Abstract
The Arg- and Lys-gingipains of Porphyromonas gingivalis are important virulence determinants in periodontal disease and may correspond to targets for immune- or drug-based treatment strategies. In this investigation we aimed to determine which of these enzymes represents the most promising molecular target for protease inhibitor-based therapy and to examine the effectiveness of the resultant compound in a murine virulence assay. Isogenic mutants with mutations in rgpA and rgpB (encoding Arg-gingipains) and in kgp (encoding Lys-gingipain) and a double mutant with mutations in rgpA and rgpB were prepared by using P. gingivalis W50. The virulence of these mutants indicated that Kgp is a promising drug target. Combinatorial chemistry was used to define the optimal substrate of Kgp, and from this information a specific slowly reversible inhibitor with a nanomolar Ki was designed and synthesized. Growth of P. gingivalis W50 in the presence of this compound resembled the phenotype of the kgp isogenic mutant; in both instances bacterial colonies failed to form pigment on blood agar, and only poor growth was obtained in a defined medium containing albumin as the sole protein source. Furthermore, pretreatment of the wild-type organism with the Kgp inhibitor led to a significant reduction in virulence in the murine assay. These data emphasize the conclusion that Kgp is an important factor for both nutrition and virulence of P. gingivalis and that inhibitors of this enzyme may have therapeutic potential for the control of P. gingivalis infections. Protease inhibitors may be a potentially novel class of antimicrobial agents with relevance to the control of other bacterial pathogens.
The need for novel approaches to combat bacterial infections has been highlighted by the increasing spread of resistance to conventional antimicrobial agents. Advances in our understanding of the molecular basis of infectious diseases allied to the power of biotechnology now herald the development of a new generation of antimicrobial agents which target key specific elements of the infectious agent. Bacterial pathogens are highly evolved organisms which have developed strategies, dependent upon the coordinated expression of appropriate genes, to facilitate the processes of colonization, acquisition of growth nutrients, evasion of host defenses, and, in some cases, invasion of host tissues prior to transmission to a new host environment. (7). In principle, the development of reagents which are designed to inhibit the function or the expression of one or more virulence determinants offers an alternative or adjunct approach to the control of infectious disease based solely on the use of antimicrobial agents which are designed to kill the target microorganism directly. An analogous approach is used in the development of vaccines which employ individual virulence determinants, such as attachment proteins and toxins, as the immunogens in order to prevent initial colonization and tissue pathology, respectively.
Since different bacteria have evolved their own particular molecular mechanisms for their individual survival strategies, it is likely that development of species-specific inhibitors of these processes is feasible. This is in marked contrast to the majority of the current generation of antimicrobial agents, which have as their targets highly conserved features of prokaryotic biology, such as cell wall synthesis and the machinery of protein synthesis. While there are considerable advantages to nonspecificity which allows a single drug to be effective against a range of pathogens, there are also significant drawbacks. Perturbation of the normal commensal flora is a frequent consequence of the systemic use of antibiotics, and there is an ensuing risk of opportunistic infection or outgrowth by potentially harmful bacteria and yeasts which are able to exploit the altered ecology. Furthermore, the use of broad-spectrum antibiotics favors the development of resistance in bacteria other than the pathogen being treated and hence increases the risk of subsequent transmission of this resistance via mobile genetic elements into the intended target.
Porphyromonas gingivalis is an anaerobic gram-negative rod which is considered to be one of the major etiological agents of chronic adult periodontal disease. This organism is asaccharolytic, uses amino acids and peptides for nutrition, and produces several extracellular proteases with different peptide bond specificities which are able to degrade a broad range of connective tissue and host defense macromolecules. Enzymes with absolute specificity for arginyl peptide bonds (referred to as R-gingipains) and lysyl bonds (K-gingipain) comprise the majority of the proteolytic activity synthesized by P. gingivalis in vitro and are the most-studied enzymes (5), (4).
The R- and K-gingipains appear to be particularly relevant to the deregulation of the inflammatory response and evasion of the host defenses through activation of the kinin cascade and fluid phase conversion of the complement C3 and C5 proteins (6, 28). Inhibition of the proteases of P. gingivalis by broad-spectrum, nonspecific inhibitors has been shown to enhance complement-mediated opsonophagocytosis of the bacterium by human neutrophils in vitro. Furthermore, the phenotypes of isogenic protease mutants (rgpA, rgpB, kgp) of P. gingivalis suggest that these enzymes have important roles in bacterial nutrition and proteolytic processing of other cell surface proteins (1). Hence, there is growing evidence that specific inhibitors of one or more P. gingivalis proteases may have significant therapeutic value in the control of this organism.
In this investigation we aimed to determine which of these enzymes is the most promising molecular target for protease inhibitor-based therapy and to examine the effectiveness of the resultant compound in a murine virulence assay.
MATERIALS AND METHODS
Bacteria and culture conditions.
P. gingivalis W50 and protease mutants were cultured in brain heart infusion (BHI) medium containing 5 mg of hemin per liter (25) or blood agar base containing 5% defibrinated horse blood in an atmosphere containing N2, H2, and CO2 (80:10:10; Don Whitley anaerobic work station). In some experiments a chemically defined medium containing bovine serum albumin as the sole protein source (18) was also employed. Escherichia coli XL-1 Blue MRF′ (Stratagene) was grown in Luria-Bertani medium (25a). If required, tetracycline was added at a concentration of 20 μg ml−1. In E. coli, plasmid pUC18 derivatives were selected by using ampicillin (50 μg ml−1). Plasmids were purified by ion-exchange chromatography (Qiagen).
Erythrocyte lysis.
