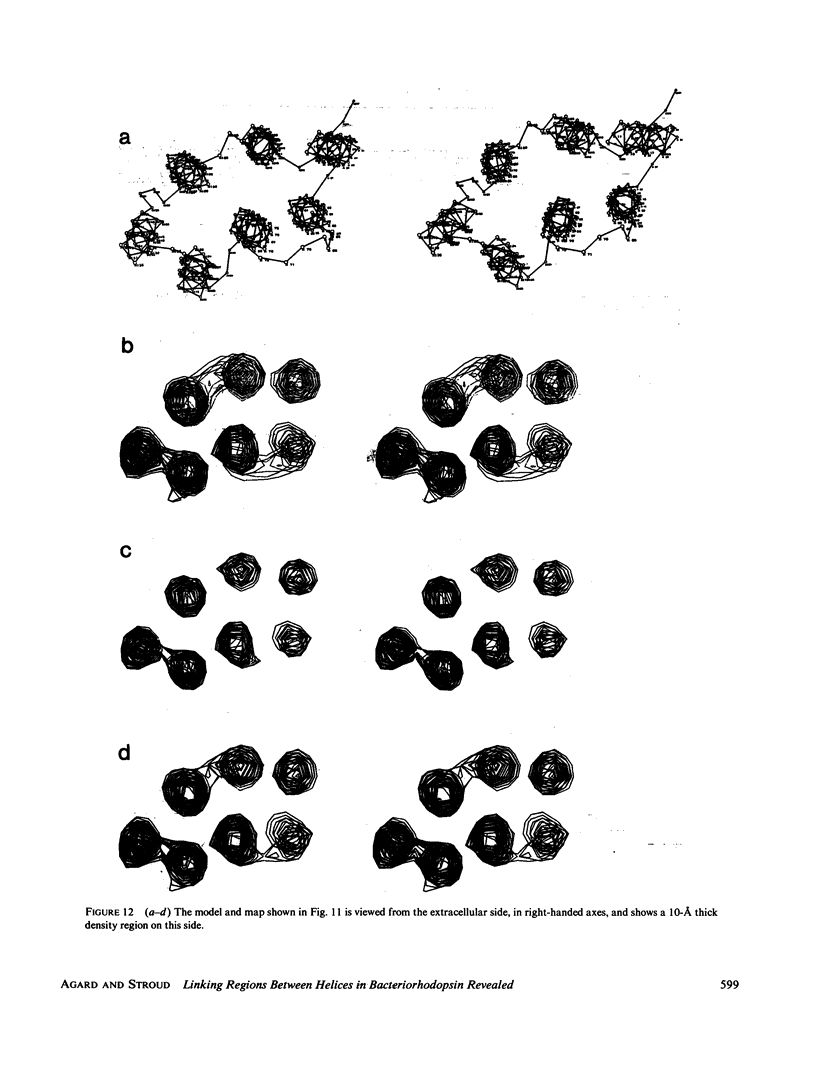
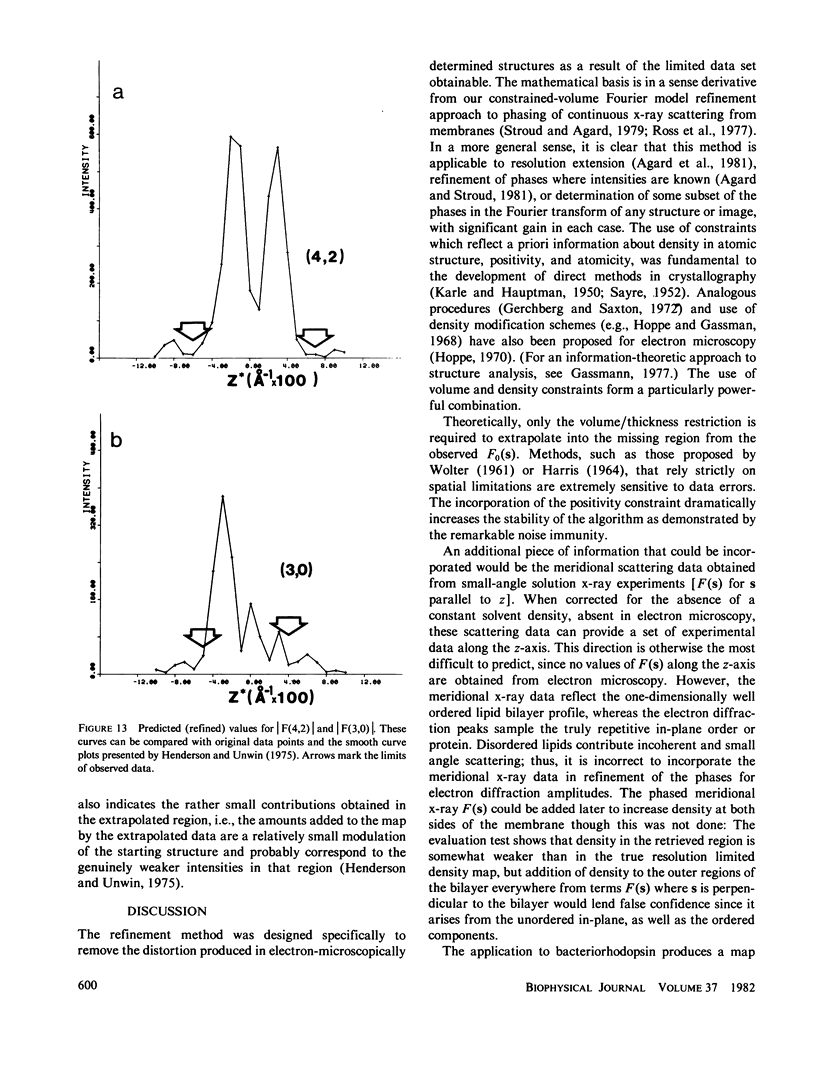
Three-dimensional electron-microscopic structural analysis requires the combination of many different tilted views of the same specimen. The relative difficulty of tilting the sample to high angles >60° without introducing severe distortion due to different focal distances across the specimen entails that the observable range of electron diffraction data is often limited to this range of angles. Thus, it is generally not possible to observe the diffraction maxima that lie within the conical region of reciprocal space around the direction perpendicular to the electron microscope grid. The absence of data in this region leads to a predictable distortion in the object, and for ±60° tilting makes the resolution essentially twice as bad in the direction perpendicular to the grid as it is for the in-plane image. Constrained density map modification and refinement methods can significantly reduce these effects. A method has been developed, tested on model cases, and applied to the electron-microscopic structure determination of bacteriorhodopsin in order to visualize the location of linking regions between helices. Electron-microscopic structural analysis of bacteriorhodopsin (Henderson and Unwin. 1975 Nature [Lond.] 257:28-32.) showed that the molecule consists of seven rods of density each nearly spanning the lipid bilayer. Owing to the distortion introduced by the missing conical region of reciprocal space data, no density was visible for the polypeptide segments linking the α-helices. Density in the refined maps indicates the location of at least five of the extrahelical segments of the polypeptide. The total number of possible ways of interconnecting the helices is reduced from 7! (5,040) to the five most consistent possibilities without recourse to other considerations. In addition, the density for the helical regions is more uniform and cylindrical throughout their length, and the length of the helices increases from 35 to 45 Å, close to the membrane thickness of 49 Å obtained for membranes dried in vacuo. Only three of the five structures consistent with the location of observed linkers place the seventh helix, onto which the chromophore can be attached by reduction in the light, at a position consistent with the main peak for deuterated retinal in the structure, as derived from neutron diffraction analysis. Two of these models are also consistent with the possible location of some of the reduced chromophore on helix B, at lys 40/41 after reduction in the dark, as well as lys 216 on helix G.