Abstract
Cryptosporidium parvum is recognized as an enteropathogen of great worldwide medical and veterinary importance, yet understanding of its pathogenesis has been hampered in part by limited knowledge of the invasion machinery of this parasite. Recently, genes containing thrombospondin type 1 (TSP1) domains have been identified in several genera of apicomplexans, including thrombospondin-related adhesive proteins (TRAPs) that have been implicated as key molecules for parasite motility and adhesion onto host cell surfaces. Previously, a large-scale random survey of the C. parvum genome conducted in our laboratory revealed the presence of multiple genomic DNA sequences with a high degree of similarity to known apicomplexan TRAP genes. In the present study, TBLASTN screening of available C. parvum genomic sequences by using TSP1 domains as queries identified a total of 12 genes possessing TSP1-like domains. All genes have putative signal peptide sequences, one or more TSP1-like domains, plus additional extracellular protein modules such as Kringle, epidermal growth factor, and Apple domains. Two genes, putative paralogs CpTSP8 and CpTSP9, contain predicted introns near their amino termini, which were verified by comparing PCR products from cDNA versus genomic DNA templates. Reverse transcription-PCR analysis of transcript levels reveals that C. parvum TSP genes were developmentally regulated with distinct patterns of expression during in vitro infection. TRAPC1, CpTSP3, and CpTSP11 were expressed at high levels during both early and late stages of infection, whereas CpTSP2, CpTSP5, CpTSP6, CpTSP8, and CpTSP9 were maximally expressed during the late stages of infection. Only CpTSP4 was highly expressed solely at an early stage of infection.
Although its association with human diseases was not perceived until 1976 (18, 27, 31), the apicomplexan parasite Cryptosporidium parvum has emerged as a significant pathogen both of humans and animals of veterinary importance. The parasite primarily infects the epithelial cells of the small intestine and causes gastrointestinal diseases in both immunocompetent and immunocompromised humans and animals. Typically self-limiting in immunologically healthy individuals, cryptosporidial infection can be persistent and life-threatening in hosts with impaired immune systems. Prolonged infection is compounded by the fact that there is currently no effective anticryptosporidial drug (19). Efforts to develop novel therapeutic strategies have been hampered by a lack of understanding of C. parvum pathogenesis and a paucity of stage- and organelle-specific markers with which to dissect the parasite's life cycle.
Thrombospondin-related adhesive proteins (TRAPs) have been identified in several genera of apicomplexans, including TRAP (38, 39, 45, 53) and CTRP (13, 54, 57, 60, 61) of Plasmodium, Etp100 of Eimeria (10, 32, 56), MIC-2 of Toxoplasma (2, 20), and NcMIC2 of Neospora (24, 46) spp. TRAPs are characterized by the presence of two adhesive modules: one or more von Willebrand factor A (vWA)-like domains, and one or more thrombospondin type 1 (TSP1)-like modules. TSP1-like domains were first identified as “region II” within the Plasmodium circumsporozoite protein (12) which, after the isolation of Plasmodium TRAP (36), was identified as a domain also present in thrombospondin and properdin. A series of studies describe a critical role for TSP1-containing proteins as mediators of host-parasite interactions and in the gliding motility of the parasite (28, 29, 47, 51). Secretion of Toxoplasma MIC-2 is associated with Toxoplasma invasion of host cells (7, 8), and disruption of the Plasmodium berghei TRAP gene resulted in sporozoites that displayed an impaired ability to invade mosquito salivary gland tissue and rat liver cells (52). TRAP genes exhibit isolate-specific and cross-species polymorphisms that might reflect the presence of immune pressure (35, 36, 37, 53). Moreover, antibodies produced against region II of CSP of P. falciparum TRAP confer some protection against the malaria parasite, identifying these proteins as potential malaria vaccine candidates (17).
The presence of TRAP family members throughout the Apicomplexa, their association with key events involved in parasite invasion and motility, and morphological and phylogenetic similarities between C. parvum and other apicomplexans suggest that TRAP may also be involved in the recognition and invasion of intestinal cells by C. parvum (22, 30). Spano et al. (49) first described and characterized a TRAP gene, TRAPC1, in C. parvum. TRAPC1 protein is localized to the apical end of sporozoites and is structurally related to the micronemal proteins MIC2, TRAP and Etp100, but with the notable lack of a vWA domain. The gene coding for a second TSP1 domain-containing C. parvum protein, TRAPC2, was partially characterized as a gene fragment (49) and contains a tandem array of TSP1 motifs. As for the TRAPs of Plasmodium sp. (36, 37), polymorphism has been observed in TRAPC1 and TRAPC2, and PCR-based polymorphism analyses of TRAPC1 and TRAPC2 have been used for genetic fingerprinting of C. parvum isolates of human and bovine origin (26, 33, 34, 48).
Previously, a large-scale random survey of the C. parvum genome conducted in our laboratory revealed the presence of multiple genomic DNA sequences similar to known apicomplexan TRAP genes, including the previously characterized TRAPC1 and TRAPC2 genes of C. parvum (23). Clone CpGR176 was identical to TRAPC2 at both the nucleic acid and the amino acid levels, whereas clones CpGR260 and CpGR493 were highly similar to TRAPC1 and C2 at the amino acid level (e value equals 10−12). To understand the extent of expansion of possible TRAP-like proteins in the Cryptosporidium genome, we initiated the present study to exhaustively identify and characterize additional C. parvum genes containing TSP1 or vWA domains. By using apicomplexan TSP1-like domains as TBLASTN queries to search our C. parvum genome sequence database, we identified a total of 12 C. parvum genes possessing one or more TSP1-like domains. Extracellular examples of the vWA-like domain were not found in the C. parvum genome, either singly or in combination with TSP1 domains. The characterization of these TSP1 containing genes will not only enhance our understanding of the phylogenetic distribution and expansion of these adhesive domains within the apicomplexa (reviewed in reference 55) but also provide insights into the general mechanism employed by C. parvum for attachment and invasion of host cells.
MATERIALS AND METHODS
Identification of C. parvum TSP1 domain-containing genes.
A local C. parvum genome database (CryptoDB) was constructed by using contig sequences assembled from a 9X random shotgun sequencing coverage generated by our ongoing C. parvum genome-sequencing project (http://www.cbc.umn.edu/ResearchProjects/AGAC/Cp/index.htm). Assembled contigs are available at http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html. To exhaustively screen for additional TSP1-like domains within C. parvum genes, various apicomplexan TSP1-like domains were used as TBLASTN queries of CryptoDB. The conservation of cysteine and tryptophan residues, and spacing of these residues, within TSP1-like domains allowed high sensitivity and high stringency in identifying putative genes. After TBLASTN searching, full-length open reading frames (ORFs) were identified within contigs of best hits by using a web-based translation program (ExPASy Translate Tool; Swiss Institute of Bioinformatics; http://www.expasy.ch/tools/dna.html). The domain structure of the resulting ORFs was determined by using reiterative PSI-BLAST (NCBI/NLM) screening of GenBank (nr database) and SMART package HMMER domain profile searches (http://www.smart.embl-heidelberg.de). Newly identified TSP1 containing genes were named to reflect, as much as possible, groupings of structural relatedness.