Hemolytic activity of whole cells was measured by using a modification of the method of Chu et al. (3). Erythrocytes were prepared from sterile horse blood by washing them four times in buffer (0.015 M NaCl, 10 mM CaCl2, 10 mM MgCl2, 5 mM sodium citrate; pH 6.8) until the supernatant was free of hemoglobin pigmentation and then resuspending them in the same buffer at a concentration of 2 × 109 erythrocytes per ml. Cells from an 18-h culture of P. gingivalis W50 or a protease mutant were harvested by centrifugation and resuspended in fresh medium to a concentration of approximately 2 × 1010 cells ml−1. When required, protease inhibitor was added to the bacterial cell suspensions at this stage, which was followed by anaerobic incubation at 37°C. Then 0.75 ml of bacterial cells was added to 0.5 ml of erythrocytes and an additional 0.25 ml of buffer, and the suspension was slowly mixed at 37°C. Samples (100 μl) were removed periodically for up to 24 h, diluted in 900 ml of buffer, and centrifuged at 3,500 × g for 5 min. The supernatants were then diluted 10-fold in buffer, and the optical density at 405 nm was measured to estimate hemoglobin release. The value for 100% hemolysis was determined by lysing 0.5 ml of an erythrocyte suspension in 1 ml of deionized water. All in vitro growth and erythrocyte lysis assays were performed in duplicate on two separate occasions, and mean values are presented below.
Protease mutant construction.
We have previously described insertional inactivation of rgpA, rgpB, rgpA plus rgpB, and kgp in P. gingivalis W50 with an Erm cassette (8) for single knockouts and a TetQ cassette (15) for the double knockout to generate W501 (rgpA::erm), D7 (rgpB::erm), E8 (rgpA::tetQ rgpB::erm), and K1A (kgp::erm), respectively,(1, 2, 24). Initial selection of mutants was performed on blood agar plates containing either clindamycin (5 μg ml−1) for Erm or clindamycin and tetracycline (1 μg ml−1) for both Erm and TetQ. The procedures used for cloning into plasmids and electrotransformation of P. gingivalis have been described previously (1, 2, 24). Following allelic exchange mutagenesis, at least six mutants in each category were characterized by enzyme assays (Arg-X and Lys-X protease activity), Western blotting, and phenotypic analysis. Southern blotting and PCR analysis were used to confirm that the mutants were isogenic. Representative mutants were also examined by Northern blotting to confirm loss of the corresponding mRNA.
Similarly, insertional inactivation of kgp was performed following interruption of the coding region in vitro approximately one-third of the way into the catalytic domain with the Erm cassette. The construct was then used as described above to generate P. gingivalis K1A (kgp) (1). All mutants (rgpA, rgpB, rgpA rgpB, and kgp) were stable upon repeated subculturing in the absence of selection pressure, and hence antibiotics were not routinely included in the growth media.
Protease purification.
RgpAcat, a soluble monomeric protease derived from rgpA, was purified from the culture supernatant of P. gingivalis W50 by using chromatographic procedures described previously (25). Kgp was purified from 24-h cultures of P. gingivalis W50 grown in BHI medium supplemented with hemin (5 mg liter−1). In a typical experiment, 2 liters of a P. gingivalis W50 culture was centrifuged at 10,000 × g for 60 min, resuspended in 50 ml of 0.025 M MOPS (morpholinepropanesulfonic acid) buffer (pH 6.0)-0.01 M EDTA-0.01 M 2-mercaptoethanol, and sonicated six times (1 min each; amplitude, 12 μm) with cooling in ice between sonication procedures. The cell extract was centrifuged at 18,000 × g for 45 min at 4°C. The supernatant containing Rgp and Kgp activities was applied to a column of Sephacryl S-200HR (length, 95 cm; inside diameter, 5 cm) equilibrated in 0.05 M sodium acetate buffer (pH 5.3). The peak fractions of Kgp were combined, the pH was adjusted to 6.0 by addition of solid sodium acetate, and the sample was applied to a column of Arg-agarose (50 ml) equilibrated in 0.025 M MOPS buffer (pH 6.0)-0.01 M CaCl2-0.2 M NaCl-0.01 M 2-mercaptoethanol. Kgp activity was selectively eluted with 0.3 M lysine in equilibration buffer, whereas Rgp activity remained bound to the column and could be eluted only with 0.1 M arginine in buffer. The fractions containing Kgp activity were pooled, dialyzed against 0.05 M Tris-HCl buffer (pH 7.5)-0.01 M 2-mercaptoethanol, and applied to a column of HA-Ultrogel (10 ml; Seprocor) equilibrated in 0.05 M Tris-HCl buffer (pH 7.5)-0.01 M 2-mercaptoethanol. Elution of Kgp was carried out in three steps, as follows: (i) equilibration buffer containing 1 M NaCl, (ii) 0.2 M sodium phosphate buffer (pH 6.0) containing 0.01 M 2-mercaptoethanol, and (iii) 2 M sodium phosphate buffer (pH 5.3) containing 0.01 M 2-mercaptoethanol. Kgp activity eluted off the column in the second step. Fractions with constant high specific activity were combined and dialyzed against 0.025 M sodium acetate buffer (pH 5.3) prior to a final purification step performed by using anion-exchange chromatography on DEAE-Sephacel and a gradient of sodium acetate buffer (pH 5.3) ranging from 0.025 to 0.25 M. The specific activities of purified RgpAcat and Kgp were 22.7 and 2.5 U/mg, respectively, when α-N-benzoyl-dl-arginine-p-nitroanilide and α-N-acetyl-l-lysine-p-nitroanilide were used as substrates for Rgp and Kgp. One unit was defined as the amount of enzyme which hydrolyzed 1 μmol of substrate under the conditions of the assay. The purity of all enzyme preparations was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and N-terminal amino acid sequencing.
Kgp inhibitor design and synthesis.
Initially, the active site of Kgp was mapped by using an in-house patented technology (Rational Proteinase Inhibitor Design [RAPiD]) (23). RAPiD provides a substrate for inhibitor testing and also defines the residues allowed at each position of substrates generally and their interrelationships. In the next step, kinetic measurements for a large number of the substrates found in the initial procedure were obtained, from which a pharmacophoric map of the active site was constructed.