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agard D. A., Steinberg R. A., Stroud R. M. Quantitative analysis of electrophoretograms: a mathematical approach to super-resolution. Anal Biochem. 1981 Mar 1;111(2):257–268. doi: 10.1016/0003-2697(81)90562-5. [DOI] [PubMed] [Google Scholar]
- Bayley H., Huang K. S., Radhakrishnan R., Ross A. H., Takagaki Y., Khorana H. G. Site of attachment of retinal in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2225–2229. doi: 10.1073/pnas.78.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaurock A. E. Bacteriorhodospin: a trans-membrane pump containing alpha-helix. J Mol Biol. 1975 Apr 5;93(2):139–158. doi: 10.1016/0022-2836(75)90124-2. [DOI] [PubMed] [Google Scholar]
- Blaurock A. E., King G. I. Asymmetric structure of the purple membrane. Science. 1977 Jun 3;196(4294):1101–1104. doi: 10.1126/science.870970. [DOI] [PubMed] [Google Scholar]
- Blaurock A. E., Stoeckenius W. Structure of the purple membrane. Nat New Biol. 1971 Sep 29;233(39):152–155. doi: 10.1038/newbio233152a0. [DOI] [PubMed] [Google Scholar]
- Dumont M. E., Wiggins J. W., Hayward S. B. Location of platinum binding sites on bacteriorhodopsin by electron diffraction. Proc Natl Acad Sci U S A. 1981 May;78(5):2947–2951. doi: 10.1073/pnas.78.5.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M., Henderson R., McLachlan A. D., Wallace B. A. Path of the polypeptide in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2023–2027. doi: 10.1073/pnas.77.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber G. E., Anderegg R. J., Herlihy W. C., Gray C. P., Biemann K., Khorana H. G. Partial primary structure of bacteriorhodopsin: sequencing methods for membrane proteins. Proc Natl Acad Sci U S A. 1979 Jan;76(1):227–231. doi: 10.1073/pnas.76.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S. B., Grano D. A., Glaeser R. M., Fisher K. A. Molecular orientation of bacteriorhodopsin within the purple membrane of Halobacterium halobium. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4320–4324. doi: 10.1073/pnas.75.9.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R. The structure of the purple membrane from Halobacterium hallobium: analysis of the X-ray diffraction pattern. J Mol Biol. 1975 Apr 5;93(2):123–138. doi: 10.1016/0022-2836(75)90123-0. [DOI] [PubMed] [Google Scholar]