Isolation of total RNA from C. parvum-infected cells.
Sterilized C. parvum oocysts (Iowa strain, originally from C. R. Sterling of Arizona State University) were used to infect confluent human adenocarcinoma (HCT-8; American Type Culture Collection, Manassas, Va.) cell monolayers at a concentration of one oocyst per cell, as previously described (42, 43, 58). Total RNA was prepared from mock-infected and C. parvum-infected HCT-8 cultures at 6, 12, 24, 48, and 72 h postinfection (p.i.) by directly lysing the cells with 4 ml of Trizol Reagent (Gibco-BRL/LifeTechnologies, Gaithersburg, Md.) according to the manufacturer's instructions. Purified RNA was resuspended in RNase-free water, and the integrity of the samples was confirmed by RNA agarose gel electrophoresis.
Expression analysis of TSP1 domain-containing genes during in vitro C. parvum development.
To investigate differential expression of the C. parvum TSP1 domain genes during in vitro C. parvum development, sequence-specific primers (Table 1) were designed for each of the TSP1 domain genes, and a semiquantitative reverse transcription-PCR (RT-PCR) analysis was carried out as previously described (1). Briefly, 2.0 μg of total RNA was mixed with 0.74 μg of random hexamer and RNase-free water (to bring up the volume to 12 μl), heated at 70°C for 10 min, and cooled on ice. To this mixture was added a 7-μl aliquot, consisting of 4.0 μl of 5× first strand buffer, 2.0 μl of 100 mM dithiothreitol, and 1.0 μl of 10 mM deoxynucleoside triphosphate mixture. The reaction was equilibrated at 42°C for 2 min on a GeneAmp PCR system 2400 (Perkin-Elmer Cetus Corp., Norwalk, Conn.), after which 1.0 μl (200 U) of SuperScript II RTase was applied. The reaction mixture was then incubated at 42°C for 50 min, heated at 70°C for 15 min, and held at 4°C. A 2.0-μl portion of the first-strand cDNA reaction was used in a 20-μl reaction that contained 1× PCR buffer with 1.5 mM MgCl2 (Perkin-Elmer, Inc.), 200 μM concentrations of each of the four deoxynucleoside triphosphates, 1.0 μM concentrations of the forward and reverse primers correspondent to each TRAP gene, 1.25 U of AmpliTaq DNA polymerase (Perkin-Elmer), and 0.4 μCi of [32P]dCTP (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.). After an initial denaturation at 94°C for 2 min, the reaction mixture underwent multiple cycles of denaturation at 94°C for 30 s, annealing at 57°C for 30 s, and extension at 72°C for 1 min. The number of cycles was preoptimized (25 cycles for amplification of CpTSP4 and 27 cycles for all other genes) so that accumulation of the PCR product was still in the exponential phase (data not shown). After an additional cycle of extension at 72°C for 5 min, the reactions were held at 4°C.
TABLE 1.
Primers used for amplification of C. parvum gene fragments
| Primer | Nucleotide sequence | Size(s) (bp) of PCR product(s) |
|---|---|---|
| TRAPC1-f | GTCAGTTTGGTCAGAATGGTC | |
| TRAPC1-r | GTGTAGCGTCTCCAGAATCTG | 395 |
| CpTSP2-f | AAGGATGCGTAGAAAGGAGAG | |
| CpTSP2-r | TGCAAGCCTTAGTCTGAAAAA | 507 |
| CpTSP3-f | TGTGTAGTTGGTCAATGGTCA | |
| CpTSP3-r | AATTTAAGCCTGTCTTCGGAG | 444 |
| CpTSP4-f | TTTTCTGAAACACACCCAGAC | |
| CpTSP4-r | ATTTTCATCAGGGTACTTGCA | 324 |
| CpTSP5-f | CTACATCAATGGCCGGATAC | |
| CpTSP5-r | GGAAAAGTGCTTCTTTCACC | 394 |
| CpTSP6-f | TAACTCATTCCCAGACAGCA | |
| CpTSP6-r | GACACAAACCCATGTTCAGA | 408 |
| CpTSP7-f | AATGGTGGTTTACCATGTCC | |
| CpTSP7-r | CACAAGACTTGGAGCAAACA | 400 |
| CpTSP8-f | GTACTTGCAATGGTGGAACA | |
| CpTSP8-r | TGATTAGATGACGGACATGC | 409 |
| CpTSP9-f | CTCTGCCGGTGGTAGTATTT | |
| CpTSP9-r | GGACCAAGAACTCCATTCTG | 391 |
| CpTSP10-f | GTCTGCATGCTCAGACACAT | |
| CpTSP10-r | CACAATCAGTTGGAACAGGA | 401 |
| CpTSP11-f | GGGAAGCCCTATGATACTGA | |
| CpTSP11-r | TGTACACCCCTGAGTTTGAA | 412 |
| CpTSP12-f | CAGTATCTGCAAATGGGTCA | |
| CpTSP12-r | CATGGACCTAATTGGCAAC | 403 |
| CpTSP8 intron-fa | TCCTTGTCATAGCATAGTGT | |
| CpTSP8 intron-ra | GACCATAATGACTCTCTTA | 790b, 564c |
| 18S rRNA-fd | CTCCACCAACTAAGAACGCC | |
| 18S rRNA-rd | AGAGATTGGAGGTTGTTCCT | 212 |
Primers spanning the region that contains the 226-bp predicted intron.
Expected length of the unspliced genomic DNA PCR product.
Expected length of the cDNA PCR product.
Primers described by Rochelle et al. (41).
As the number of developing C. parvum life stages within infected cells changes over time, primers specific for C. parvum 18S rRNA (41; see Table 1) were used to normalize the amount of cDNA product of the TSP1 domain genes to that of C. parvum rRNA in the same sample. Since rRNA is much more abundant than any specific mRNAs, 2.0 μl of a 1:25 dilution of the cDNA samples were used and reactions underwent only 23 cycles of amplification (1). Furthermore, to monitor PCR efficiency and possible cross-contamination between reactions, a negative control with 2.0 μl of cDNA made from mock-infected HCT-8 cells and a positive control with 2.0 ng of C. parvum genomic DNA were also included in each batch of PCRs.
The PCR products were separated on a 4.0% nondenaturing polyacrylamide gel and signals from specific products were captured and quantified by using a phosphorimaging system (Molecular Dynamics, Inc., Sunnyvale, Calif.). The expression level of TSP1 domain genes at each time point was calculated as the ratio of its RT-PCR product signal to that of the C. parvum 18S rRNA (1). Three independent time course experiments were used in the analysis. Each of the RNA samples from C. parvum-infected HCT-8 cells was demonstrated to be free of contaminating C. parvum genomic DNA by the lack of amplification product from a sample not subjected to RT (data not shown). The mean expression level of each of the TSP1 domain genes was calculated, and the standard deviation was determined. The percentage of expression of each TSP1 domain gene at each time point was also reported as a percentage of the highest level of expression.