RAPiD screening in substrate mode.
The 10 mM RAPiD mother plates were sequentially diluted to 1 mM and then to 100 μM (both in 100% dimethyl sulfoxide [DMSO]) and finally to 10 μM (in 1 mM Tris [pH 7.0] containing 10% [vol/vol] DMSO). Assays were carried out by dispensing 10 μl of the 10 μM library per well and adding 90 μl of the protease solution (0.5 nM Kgp in 50 mM bis-Tris propane [pH 8.0] containing 1% [vol/vol] Triton X-100 and 5 mM 2-mercaptoethanol) to initiate the reaction. Protease activity was monitored over time by collecting data at zero time and at 1, 2, 4, and 24 h. The data were saved as text files and imported, arranged, and presented by using the FOCUS software (written by Peptide Therapeutics Group, plc.).
Measurement of the inhibition constant (Ki).
Fifty microliters of Kgp (1 nM in 50 mM bis-Tris propane [pH 8.0] containing 1% [vol/vol] Triton X-100 and 5 mM 2-mercaptoethanol) was added to columns 1 to 11 of a 96-well plate, and 100 μl was added to column 12. Two microliters of inhibitor (100 μM in 100% DMSO) was added to column 12, the sample was mixed three times by pipetting, and a doubling dilution was prepared across the plate by serial transfer into adjacent wells. Fifty microliters of succinyl-Ala-Phe-Lys-(7-amido-4-methylcoumarin) (40 μM in buffer) was added to all wells, and the contents were mixed. The reaction was monitored for AMC fluorescence for 15 min at 25°C, and the progress curves were automatically converted to rates by the Fluoroskan Ascent software.
Arg-gingipain, trypsin, and cathepsin B were assayed by using essentially the same procedure, except that for Arg-gingipain the substrate was Z-Arg-AMC, for trypsin the buffer was 10 mM Tris-10 mM CaCl2 (pH 8.0) and the substrate was Z-Gly-Gly-Arg-AMC, and for cathepsin B the buffer was 50 mM sodium phosphate-1 mM EDTA (pH 6.25)-10 mM 2-mercaptoethanol and the substrate was Z-Arg-Arg-AMC. Trypsin (human, iodination grade) was obtained from Sigma, and cathepsin B was obtained from Athens Research and Technology (Athens, Ga.)
The inhibition constants were calculated by using the following equation with the assumption that inhibition was fully competitive.
 |
where vi is the observed residual activity, [S] is the substrate concentration used in the assay, Vmax is the maximal velocity at an inhibitor concentration of zero, Ki is the inhibitor dissociation constant, and [I] is the inhibitor concentration. Curves were fitted by nonlinear regression analysis by using fixed values for the substrate concentration and the value of the Michaelis constant (Km). Data analysis was carried out by using Prism v 2.01 (GraphPad, San Diego, Calif.)
General description and synthesis of Kgp inhibitor A71561.
The International Union of Pure and Applied Chemistry name for the inhibitor A71561 molecule is 1-(3-phenylpropionyl)piperidine-3-(R,S)-carboxylic acid-[4-amino-1(S)-(benzothiazole-2-carbonyl)butyl] amide. A71561 was prepared in solution by initially coupling 3-phenylpropionic acid to (R,S)-nipecotic acid. This was followed by coupling the resulting product to (N-carbobenzyloxy)-lysine ketobenzothiazole by standard carbodiimide activation. As a final step, the Nd-Cbz protection was removed by HBr in acetic acid, and the resulting crude product was purified by reverse-phase high-performance liquid chromatography (C18, 5μ; Phenomenex Jupiter, Macclesfield, Cheshire, United Kingdom). Column elution was carried out by using a linear gradient of water-0.1% trifluoracetic acid to acetonitrile-0.1% trifluoracetic acid over 30 min. The purified molecule was characterized by using high-performance liquid chromatography, electrospray mass spectrometry, and 1H and 13C individual and correlation nuclear magnetic resonance spectra.
Murine virulence model.
The virulence potentials of P. gingivalis W50 and protease mutants and the effect of protease inhibition were examined by using the murine lesion model as described by Kesavalu et al. (12). Briefly, bacterial cells from an 18-h culture in BHI medium containing 5 mg of hemin per liter were harvested by centrifugation and resuspended to a final density of 1011 cells ml−1 in the culture medium. Cell suspensions were then prepared at the required cell density for inoculation into the mice. Protease inhibitor stock solutions (final concentration, 40 mM in DMSO) were added to the cell suspensions to obtain the desired final concentrations, and the suspensions were then incubated anaerobically for 30 min. Aliquots were then removed to determine total viable counts and Arg-gingipain and Lys-gingipain enzyme activities. Control bacterial cell suspensions were incubated with DMSO alone.
Groups of BALB/c mice (usually 8 or 12 mice) were inoculated subcutaneously twice with 100-μl portions of bacterial cell suspensions on the midpoint of each flank towards the dorsal surface. In each experiment, three bacterial doses were usually employed, equivalent to 2 × 1010, 1 × 1010, or 5 × 109 bacterial cells per mouse in each group. The animals were then monitored twice daily for 10 days for the development of lesions and to assess their general condition according to a standardized appearance and behavior scoring system which was approved by the local ethics committee and the United Kingdom Home Office animal experimentation licensing authority. The dimensions of lesions were measured with calipers. Moribund animals and animals with lesions that were more than 15 mm in any dimension were sacrificed and recorded as deaths. Only the data for time of survival are shown below. All animal experiments were conducted on at least two separate occasions.
Statistical analysis.
We used standard methods for survival analysis, allowing for right-censoring of time to death when animals survived to the scheduled end of the experiment. Life table methods were used to display cumulative survival probabilities during the experiment, while Cox proportional hazard models were used to estimate hazard ratios for comparisons of interest, controlling for the effects of bacterial dose and for between-experiment variation in cumulative survival.