- Henderson R., Unwin P. N. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975 Sep 4;257(5521):28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- Katre N. V., Wolber P. K., Stoeckenius W., Stroud R. M. Attachment site(s) of retinal in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4068–4072. doi: 10.1073/pnas.78.7.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. I., Mowery P. C., Stoeckenius W., Crespi H. L., Schoenborn B. P. Location of the chromophore in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4726–4730. doi: 10.1073/pnas.77.8.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke H. D., Oesterhelt D. Lysine 216 is a binding site of the retinyl moiety in bacteriorhodopsin. FEBS Lett. 1981 Jun 15;128(2):255–260. doi: 10.1016/0014-5793(81)80093-2. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Abdulaev N. G., Feigina M. Y., Kiselev A. V., Lobanov N. A. The structural basis of the functioning of bacteriorhodopsin: an overview. FEBS Lett. 1979 Apr 15;100(2):219–224. doi: 10.1016/0014-5793(79)80338-5. [DOI] [PubMed] [Google Scholar]
- Ross M. J., Klymkowsky M. W., Agard D. A., Stroud R. M. Structural studies of a membrane-bound acetylcholine receptor from Torpedo californica. J Mol Biol. 1977 Nov;116(4):635–659. doi: 10.1016/0022-2836(77)90264-9. [DOI] [PubMed] [Google Scholar]
- Stroud R. M., Agard D. A. Structure determination of asymmetric membrane profiles using an iterative Fourier method. Biophys J. 1979 Mar;25(3):495–512. doi: 10.1016/S0006-3495(79)85319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K. A. Structure determination of frozen, hydrated, crystalline biological specimens. J Microsc. 1978 Jan;112(1):115–125. doi: 10.1111/j.1365-2818.1978.tb01159.x. [DOI] [PubMed] [Google Scholar]
- Unwin P. N., Henderson R. Molecular structure determination by electron microscopy of unstained crystalline specimens. J Mol Biol. 1975 May 25;94(3):425–440. doi: 10.1016/0022-2836(75)90212-0. [DOI] [PubMed] [Google Scholar]