RESULTS
C. parvum genome possesses a superfamily of TSP1-like domain genes.
In the present study a local Cryptosporidium genome database (CryptoDB) was constructed and screened via standalone TBLASTN by using various apicomplexan TSP1 and vWA domains as queries. TSP1-like domains are excellent queries by virtue of two conserved tryptophan residues and either four or six conserved cysteine residues. An exhaustive screening process, in which each newly identified gene was in turn used as a TBLASTN query of CryptoDB, revealed the presence of TSP1-like domains within 12 C. parvum genes. TSP1 domains are therefore greatly expanded within the C. parvum genome. In contrast, vWA domains were not identified within any of the 12 TSP1 domain-containing genes, as assessed by PSI-BLAST screening of GenBank (nr) by using each gene, and fragments thereof, as queries. Additionally, vWA domains were not identified after screening of each TSP1 domain-containing gene by using an extracellular vWA HMMER profile (SMART database; http://smart.embl-heidelberg.de). Further extensive screening of CryptoDB by using as queries all known apicomplexan and numerous vertebrate vWA domains did not identify a vWA domain, either singly or in combination with TSP1 domains. We therefore propose that Cryptosporidium does not utilize vWA in extracellular adhesive interactions and that a capacity for gliding motility and invasion does not require this adhesive module.
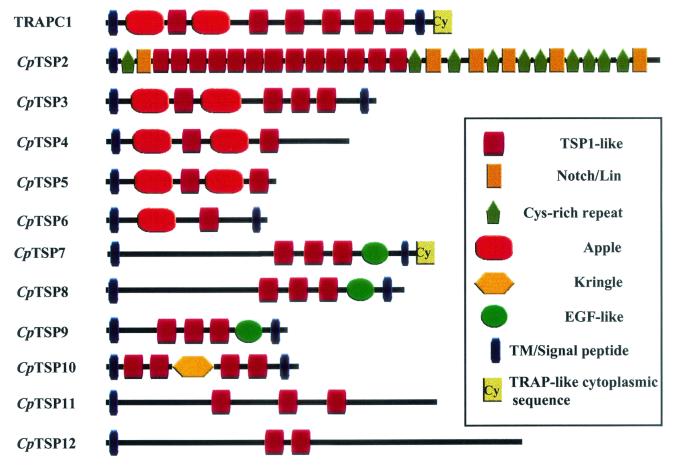
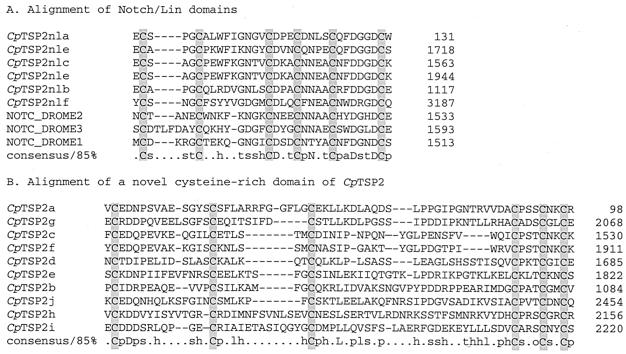
Cryptosporidium TSP1 domains were previously described in TRAPC1 (49) and within two gene fragments, TRAPC2 (50) and TRAPC3 (49; GenBank accession number AF073838). Genome analysis in the present study revealed that TRAPC2 and TRAPC3 are fragments of a single ORF and are herein named CpTSP2. This departure from the previous Cryptosporidium TRAP nomenclature reflects the observation that, unlike TRAPC1, CpTSP2 does not possess “TRAP/MIC2”-like motifs, namely, a transmembrane region combined with a short, acidic cytoplasmic domain having a conserved carboxy-terminal proximal tryptophan residue. The newly identified TSP1-like domain-containing genes are similarly herein termed CpTSP3 through CpTSP12. TSP1 domains are present as multiple copies within all C. parvum TSP1 domain genes, except for a single domain within CpTSP6, and are notably present as 13 tandemly arrayed copies within CpTSP2 (see schematic, Fig. 1). The CpTSP2 gene encodes a large, 3,530-amino-acid protein that has a unique multidomain structure additionally composed of multiple Notch/Lin-like domains and a novel cysteine-rich domain that is repeated 10 times (alignments shown in Fig. 2).
FIG. 1.
Schematic of C. parvum TSP1 domain-containing genes.
FIG. 2.
Alignment of Notch/Lin-like domains (A) and a novel adjacent repeated domain (B) from C. parvum CpTSP2. Conserved cysteine residues are shaded (NOTC_DROME; GenBank accession number P07207).
The domain structure of the C. parvum TSP1 domain genes was analyzed by using the web-based SMART package (http://smart.embl-heidelberg.de) of HMMER domain consensus profiles. Domains were additionally identified or confirmed via reiterative PSI-BLAST analysis of GenBank (nr database) by using TSP1 domain genes, and fragments thereof, as queries. In this manner additional domains were identified and the domain structure was addressed for all 12 Cryptosporidium TSP1 domain genes (Fig. 1). All TSP1 domain genes possess a putative signal peptide sequence near the amino terminus. CpTSP3 and CpTSP6 through CpTSP10, as well as the previously described TRAPC1, have putative transmembrane regions near their carboxy termini. CpTSP8 and CpTSP10 have short cytoplasmic domains less than 10 amino acids in length, whereas the other transmembrane proteins have cytoplasmic domains of greater than 40 amino acids. The latter proteins possess tyrosine-rich motifs within their cytoplasmic tails that might represent recently described microneme or rhoptry targeting sequences (14, 21) and warrant further study to address protein localization. The cytoplasmic tail of CpTSP7 has an acidic nature and tryptophan residue near the -COOH terminus reminiscent of Plasmodium TRAP and Toxoplasma MIC2. CpTSP7 is thus a candidate for a “TRAP family” gene and is therefore potentially involved in extracellular adhesive events coupled with cytoplasmic interaction, either directly or indirectly, with an actin/myosin motility apparatus.
TRAPC1 and CpTSP3 are located on opposite strands within the same genomic contig and likely have a paralogous relationship via gene duplication. Similarities in domain structure with CpTSP4 through CpTSP6 suggests that they might also have an ancestral relationship via gene duplication.
TRAPC1 and CpTSP3 through CpTSP6 contain divergent Apple domains.