RESULTS
Growth characteristics of P. gingivalis protease mutants.
As previously reported, the growth rates and final cell yields of the rgpA and rgpB single and double mutants and the kgp mutant are equivalent to those of parent strain W50 during growth in BHI medium (1, 24). However, growth of K1A (kgp) is significantly reduced compared to growth of the wild type in a chemically defined medium in which bovine serum albumin is the sole source of amino acids (18). No gross differences in the rates of growth on blood agar were observed for the mutants. In the case of the parent strain and the rgpA rgpB mutants, the colonies were pigmented by day 7, and clear zones of hemolysis were evident by day 10. However, K1A (kgp) failed to show signs of pigmentation or hemolysis by day 10.
Virulence phenotype P. gingivalis protease mutants.
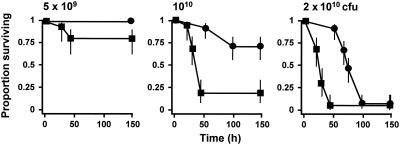
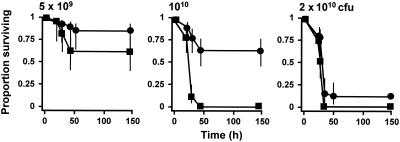
Single insertional inactivation of either the rgpA or rgpB gene resulted in no significant difference between the virulence of the resultant mutant and the virulence of the parent strain on the basis of lesion development and survival times of treated animals (data not shown). However, mutant E8 (rgpA::tetQ rgpB::erm) was significantly less virulent in the murine model (Fig. 1). At the lowest dose of bacterial cells (5 × 109 CFU/mouse) 80% of the mice inoculated with the parent strain survived over the course of the experiment. In comparison, all mice inoculated with E8 at this dose survived, although there was no significant reduction in the hazard ratio (data not shown). However, at higher doses, 1010 and 2 × 1010 bacterial cells/mouse, the hazard ratios of the rgpA rgpB double mutant compared to the parent strain were 0.159 (95% confidence interval, 0.078 to 0.319; P < 0.001) and 0.206 (95% confidence interval, 0.114 to 0.643; P < 0.001), respectively. Hence, in this model system inactivation of both Arg-gingipain-encoding genes was required to attenuate the virulence of P. gingivalis W50.
FIG. 1.
Virulence of P. gingivalis W50 and rgpA rgpB mutant: survival curves for animals in the murine model of P. gingivalis virulence. Groups of mice (n = 12) were inoculated with 5 × 109, 1010, or 2 × 1010 CFU of P. gingivalis W50 (▪) or the rgpA rgpB mutant (•). The vertical lines indicate standard deviations.
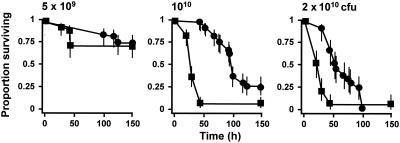
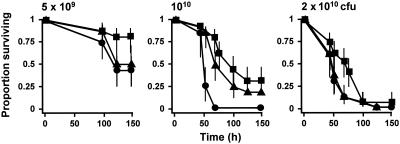
Loss of function of kgp also led to a reduction in the virulence of the resultant mutant (Fig. 2). The reduction was significant at doses of 1010 and 2 × 1010 CFU/mouse, at which the hazard ratios for survival were 0.175 (95% confidence interval, 0.111 to 0.276; P < 0.001) and 0.372 (95% confidence interval, 0.229 to 0.608; P < 0.001), respectively, relative to the survival of parent strain W50. Since hemin accumulation and the characteristic pigmentation of P. gingivalis do not occur in the kgp mutant, presumably as a consequence of an inability to degrade hemoglobin and hence harvest hemin from erythrocytes, we examined whether coadministration of free hemin with the P. gingivalis (kgp) inoculum into the mice was able to restore the virulence of this mutant (Fig. 3). Allowing for bacterial dose, inclusion of hemin at a concentration of 5 mg liter−1 in the two 100-μl inocula resulted in an increase in the hazard ratio to 1.691 (95% confidence interval, 1.199 to 2.385; P = 0.003) relative to the mutant cells without hemin, and the ratio increased to 2.534 (95% confidence interval, 1.624 to 3.954; P < 0.001) when 50 mg of hemin per liter was present in the inoculum. Hence, high concentrations of free hemin are able to partially complement the kgp mutation in P. gingivalis in this virulence model.
FIG. 2.
Virulence of P. gingivalis W50 (▪) and the kgp mutant (•): survival curves for animals in the murine model of P. gingivalis virulence. See the legend to Fig. 1 for an explanation of the bacterial doses.
FIG. 3.
Influence of coadministration of hemin on the virulence of P. gingivalis (kgp). Bacterial cell suspensions were mixed with hemin at a final concentration of 5 mg liter−1 (▴) or 50 mg liter−1 (•), and the effects were compared with the effects of untreated cells (▪). See the legend to Fig. 1 for an explanation of the bacterial doses.
Influence of leupeptin on virulence of P. gingivalis W50.
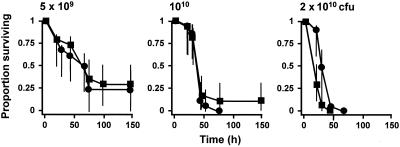
In order to examine the effect of a commercially available inhibitor of the Arg-gingipains on the virulence of P. gingivalis in the murine model, cells of the W50 parent strain were pretreated with 2 mM leupeptin, a potent inhibitor of these enzymes, prior to subcutaneous inoculation. No residual Arg-gingipain activity was detected in cell suspensions at any of the three bacterial doses employed following leupeptin treatment, while the Lys-gingipain activity was unaffected (data not shown). However, we observed no effect of leupeptin inhibition on the virulence of the parent strain at any bacterial dose (Fig. 4). When bacterial dose was controlled for, the overall hazard ratio when leupeptin was included was 0.836 (95% confidence interval, 0.528 to 1.324; P = 0.445).
FIG. 4.
Virulence of P. gingivalis W50 pretreated with leupeptin in the murine model. Bacterial cell suspensions were preincubated with leupeptin (2 mM) for 30 min under anaerobic conditions prior to inoculation. Symbols: ▪, untreated cells; •, leupeptin-treated cells. See the legend to Fig. 1 for an explanation of the bacterial doses.
In light of these data and the major influence of insertional inactivation of kgp on the virulence of P. gingivalis (coupled to a plausible biological explanation for the effect based on an inability to sequester hemin), we concentrated on the development and application of a specific inhibitor of Lys-gingipain.
Properties of A71561, a specific inhibitor of Kgp.
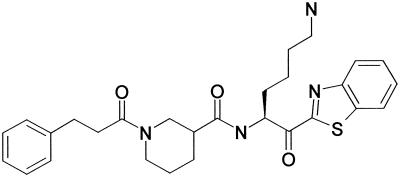
Guided by the data from the RAPiD protease prescreening which indicated an absolute requirement for Lys in P1 and little specificity for P2 to P4 residues in the substrate, we screened Kgp against an existing inhibitor library in order to identify inhibitors that were selective for Kgp over Rgps, and inhibition constants were calculated. Based on this analysis, the following inhibitor was selected: 1-(3-phenylpropionyl)piperidine-3(R,S)-carboxylic acid-[4-amino-1(S)-(benzothiazole-2-carbonyl)butyl] amide (A71561), which was known to not inhibit several mammalian proteases. A71561 is a slowly reversible inhibitor of Kgp with a Ki of 0.9 nM and shows no inhibition of the Arg-gingipains at concentrations up to 100 μM. It displays 30-fold selectivity over trypsin (Ki = 30 nM) and 120,000-fold selectivity over cathepsin B (Ki = 115 μM), two enzymes with substrate specificity related to that of Kgp. The structure of A71561 is shown in Fig. 5.
FIG. 5.
Structure of the Kgp inhibitor 1-(3-phenylpropionyl)piperidine-3(R,S)-carboxylic acid-[4-amino-1(S)-(benzothiazole-2-carbonyl)butyl] amide (A71561) used in this study.
Effects of specific inhibition of Kgp on the in vitro phenotype of P. gingivalis W50: growth of P. gingivalis in the presence of A71561.
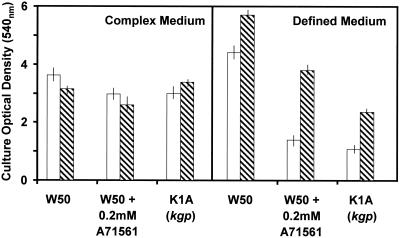
Incorporation of A71561 into BHI medium at a concentration of 2 mM had no effect on the growth of P. gingivalis W50 as determined by culture optical density at 24 and 48 h (Fig. 6). No Kgp activity was detectable in the culture at either time. The growth of P. gingivalis W50 in the presence of A71561 in BHI medium is analogous to the growth of the kgp mutant in this complex growth medium (1) (Fig. 6). In contrast, in the defined medium containing bovine serum albumin as the sole source of peptides and amino acids, the presence of the Kgp inhibitor led to a significant reduction in the growth of strain W50 similar to the effect of insertional inactivation of kgp on the growth of the mutant strain in this medium (Fig. 6)
FIG. 6.
Influence of incorporation of the synthetic Kgp inhibitor (A71561) into complex or defined medium on the growth of P. gingivalis W50. The open bars indicate culture optical densities after 24 h, and the cross-hatched bars indicate culture optical densities after 48 h. No Kgp activity was detectable in the cultures with inhibitor up to 48 h. The vertical lines indicate standard deviations.
Effect of Kgp inhibition on pigmentation and hemolysis of P. gingivalis W50.
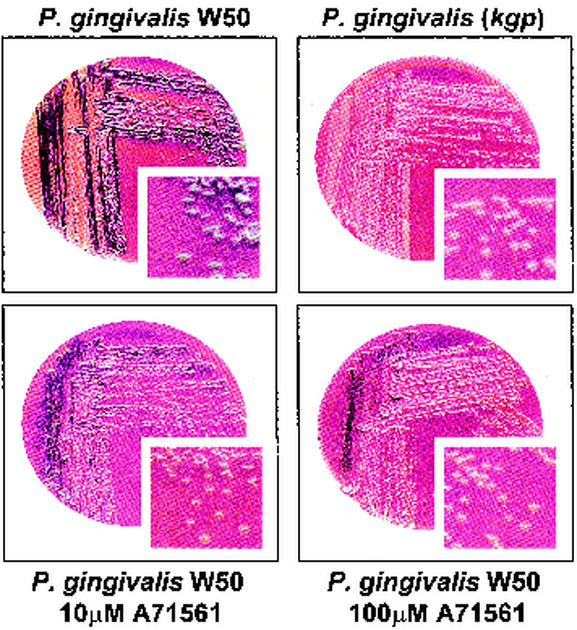
Growth of P. gingivalis W50 on blood agar plates containing 10 μM A71561 led to a reduction in the pigmentation of the colonies after 10 days compared to the pigmentation of the parent strain and to the absence of a clear zone of hemolysis. In the presence of 100 μM A71561, the W50 colonies remained creamy white after 10 days of growth, showed no hemolytic activity, and were similar in appearance to colonies of the kgp mutant strain (Fig. 7).
FIG. 7.
Effect of incorporation into blood agar of the synthetic Kgp inhibitor (A71561) on the pigmentation and hemolysis of P. gingivalis W50 and the kgp mutant strain.