TRAPC1 and CpTSP3 through CpTSP5 share an amino-terminal region architecture consisting of two Apple domains intervened with a single TSP1-like domain. The second Apple domain is followed by a variable number of additional TSP1-like domains. Apple domains were initially described in the human kallikrein gene (9) and have likely arisen in Apicomplexa via lateral capture, present in Toxoplasma (3) and Eimeria (4). Their presence in Cryptosporidium suggests that, like TSP1 domains, their lateral transfer preceded species divergence of the apicomplexan species. Alignment of C. parvum Apple domains (Fig. 3) delineates two motifs differing by a pair of cysteines, presumably participating in an additional disulfide bond. Although Apple domains containing the extra pair of cysteines are not readily identified by PSI-BLAST or SMART analysis, BLAST screening of CryptoDB with kallikrein and other Apple domain-containing queries identifies the putative Apple domains in CpTSP4 and CpTSP6. Alignments show that this domain is repeated twice in TRAPC1 and CpTSP3 through CpTSP5 and present as a single copy in CpTSP6. Based upon the conservation of structure and repetition within TSP1 domain genes, we propose that this is a discrete globular domain and is probably a divergent Apple domain.
FIG. 3.
Alignment of Apple-like domains from C. parvum genes with Apple domains from human kallikrein (GenBank accession number P03952.) CpApp5 through CpApp8 are Apple domain-containing genes within CryptoDB identified via TBLASTN analysis and have not been previously described. Cysteine residues are shaded.
Introns within CpTSP8 and CpTSP9.
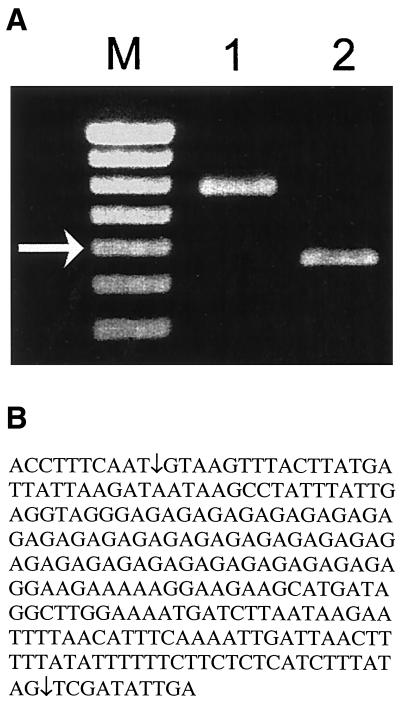
CpTSP7, CpTSP8, and CpTSP9 share a domain structure composed of three contiguous TSP1-like domains, followed by a single epidermal growth factor-like domain adjacent a putative transmembrane domain. The domain structure and conservation of cysteine residue spacing suggests that they also have a parologous relationship arising via gene duplication. Comparison of the CpTSP8 ORF with Crytosporidium EST databases (http://www.ebi.ac.uk/parasites/cparvEST.html) revealed the presence of a 266-bp intron (nucleotide positions 961 to 1186 relative to the start codon; see schematic in Fig. 4). Comparison of the domain structure between CpTSP8 and CpTSP9 identified a 77-bp intron (nucleotide positions 132 to 208) with the CpTSP9 ORF. The presence of an intron has only one precedent within Cryptosporidium, the beta-tubulin gene (5). Functional splicing of the CpTSP8 (Fig. 4) and CpTSP9 (data not shown) introns were confirmed by comparison of PCR product lengths from genomic DNA versus reverse-transcribed RNA templates (Fig. 4A) and sequencing of the resulting PCR product derived from cDNA (Fig. 4B). CpTSP8 and CpTSP9 are located on the same contig on opposite strands, supporting a paralogous relationship. CpTSP7 and CpTSP8 both contain amino-terminal regions of roughly 40 kDa which do not have any similarity to each other nor to any gene in GenBank as queried by using PSI-BLAST.
FIG. 4.
PCR analysis and sequencing of the C. parvum CpTSP8 region that contains a predicted intron. (A) C. parvum genomic DNA templates (lane 1) and reverse-transcribed RNA of C. parvum-infected HCT-8 cells (48 h p.i.; lane 2) were amplified by using sequence-specific primers spanning the 226-bp intron (Table 1), and the resulting PCR products were separated on 1.0% agarose gel. M, 100-bp DNA ladder. The arrowed band is 600 bp. (B) The RT-PCR product illustrated in lane 2 of panel A was TA cloned, and the insert of a positive colony was sequenced from both directions by using PCR primers. The functional splicing sites are indicated by arrows. Also included are 10 nucleotides of the exon flanking each side of the intron.
Additional C. parvum TSP1 domain genes.
CpTSP10 through CpTSP12 show no relation in their domain superstructure, either to each other or to the other TSP1 domain genes and yield no clues that they may have arisen via recent gene duplication. CpTSP10 is notable for the presence of a kringle domain, whereas CpTSP11 and CpTSP12 do not contain recognizable modules other than TSP1-like domains. Kringle domains are another example of lateral capture of an extracellular protein module and have not been previously described within apicomplexan genes. Complete annotation of the Plasmodium and Toxoplasma genomes will give an indication of the extent of expansion of this domain in Apicomplexa. Preliminary BLAST screens of the CryptoDB did not identify additional genes possessing kringle domains. The validity of the kringle domain in CpTSP10 was demonstrated first by its identification with the SMART package of HMMER consensus profiles (e value equals 10−7), and the fact that the C. parvum kringle domain readily identifies plasminogen activator and other kringle domain-containing genes after a PSI-BLAST screen of GenBank (e value equals 10−6).
Expression analysis of TSP1 domain genes.
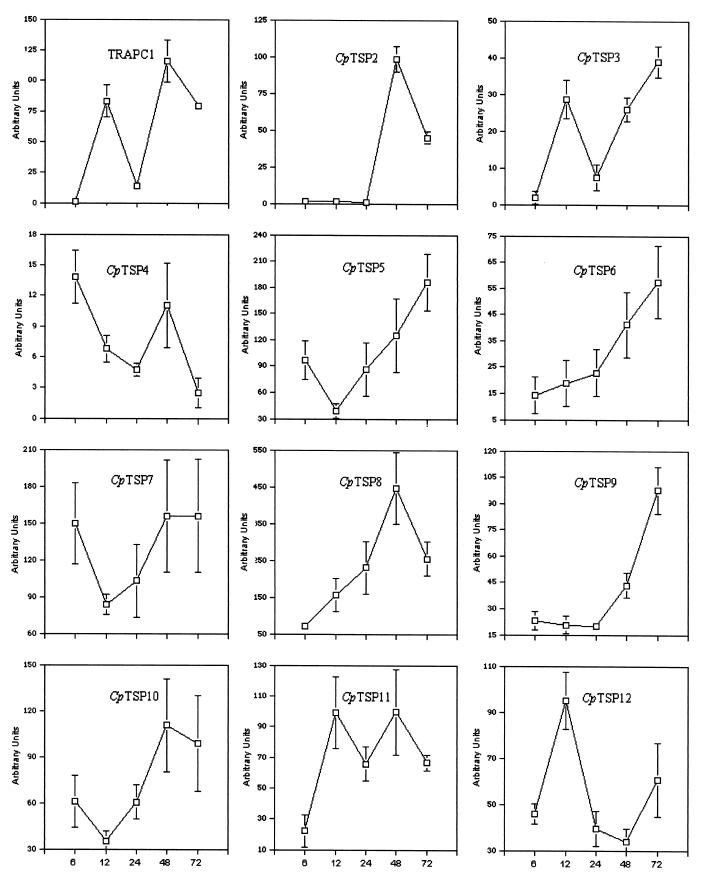
RT-PCR analysis of gene expression of TRAPC1 and CpTSP2 thru CpTSP12 is presented in Fig. 5 and 6. As the number of developing C. parvum life stages within infected cells changes over time, primers specific for C. parvum 18S rRNA (41; see Table 1) were used to normalize the amount of cDNA product of the TSP1 domain genes to that of C. parvum rRNA in the same sample. The mRNA level of TRAPC1 in infected cultures display a high degree of variability, demonstrating that TRAPC1 expression is clearly regulated as C. parvum progresses through its life cycle. The mRNA levels of CpTSP2 thru CpTSP12 in infected HCT-8 cells are also illustrated in Fig. 6, and the percent expression of TSP1 domain genes in infected HCT-8 cells at each sampled time point is presented in Table 2.
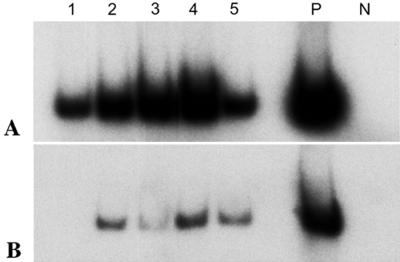
FIG. 5.
RT-PCR analysis of C. parvum TRAPC1 expression during in vitro development in HCT-8 cells. (A) Total RNA (2.0 μg) isolated from C. parvum-infected HCT-8 cells at 6, 12, 24, 48, and 72 h p.i. (lanes 1 to 5, respectively) was reverse transcribed by using random primers, and 2.0 μl of a 1:50 dilution of each cDNA reaction was subjected to 23 cycles of amplification in the presence of [32P]dCTP and primers specific for C. parvum 18S rRNA (Table 1). P, positive control (C. parvum genomic DNA used as a template in PCR); N, negative control (total RNA from mock-infected cells used in RT). (B) Two microliters of each cDNA reaction was amplified by using primers specific for C. parvum TRAPC1 (Table 1).
FIG. 6.
Expression profiles of C. parvum TRAPC1 and CpTSP2 to CpTSP12 during in vitro C. parvum development in HCT-8 cells. The data are expressed as the ratio of TRAPC1 and CpTSP2 thru CpTSP12 signal to C. parvum 18S rRNA signal present in each of the infected samples, as determined by using a phosphorimaging system. Error bars indicate the standard deviation of the mean of three independent experiments.
TABLE 2.
Expression of C. parvum TSP1 domain-containing genes during development in HCT-8 cells
| Gene | Relative expression (%) at different time points
|
||||
|---|---|---|---|---|---|
| 6 h | 12 h | 24 h | 48 h | 72 h | |
| TRAPC1 | 1 | 72 | 12 | 100 | 68 |
| CpTSP2 | 2 | 1 | 1 | 100 | 46 |
| CpTSP3 | 5 | 74 | 19 | 67 | 100 |
| CpTSP4 | 100 | 49 | 34 | 80 | 18 |
| CpTSP5 | 52 | 21 | 53 | 67 | 100 |
| CpTSP6 | 25 | 33 | 40 | 72 | 100 |
| CpTSP7 | 96 | 54 | 66 | 100 | 100 |
| CpTSP8 | 16 | 35 | 52 | 100 | 57 |
| CpTSP9 | 24 | 21 | 20 | 44 | 100 |
| CpTSP10 | 55 | 30 | 55 | 100 | 90 |
| CpTSP11 | 22 | 100 | 66 | 100 | 67 |
| CpTSP12 | 48 | 100 | 43 | 36 | 64 |
As illustrated in Fig. 6, expression of the TSP1 domain genes is regulated in a complex fashion. Some of TSP1 domain genes are expressed at high levels at both early and late stages of infection (TRAPC1, CpTSP3, and CpTSP11), whereas others are expressed more abundantly at late stages (CpTSP2, CpTSP5, CpTSP6, CpTSP8, and CpTSP9) or only at early stages of infection (CpTSP4).
DISCUSSION
Accumulating evidence suggests that the molecular machinery for gliding motility and cell invasion is conserved across apicomplexan genera (22). TRAP homologues in different apicomplexans share structural and functional domains, indicating that they constitute a family of functionally homologous proteins playing key roles in the ability of the parasites to recognize and invade host cells. TRAP proteins localize to apical region microneme secretory vesicles and are released via calcium-mediated signals to the cell surface at the apical pole (30, 55). A capacity for gliding motility is proposed to involve extracellular adhesion to host tissue substrates, coupled across the transmembrane region to cytoplasmic interaction with a subpellicular myosin/actin microfilamentous motor apparatus. In this fashion, adhesive receptors stream from the apical to posterior poles of the parasite surface, propelling the parasite forward either on host cell surfaces or during invasion of host cells. Cell invasion is thought to involve additional formation of a tight junction forming between the parasite and host membranes, with anterior-to-posterior migration of this junction driven by the subpellicular motor filaments (15, 16).
The TRAPC1 gene isolated by Spano et al. (49) encodes a protein architecturally similar to Toxoplasma MIC2, with the notable absence of a vWA-like domain. Despite the lack of a vWA module, TRAPC1 is a prominent candidate for a functional Cryptosporidium homologue of MIC2 by virtue of possessing a signal peptide sequence; multiple tandemly arrayed TSP1 domains; a transmembrane region; and a characteristic short, acidic cytoplasmic domain containing a carboxy-terminal tryptophan residue conserved in TRAP, CTRP, MIC2, and Etp100. In addition, TRAPC1 protein is localized within the apical complex of excysted C. parvum sporozoites, similar to the micronemal destination of TRAP, CTRP, MIC2, and Etp100. TRAPC1 is herein described to possess Apple domains and thereby is further distinguished from Plasmodium TRAP and Toxoplasma MIC2, which lack Apple domains. In the present study the identification of a superfamily of C. parvum TSP1 domain-containing genes adds new members to the growing family of TRAP-like proteins (55) and provides information on the phylogenetic distribution of this protein family in the C. parvum genome. Of these genes only CpTSP7 shares with TRAPC1 and other apicomplexan TRAP family members a transmembrane domain followed by a short, acidic cytoplasmic domain and a conserved COOH-terminal tryptophan residue. Although the functional roles of TRAPC1 and CpTSP2 through CpTSP12 have yet to be elucidated, the similar invasion machinery of C. parvum to that of other apicomplexans suggest that C. parvum TSP1 domain containing genes may also be involved in the invasion process of host intestinal cells by C. parvum zoites.