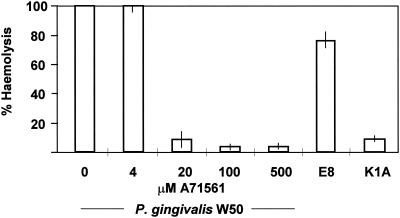
A more quantitative assessment of hemolysis was obtained by the erythrocyte lysis assay. This assay demonstrated that there was a >90% reduction in the hemolytic activity of K1A cells from 18-h broth cultures compared to the activity of parent strain W50. Single mutations in rgpA and rgpB resulted in <10% reduction in the hemolytic activity, while mutations in both rgpA and rgpB led to a reduction of approximately 20%. Inclusion of A71561 at concentrations greater than 0.02 mM in the erythrocyte lysis assay mixture with P. gingivalis W50 whole cells reduced the hemolytic activity to approximately 10% of the activity of the cells in the absence of inhibitor (Fig. 8).
FIG. 8.
Inhibition of P. gingivalis erythrocyte lysis by the synthetic Kgp inhibitor (A71561). Washed cells from an overnight culture of P. gingivalis were resuspended in buffer containing 0 to 500 μM A71561 before incubation with horse erythrocytes for 24 h. The lysis induced by P. gingivalis E8 (rgpA rgpB) and the lysis induced by P. gingivalis K1A (kgp) are shown for comparison. The vertical lines indicate standard deviations.
Hence, as determined by growth, pigmentation, and hemolytic activity analyses, inhibition of the Kgp activity of P. gingivalis W50 with a specific protease inhibitor resulted in a phenotype that mimicked the phenotype of the kgp mutant.
Effects of specific inhibition of Kgp on P. gingivalis virulence.
Treatment of cell suspensions P. gingivalis W50 containing 2.5 × 1010, 5 × 1010, and 1011 CFU/ml with 2 mM A71561 led to complete inhibition of Kgp activity but had no influence on the Rgp activity of these suspensions (data not shown). The treated cell suspensions were significantly less virulent in the murine model than uninhibited cells. This was particularly evident at a concentration of 1010 CFU/mouse, at which the hazard ratio was 0.134 (95% confidence interval, 0.061 to 0.293; P < 0.001), but statistical significance was also observed with the 2 × 1010-CFU/ml dose (hazard ratio, 0.526; 95% confidence interval, 0.286 to 0.969; P < 0.05). After bacterial dose and experiment were controlled for, the Kgp inhibitor caused significant attenuation in this model (overall hazard ratio, 0.276; 95% confidence interval, 0.177 to 0.430; P < 0.001) (Fig. 9). Use of the Kgp inhibitor at concentrations lower than 2 mM did not lead to a reduction in the virulence of P. gingivalis even though A71561 concentrations as low as 0.1 mM were sufficient to block all of the Kgp activity of the initial inoculum.
FIG. 9.
Virulence of P. gingivalis W50 pretreated with the Kgp inhibitor (A71561) in the murine model. Bacterial cell suspensions were preincubated with A71561 for 30 min under anaerobic conditions prior to inoculation. Symbols: ▪, untreated cells; •, A71561-treated cells. See the legend to Fig. 1 for an explanation of the bacterial doses.
DISCUSSION
The production of extracellular proteases by many bacterial pathogens is considered to be an important characteristic which contributes to the disease process. The pathological effects can be attributed to direct action on host connective tissue proteins, including the collagenases of Clostridium spp., and the extracellular metallo- and elastase-like proteases of Serratia spp. and Pseudomonas spp., which are able to degrade several host proteins, including fibronectin and elastin. Alternatively, potentiation of inflammatory processes may cause pathology indirectly. Activation of proinflammatory cytokines, the bradykinin generation system, and other endogenous host protease systems, as well as inactivation of host protease inhibitors, immunoglobulins, and complement, are well-established examples (27).
The production of extracellular proteases may be beneficial to the organism through the development of new sites for colonization within the host, for the acquisition of carbon and nitrogen sources and micronutrients, such as metal ions for anabolic and catabolic purposes, and for evasion of host defenses. In principle, therefore, the inhibition of bacterial extracellular protease function may have therapeutic value not only because it limits the direct and indirect damage to the host caused by the protease in question but also because it interferes with a critical component of the pathogen's survival strategy. This may be particularly relevant when the physiological function of the protease is an important component of its strategy to evade host defenses. In this instance, loss of protease function may render the organism more susceptible to control by the endogenous defense mechanisms of the host.
In this investigation we tried to identify a target protease of P. gingivalis for inhibitor design and synthesis based on a virulence assay performed with isogenic mutants and a murine model of soft-tissue destruction. While the model used differs significantly from the natural history of the chronic disease associated with this organism in human oral infections, it represents a simple and readily quantifiable means of determining the invasive capacity of the strains examined and was considered the most practical first screening procedure for target enzymes and inhibitors.
Insertional inactivation of both Arg-gingipain-encoding genes, rgpA and rgpB, led to a significant reduction in the virulence of P. gingivalis W50 in the present study. This is in accord with previous studies which demonstrated the pathogenic importance of the Arg-gingipains in animal models. For example, immunization of both mice and primates with preparations containing these enzymes or synthetic peptides corresponding to sequences from the catalytic domain can protect animals from subsequent challenge by live bacteria (9, 10, 19) Inactivation of either rgpA or rgpB alone was insufficient to reduce the virulence of P. gingivalis significantly in this study. Other workers have reported that loss of function of only rgpA or rgpB may lead to attenuation of the virulence of P. gingivalis in a similar murine model (8, 21). The differences between the previous studies and the present study may reflect strain differences and/or the sensitivities of the models.