An interesting result of this Cryptosporidium genome-wide screen is the failure to identify a vWA domain, either alone or in tandem with a TSP1 domain. All known apicomplexan vWA domains were tried as TBLASTN queries, as well as various vWA domains of higher eukaryotes—all without success. Although it is possible that Cryptosporidium possesses vWA domains that have diverged beyond recognition by TBLASTN screens, it is unlikely that any vWA domain would be missed by SMART profile screening or reiterative PSI-BLAST analysis if it were to reside within any of the 12 identified TSP1-containing genes. Because TSP1 domains are superb TBLASTN queries, and it is unlikely that TSP1 domains were overlooked in the present study, it is therefore assumed that no gene exists in the current Cryptosporidium genome databases possessing both a TSP1 domain and a vWA domain. Thus, Cryptosporidium is fundamentally distinct in this regard in comparison with all apicomplexans analyzed to date: Plasmodium, Toxoplasma, Eimeria, and Neospora. Whether vestigial vWA domains were lost in Cryptosporidium or acquired in other apicomplexans after the split into species is unknown. In Plasmodium spp., an intact vWA domain is essential for TRAP-mediated invasion of mosquito salivary glands but is not required for gliding motility (25, 59). A simple model would propose that vWA domains are involved solely in the insect stage of parasite life cycles, but Toxoplasma MIC2 presumably requires a vWA domain whereas the Toxoplasma parasite does not utilize an insect vector. Regardless of whether vWA domains never entered the Cryptosporidum genome or became vestigial and lost, it can be proposed that, if Cryptosporidium TSP superfamily proteins are involved in the motility and invasion of intestinal epithelial cells, then vWA domains are not essential for this process.
Due to its critical role in parasite motility and attachment to host cells, P. falciparum TRAP has been suggested as a candidate vaccine gene, and it was shown that the disruption of TRAP dramatically blocked parasite motility and resulted in much-reduced infectivity (17, 44). Previously, mouse monoclonal antibodies to C. parvum TRAPC1 functional domains were used in an in vitro inhibition assay to assess their abilities to inhibit parasite infection and growth in host cells. However, only limited reductions on the number of C. parvum development stages were observed at 48 h p.i. (6). Since it is now clear that that there are several additional TSP1 domain genes and that at any stage of infection process there is one or more TSP1 domain genes expressed at high levels (Fig. 6), antibodies against one TSP containing protein may not be sufficient to dramatically inhibit C. parvum's ability to invade host cells. Alternatively, TSP1 domain genes may represent only one of the multiple invasion pathways used by C. parvum.
Previous studies found that TRAP homologues from different Plasmodium species were expressed exclusively at the sporozoite developmental stage and that the expression pattern of P. falciparum TRAP parallels the development of P. falciparum infectivity during sporozoite ontogeny (11, 40). In the present study, it was found that C. parvum TSP1 domain gene homologues display distinct expression profiles; some of them are expressed highly at early stages of infection (CpTSP4, CpTSP7, and CpTSP11), whereas some others are expressed mostly at late stages. The overall expression of all TRAPs peaks at 48 h p.i., although expression of specific TSP1 domain genes is higher at earlier stages than at later stages (i.e., CpTSP2 and CpTSP4). However, since C. parvum has a multistage life cycle and the in vitro development is not synchronous, it remains to be elucidated whether the expression of specific TSP1 domain genes is restricted to a specific developmental stage.
Acknowledgments
This work was supported in part by grants AI35479 and AI46397 from the National Institutes of Health to M.S.A.
We thank Cheryl A. Lancto for invaluable assistance in cell culture.
Editor: J. M. Mansfield
REFERENCES
- 1.Abrahamsen, M. S., and A. A. Schroeder. 1999. Characterization of intracellular Cryptosporidium parvum gene expression. Mol. Biochem. Parasitol. 104:141-146. [DOI] [PubMed] [Google Scholar]
- 2.Achbarou, A., O. Mercereau-Puijalon, J. M. Autheman, B. Fortier, D. Camus, and J. F. Dubremetz. 1991. Characterization of microneme proteins of Toxoplasma gondii. Mol. Biochem. Parasitol. 47:223-233. [DOI] [PubMed] [Google Scholar]
- 3.Brecht, S., V. B. Carruthers, D. J. Ferguson, O. K. Giddings, G. Wang, U. Jakle, J. M. Harper, L. D. Sibley, and D. Soldati. 2001. The Toxoplasma micronemal protein MIC4 is an adhesin composed of six conserved apple domains. J. Biol. Chem. 276:4119-4127. [DOI] [PubMed] [Google Scholar]
- 4.Brown, P. J., K. J. Billington, J. M. Bumstead, J. D. Clark, and F. M. Tomley. 2000. A microneme protein from Eimeria tenella with homology to the Apple domains of coagulation factor XI and plasma pre-kallikrein. Mol. Biochem. Parasitol. 107:91-102. [DOI] [PubMed] [Google Scholar]
- 5.Cacciò, S., G. La Rosa, and E. Pozio. 1997. The beta-tubulin gene of Cryptosporidium parvum. Mol. Biochem. Parasitol. 89:307-311. [DOI] [PubMed] [Google Scholar]
- 6.Camero, L., M. Arrowood, Y. Shulaw, A. A. Lal, and L. H. Xiao. 1999. Characterization of new monoclonal antibodies against Cryptosporidium parvum sporozoites. J. Eukaryot. Microbiol. 46:58S-59S. [PubMed]
- 7.Carruthers, V. B., and L. D. Sibley. 1997. Sequential protein secretion from three distinct organells of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur. J. Cell Biol. 73:114-123. [PubMed] [Google Scholar]
- 8.Carruthers, V. B., O. K. Giddings, and L. D. Sibley. 1999. Secretion of micronemal proteins is associated with Toxoplasma invasion of host cells. Cell Microbiol. 1:225-235. [DOI] [PubMed] [Google Scholar]
- 9.Chung, D. W., K. Fujikawa, B. A. McMullen, and E. W. Davie. 1986. Human plasma prekallikrein, a zymogen to a serine protease that contains four tandem repeats. Biochemistry 25:2410-2417. [DOI] [PubMed] [Google Scholar]
- 10.Clarke, L. E., F. M. Tomley, M. H. Wisher, I. J. Foulds, and M. E. Boursnell. 1990. Regions of an Eimeria tenella antigen contain sequences which are conserved in circumsporozoite proteins from Plasmodium spp. and which are related to the thrombospondin gene family. Mol. Biochem. Parasitol. 41:269-280. [DOI] [PubMed] [Google Scholar]
- 11.Cowan, G., S. Krishna, A. Crisanti, and K. Robson. 1992. Expression of thrombospondin-related anonymous protein in Plasmodium falciparum sporozoites. Lancet 339:1412-1413. [DOI] [PubMed] [Google Scholar]
- 12.Dame, J. D., J. L. Williams, T. F. McCutchan, J. L. Webber, R. A. Wirtz, W. T. Hockmeyer, W. L. Maloy, J. D. Haynes, I. Schneider, D. D. Roberts, G. S. Sanders, E. P. Reddy, C. L. Digs, and L. H. Miller. 1984. Structure of the gene encoding the immunodominant surface antigen on the sporozoite of human malaria parasite Plasmodium falciparum. Science 225:593-599. [DOI] [PubMed] [Google Scholar]
- 13.Dessens, J. T., A. L. Beetsma, G. Dimopoulos, K. Wengelnik, A. Crisanti, F. C. Kafatos, and R. E. Sinden. 1999. CTRP is essential for mosquito infection by malaria ookinetes. EMBO J. 18:6221-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Cristina, M., R. Spaccapello, D. Soldati, F. Bistoni, and A. Crisanti. 2000. Two conserved amino acid motifs mediate protein targeting to the micronemes of the apicomplexan parasite Toxoplasma gondii. Mol. Cell. Biol. 20:7332-7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobrowolski, J. M., and L. D. Sibley. 1996. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell 84:933-939. [DOI] [PubMed] [Google Scholar]
- 16.Dobrowolski, J. M., V. B. Carruthers, and L. D. Sibley. 1997. Participation of myosin in gliding motility and host cell invasion by Toxoplasma gondii. Mol. Microbiol. 26:163-173. [DOI] [PubMed] [Google Scholar]
- 17.Dolo, A., D. Modiano, O. Doumbo, A. Bosman, T. Sidibe, M. M. Keita, S. Naitza, K. J. Robson, and A. Crisanti. 1999. Thrombospondin related adhesive protein (TRAP), a potential malaria vaccine candidate. Parasitologia 41:425-428. [PubMed] [Google Scholar]
- 18.Fayer, R., C. A. Speer, and J. P. Dubey. 1997. General biology of Cryptosporidium, p. 1-42. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Inc., Boca Raton, Fla.
- 19.Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 20.Fourmaux, M. N., A. Achbarou, O. Mercereau-Puijalon, C. Biderre, I. Briche, A. Loyens, C. Odberg-Ferragut, D. Camus, and J. F. Dubremetz. 1996. The MIC1 microneme protein of Toxoplasma gondii contains a duplicated receptor-like domain and binds to host cell surface. Mol. Biochem. Parasitol. 83:201-210. [DOI] [PubMed] [Google Scholar]
- 21.Hoppe, H. C., H. M. Ngo, M. Yang, and K. A. Joiner. 2000. Targeting to rhoptry organelles of Toxoplasma gondii involves evolutionary conserved mechanisms. Nat. Cell Biol. 2:449-456. [DOI] [PubMed] [Google Scholar]
- 22.Kappe, S., T. Bruderer, S. Gantt, H. Fujioka, V. Nussenzweig, and R. Menard. 1999. Conservation of a gliding motility and cell invasion machinery in apicomplexan parasites. J. Cell Biol. 147:937-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, C., V. Vigdorovich, V. Kapur, and M. S. Abrahamsen. 1999. A random survey of the Cryptosporidium parvum genome. Infect. Immun. 67:3960-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovett, J. L., D. K. Howe, and L. D. Sibley. 2000. Molecular characterization of a thrombospondin-related anonymous protein homologue in Neospora canium. Mol. Biochem. Parasitol. 15:33-43. [DOI] [PubMed] [Google Scholar]
- 25.Matuschewski, K., A. C. Nunes, V. Nussenzweig, and R. Menard. 2002. Plasmodium sporozoite invation into insect and mammalian cells is directed by the same dual binding system. EMBO J. 21:1597-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLauchlin, J., S. Pedraza-Diaz, C. Amar-Hoetzeneder, and G. L. Nichols. 1999. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J. Clin. Microbiol. 37:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meisel, J. L., D. R. Perera, C. Meligro, and C. E. Rubin. 1976. Overwhelming watery diarrhea associated with Cryptosporidium in an immunosuppressed patient. Gastroenterology 70:1156-1160. [PubMed] [Google Scholar]
- 28.Menard, R. 2000. The journey of the malaria sporozoite through its hosts: two parasite proteins lead the way. Microbes Infect. 2:633-642. [DOI] [PubMed] [Google Scholar]
- 29.Müller, H. M., E. Scarselli, and A. Crisanti. 1993. Thrombospondin related anonymous protein (TRAP) of Plasmodium falciparum in parasite-host cell interactions. Parasitologia 35(Suppl.):69-72. [PubMed] [Google Scholar]
- 30.Naitza, S., F. Spano, K. J. H. Robson, and A. Crisanti. 1998. The thrombospondin-related protein family of apicomplexan parasites: the gears of the cell invasion machinery. Parasitol. Today 14:479-484. [DOI] [PubMed] [Google Scholar]
- 31.Nime, F. A., J. D. Burek, D. L. Page, M. A. Holscher, and J. H. Yardley. 1976. Acute enterocolitis in a human being infected with the protozoan Cryptosporidium. Gastroenterology 70:592-598. [PubMed] [Google Scholar]
- 32.Pasamontes, L., D. Hug, M. Humbelin, and G. Weber. 1993. Sequence of a major Eimeria maxima antigen homologous to the Eimeria tenella microneme protein Etp100. Mol. Biochem. Parasitol. 57:171-174. [DOI] [PubMed] [Google Scholar]
- 33.Pedraza-Diaz, S., C. Amar, and J. McLauchlin. 2000. The identification and characterization of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol. Lett. 189:189-194. [DOI] [PubMed] [Google Scholar]
- 34.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. Ong, W. R. MacKenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Putaporntip, C., S. Jongwutiwes, T. Tia, M. U. Ferreira, H. Kanbara, and K. Tanabe. 2001. Diversity in the thrombospondin-related adhesive protein gene (TRAP) of Plasmodium vivax. Gene 268:97-104. [DOI] [PubMed] [Google Scholar]
- 36.Robson, K. J., A. Dolo, I. R. Hackford, O. Doumbo, M. B. Richards, M. M. Keita, T. Sidibe, A. Bosman, D. Modiano, and A. Crisanti. 1998. Natural polymorphism in the thrombospondin-related adhesive protein of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 58:81-89. [DOI] [PubMed] [Google Scholar]
- 37.Robson, K. J., J. R. Hall, L. C. Davies, A. Crisanti, A. V. Hill, and T. E. Wellems. 1990. Polymorphism of the TRAP gene of Plasmodium falciparum. Proc. R. Soc. Lond. B Biol. Sci. 242:205-216. [DOI] [PubMed] [Google Scholar]