Although loss of function of the Arg-gingipains through gene inactivation led to a significant reduction in virulence, we observed no such attenuation following chemical inhibition of these enzymes by leupeptin treatment of the wild-type strain prior to inoculation. There may be a number of reasons for this discrepancy. First, in addition to their extracellular functions, the Arg-gingipains are known to play a housekeeping role in the proteolytic processing and maturation of other P. gingivalis surface proteins, including fimbrillin, a 75-kDa outer membrane protein, and Kgp ((11, 20). Hence, in the case of the rgpA rgpB mutant, the cells may display a number of pleiotropic alterations, and this may contribute to their reduced invasive capabilities in the murine model. Conversely, wild-type cells treated with a chemical inhibitor of the Arg-gingipains may be compromised solely with respect to the extracellular functions of these enzymes, and this may be insufficient to attenuate the virulence of the organism in this system. Second, sustained inhibition of the Arg-gingipains may be required throughout the course of infection by the wild-type organism in this model in order to overcome the activity of newly formed enzymes following bacterial growth.
Insertional inactivation of kgp was shown to generate a distinctive phenotype in the resultant mutant. First, as reported previously (22), the mutant fails to produce black pigment and instead produces creamy white colonies on blood agar. This is likely to be attributable to the inability of the mutant to lyse erythrocytes, degrade hemoglobin, and hence release hemin for accumulation at the cell surface; hence, Kgp is also referred to as a hemoglobinase (14) and is capable of degrading a wide range of hemin carrier proteins (26). Second, growth of the kgp mutant is significantly compromised compared to growth of the wild type in defined media in which the sole amino acid source is a high-molecular-mass protein, whereas growth in complex medium is unaffected, probably because the medium contains a high concentration of low-molecular-weight peptides. Hence, Kgp appears to be an important enzyme in both the micronutrient assimilation strategy and the carbon and nitrogen assimilation strategy in this organism. Third, inactivation of kgp leads to a significant reduction in virulence in the murine model. The attenuation may reflect an impaired nutritional capability in vivo and/or a greater susceptibility to oxygen radical damage due to the absence of hemin accumulation at the cell surface. Alternatively, as hemin is believed to be a virulence regulator in P. gingivalis (16, 17), the inability to sequester this iron source may lead to the down regulation of a number of traits required for maximal expression of virulence in vivo. In the present study the close interrelationship among Kgp, hemin release or acquisition, and virulence was further emphasized by the observation that coadministration of hemin with the kgp mutant in the murine model was able to partially restore the virulence of the mutant to wild-type levels.
These observations made with Arg-gingipain and Lys-gingipain mutants suggested that Kgp is the most promising target protease for inhibitor design and synthesis. The compound selected for further study, 1-(3-phenylpropionyl)piperidine-3(R,S)-carboxylic acid-[4-amino-1(S)-(benzothiazole-2-carbonyl)butyl] amide (A71561), is a slowly reversible Kgp inhibitor with a low Ki and high specificity for the target enzyme over the Arg-gingipains and other proteases and with preference for Arg and/or Lys in P1. Growth of the wild-type organism in vitro on solid medium or in liquid broth containing A71561 led to a phenotype very similar to that of the kgp mutant. The wild type failed to produce pigment on blood agar containing 100 μM inhibitor and grew significantly less well in defined media, but it was unaffected in complex media containing A71561. Furthermore, pretreatment of wild-type cells with A71561 at a concentration of 2 mM prior to inoculation into the mice led to an overall significant reduction in virulence, again mimicking the P. gingivalis kgp mutant phenotype.
Treatment of the wild-type cells with a high concentration (2 mM) of A71561 was required to demonstrate a significant reduction in virulence despite the fact that complete inhibition of all Kgp in the inoculum was achieved by using concentrations of the inhibitor less than 0.1 mM. The excess inhibitor may be required to effectively inhibit newly generated enzyme following growth of the bacterial dose administered. High concentrations of a broad-spectrum inhibitor of thiol enzymes, tosyl lysyl chloromethyl ketone (TLCK), were also required to attenuate the virulence of P. gingivalis in a similar murine model (13). In this instance, treatment with 2 mM TLCK provided complete protection from mortality, whereas treatment with 0.2 mM TLCK provided only 20% protection. In the future, it may be possible to significantly reduce the concentration of inhibitor if the inhibitor is applied continuously via systemic administration or local delivery rather than by ex vivo treatment of the inoculum. However, the experiments are dependent upon on-going work involving the design and synthesis of compounds with appropriate pharmacokinetic properties to ensure an extended in vivo half-life.
The therapeutic success of specific inhibitors of the human immunodeficiency virus aspartyl protease, which is essential for maturation of the viral polyprotein and hence replication, has led to increased awareness of the potential value of protease inhibitors in the treatment of infectious disease. Hence, there is growing interest in the development of inhibitors of proteases of other pathogenic viruses, yeasts, protozoans, and helminths. The investigations in this study are, to our knowledge, the first example in which targeted protease inhibition of a bacterial enzyme shows promise as a novel antimicrobial strategy.
Acknowledgments
This work was supported by the Medical Research Council (grants PG9318173 and G9719660).