- 38.Robson, K. J., J. R. Hall, M. W. Jennings, T. J. Harris, K. Marsh, C. I. Newbold, V. E. Tate, and D. J. Weatherall. 1988. A highly conserved amino-acid sequence in thrombospondin, properdin and in proteins from sporozoites and blood stages of a human malaria parasite. Nature 335:79-82. [DOI] [PubMed] [Google Scholar]
- 39.Robson, K. J., S. Naitza, G. Barker, R. E. Sinden, and A. Crisanti. 1997. Cloning and expression of the thrombospondin related adhesive protein gene of Plasmodium berghei. Mol. Biochem. Parasitol. 84:1-12. [DOI] [PubMed] [Google Scholar]
- 40.Robson, K. J., U. Frevert, I. Reckmann, G. Cowan, J. Beier, I. G. Scragg, K. Takehara, D. H. Bishop, G. Pradel, R. Sinden, et al. 1995. Thrombospondin-related adhesive protein (TRAP) of Plasmodium falciparum: expression during sporozoite ontogeny and binding to human hepatocytes. EMBO J. 14:3883-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rochelle, P. A., R. DeLeon, M. Stewart, and R. Wolfe. 1997. Comparison of primers and optimization of PCR conditions for detection of Cryptosporidium parvum and Giardia lamblia in water. Appl. Environ. Microbiol. 63:106-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schroeder, A. A., A. M. Brown, and M. S. Abrahamsen. 1998. Identification and cloning of a developmentally regulated Cryptosporidium parvum gene by differential mRNA display PCR. Gene 216:327-334. [DOI] [PubMed] [Google Scholar]
- 43.Schroeder, A. A., C. E. Lawrence, and M. S. Abrahamsen. 1999. Differential mRNA display cloning and characterization of a Cryptosporidium parvum gene expressed during intracellular development. J. Parasitol. 85:213-220. [PubMed] [Google Scholar]
- 44.Sharma, P., A. Bharadwaj, V. K. Bhasin, V. N. Sailaja, and V. S. Chauhan. 1996. Antibodies to a conserved-motif peptide sequence of the Plasmodium falciparum thrombospondin-related anonymous protein and circumsporozoite protein recognize a 78-kilodalton protein in the asexual blood stages of the parasite and inhibit merozoite invasion in vitro. Infect. Immun. 64:2172-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sijwali, P. S., P. Malhotra, S. K. Puri, and V. S. Chauhan. 1997. Cloning and sequence analysis of the thrombospondin-related adhesive protein (TRAP) gene of Plasmodium cynomolgi bastianelli. Mol. Biochem. Parasitol. 90:371-376. [DOI] [PubMed] [Google Scholar]
- 46.Sonda, S., N. Fuchs, B. Gottstein, and A. Hemphill. 2000. Molecular characterization of a novel microneme antigen in Neospora caninum. Mol. Biochem. Parasitol. 108:39-51. [DOI] [PubMed] [Google Scholar]
- 47.Spaccapelo, R., S. Naitza, K. J. H. Robson, and A. Crisanti. 1997. Thrombospondin-related adhesive protein (TRAP) of Plasmodium berghei and parasite motility. Lancet 350:335.. [DOI] [PubMed] [Google Scholar]
- 48.Spano, F., L. Putignani, S. Guida, and A. Crisanti. 1998. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp. Parasitol. 90:195-198. [DOI] [PubMed] [Google Scholar]
- 49.Spano, F., L. Putignani, S. Naitza, C. Puri, S. Wright, and A. Crisanti. 1998. Molecular cloning and expression analysis of a Cryptosporidium parvum gene encoding a new member of the thrombospondin family. Mol. Biochem. Parasitol. 92:147-162. [DOI] [PubMed] [Google Scholar]
- 50.Sulaiman, I. M., L. Xiao, C. Yang, L. Escalante, A. Moore, C. B. Beard, M. J. Arrowood, and A. A. Lal. 1998. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg. Infect. Dis. 4:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sultan, A. A. 1999. Molecular mechanisms of malaria sporozoite motility and invasion of host cells. Int. Microbiol. 2:155-160. [PubMed] [Google Scholar]
- 52.Sultan, A. A., V. Thathy, U. Frevert, K. J. H. Robson, A. Crisanti, R. S. Nussenzweig, and R. Ménard. 1997. TRAP is necessary for gliding motility and infectivity of Plasmodium sporozoites. Cell 90:511-522. [DOI] [PubMed] [Google Scholar]
- 53.Templeton, T. J., and D. C. Kaslow. 1997. Cloning and cross-species comparison of the thrombospondin-related anonymous protein (TRAP) gene from Plasmodium knowlesi, Plasmodium vivax, and Plasmodium gallinaceum. Mol. Biochem. Parasitol. 84:13-24. [DOI] [PubMed] [Google Scholar]
- 54.Templeton, T. J., D. C. Kaslow, and D. A. Fidock. 2000. Developmental arrest of the human malaria parasite Plasmodium falciparum within the mosquito midgut via CTRP gene disruption. Mol. Microbiol. 36:1-9. [DOI] [PubMed] [Google Scholar]
- 55.Tomley, F. M., and D. S. Soldati. 2001. Mix and match modules: structure and function of microneme proteins in apicomplexan parasites. Trends Parasitol. 17:81-88. [DOI] [PubMed] [Google Scholar]
- 56.Tomley, F. M., L. E. Clarke, U. Kawazoe, R. Dijkema, and J. J. Kok. 1991. Sequence of the gene encoding an immunodominant microneme protein of Eimeria tenella. Mol. Biochem. Parasitol. 49:277-288. [DOI] [PubMed] [Google Scholar]
- 57.Trottein, F., T. Triglia, and A. F. Cowman. 1995. Molecular cloning of a gene from Plasmodium falciparum that codes for a protein sharing motifs found in adhesive molecules from mammals and plasmodia. Mol. Biochem. Parasitol. 74:129-141. [DOI] [PubMed] [Google Scholar]
- 58.Upton, S. J., T. Tilley, and D. B. Brillhart. 1995. Effects of select medium supplements on in vitro development of Cryptosporidium parvum in HCT-8 cells. J. Clin. Microbiol. 33:371-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wengelnik, K., R. Spaccapelo, S. Naitza, K. J. H. Robson, C. J. Janse, F. Bistoni, A. P. Waters, and A. Crisanti. 1999. The A-domain and the thrombospondin-related motif of Plasmodium falciparum TRAP are implicated in the invasion process of mosquito salivary glands. EMBO J. 18:5195-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuda, M., H. Sakaida, and Y. Chinzei. 1999. Targeted disruption of the Plasmodium berghei CTRP gene reveals its essential role in malaria infection of the vector mosquito. J. Exp. Med. 190:1711-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuda, M., T. Sawai, and Y. Chinzei. 1999. Structure and expression of an adhesive protein-like molecule of mosquito invasive-stage malarial parasite. J. Exp. Med. 189:1947-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]