Editor: B. B. Finlay
REFERENCES
- 1.Aduse-Opoku, J., N. N. Davies, A. Gallagher, A. Hashim, H. E. Evans, M. Rangarajan, J. M. Slaney, and M. A. Curtis. 2000. Generation of Lys-gingipain protease activity in Porphyromonas gingivalis W50 is independent of Arg-gingipain protease activities. Microbiology 146:1933-1940. [DOI] [PubMed] [Google Scholar]
- 2.Aduse-Opoku, J., M. Rangarajan, K. A. Young, and M. A. Curtis. 1998. Maturation of the arginine-specific proteases of Porphyromonas gingivalis W50 is dependent on a functional prR2 protease gene. Infect. Immun. 66:1594-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu, L., T. E. Bramanti, J. L. Ebersole, and S. C. Holt. 1991. Hemolytic activity in the periodontopathogen Porphyromonas gingivalis: kinetics of enzyme release and localization. Infect. Immun. 59:1932-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtis, M. A., J. Aduse-Opoku, and M. Rangarajan. 2001. Cysteine proteases of Porphyromonas gingivalis. Crit. Rev. Oral Biol. Med. 12:192-216. [DOI] [PubMed] [Google Scholar]
- 5.Curtis, M. A., H. K. Kuramitsu, M. Lantz, F. L. Macrina, K. Nakayama, J. Potempa, E. C. Reynolds, and J. Aduse-Opoku. 1999. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J. Periodontal Res. 34:464-472. [DOI] [PubMed] [Google Scholar]
- 6.Discipio, R. G., P. J. Daffern, M. Kawahara, R. Pike, J. Travis, T. E. Hugli, and J. Potempa. 1996. Cleavage of human complement component C5 by cysteine proteinases from Porphyromonas (Bacteroides) gingivalis. Prior oxidation of C5 augments proteinase digestion of C5. Immunology 87:660-667. (Erratum, 88:657.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher, H. M., H. A. Schenkein, R. M. Morgan, K. A. Bailey, C. R. Berry, and F. L. Macrina. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63:1521-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genco, C. A., B. M. Odusanya, J. Potempa, J. Mikolajczyk-Pawlinska, and J. Travis. 1998. A peptide domain on gingipain R which confers immunity against Porphyromonas gingivalis infection in mice. Infect. Immun. 66:4108-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genco, C. A., J. Potempa, J. Mikolajczyk-Pawlinska, and J. Travis. 1999. Role of gingipains R in the pathogenesis of Porphyromonas gingivalis-mediated periodontal disease. Clin. Infect. Dis. 28:456-465. [DOI] [PubMed] [Google Scholar]
- 11.Kadowaki, T., K. Nakayama, F. Yoshimura, K. Okamoto, N. Abe, and K. Yamamoto. 1998. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 273:29072-29076. [DOI] [PubMed] [Google Scholar]
- 12.Kesavalu, L., J. L. Ebersole, R. L. Machen, and S. C. Holt. 1992. Porphyromonas gingivalis virulence in mice: induction of immunity to bacterial components. Infect. Immun. 60:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kesavalu, L., S. C. Holt, and J. L. Ebersole. 1996. Trypsin-like protease activity of Porphyromonas gingivalis as a potential virulence factor in a murine lesion model. Microb. Pathog. 20:1-10. [DOI] [PubMed] [Google Scholar]
- 14.Lewis, J. P., J. A. Dawson, J. C. Hannis, D. Muddiman, and F. L. Macrina. 1999. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J. Bacteriol. 181:4905-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maley, J., N. B. Shoemaker, and I. S. Roberts. 1992. The introduction of colonic-Bacteroides shuttle plasmids into Porphyromonas gingivalis: identification of a putative P. gingivalis insertion-sequence element. FEMS Microbiol. Lett. 72:75-81. [DOI] [PubMed] [Google Scholar]
- 16.Marsh, P. D., A. S. McDermid, A. S. McKee, and A. Baskerville. 1994. The effect of growth rate and haemin on the virulence and proteolytic activity of Porphyromonas gingivalis W50. Microbiology 140:861-865. [DOI] [PubMed] [Google Scholar]
- 17.McKee, A. S., A. S. McDermid, A. Baskerville, A. B. Dowsett, D. C. Ellwood, and P. D. Marsh. 1986. Effect of hemin on the physiology and virulence of Bacteroides gingivalis W50. Infect. Immun. 52:349-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milner, P., J. E. Batten, and M. A. Curtis. 1996. Development of a simple chemically defined medium for Porphyromonas gingivalis: requirement for alpha-ketoglutarate. FEMS Microbiol. Lett. 140:125-130. [DOI] [PubMed] [Google Scholar]
- 19.Moritz, A. J., D. Cappelli, M. S. Lantz, S. C. Holt, and J. L. Ebersole. 1998. Immunization with Porphyromonas gingivalis cysteine protease: effects on experimental gingivitis and ligature-induced periodontitis in Macaca fascicularis. J. Periodontol. 69:686-697. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama, K., F. Yoshimura, T. Kadowaki, and K. Yamamoto. 1996. Involvement of arginine-specific cysteine proteinase (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J. Bacteriol. 178:2818-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Brien-Simpson, N. M., R. A. Paolini, B. Hoffmann, N. Slakeski, S. G. Dashper, and E. C. Reynolds. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto, K., K. Nakayama, T. Kadowaki, N. Abe, D. B. Ratnayake, and K. Yamamoto. 1998. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J. Biol. Chem. 273:21225-21231. [DOI] [PubMed] [Google Scholar]
- 23.Quibell, M. 1999. The RAPiD approach to library design. Innovat. Pharm. Technol. 2:16-21. [Google Scholar]
- 24.Rangarajan, M., J. Aduse-Opoku, J. M. Slaney, K. A. Young, and M. A. Curtis. 1997. The prpR1 and prR2 arginine-specific protease genes of Porphyromonas gingivalis W50 produce five biochemically distinct enzymes. Mol. Microbiol. 23:955-965. [DOI] [PubMed] [Google Scholar]
- 25.Rangarajan, M., S. J. Smith, S. U, and M. A. Curtis. 1997. Biochemical characterization of the arginine-specific proteases of Porphyromonas gingivalis W50 suggests a common precursor. Biochem. J. 323:701-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Sroka, A., M. Sztukowska, J. Potempa, J. Travis, and C. A. Genco. 2001. Degradation of host heme proteins by lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis. J. Bacteriol. 183:5609-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Travis, J., J. Potempa, and H. Maeda. 1995. Are bacterial proteinases pathogenic factors? Trends Microbiol. 3:405-407. [DOI] [PubMed] [Google Scholar]
- 28.Wingrove, J. A., R. G. Discipio, Z. Chen, J. Potempa, J. Travis, and T. E. Hugli. 1992. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J. Biol. Chem. 267:18902-18907. [PubMed] [Google Scholar]