Abstract
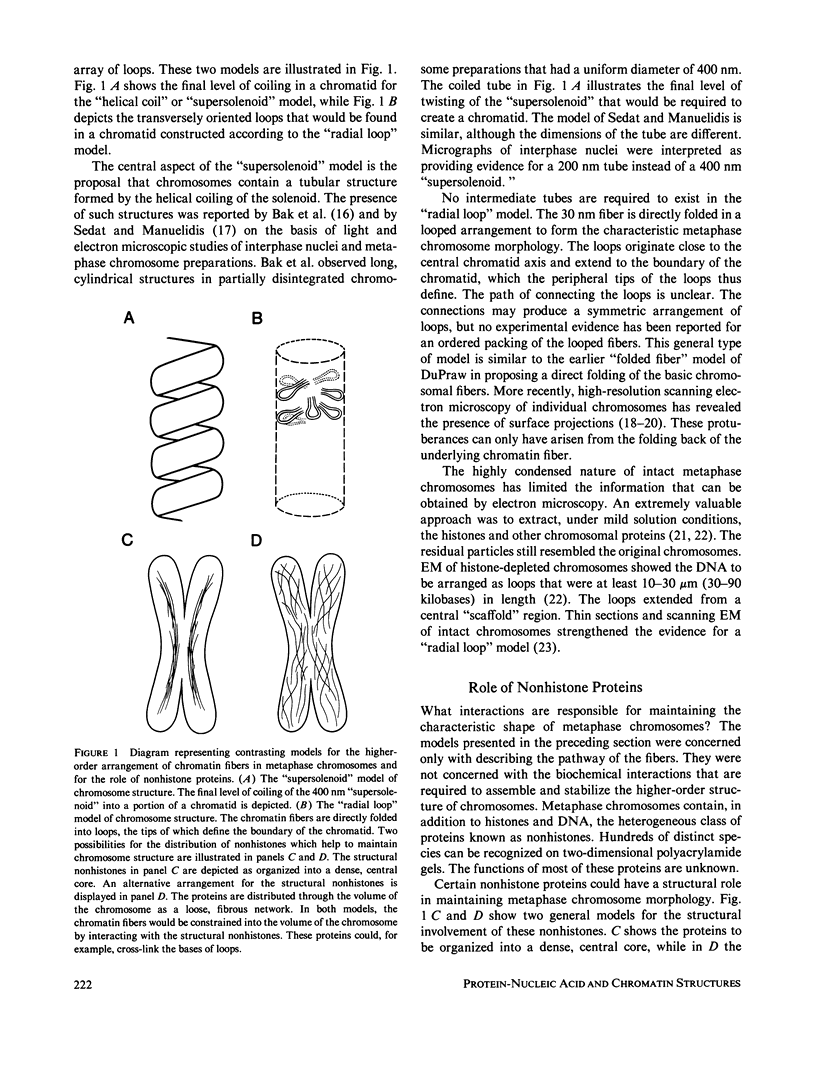
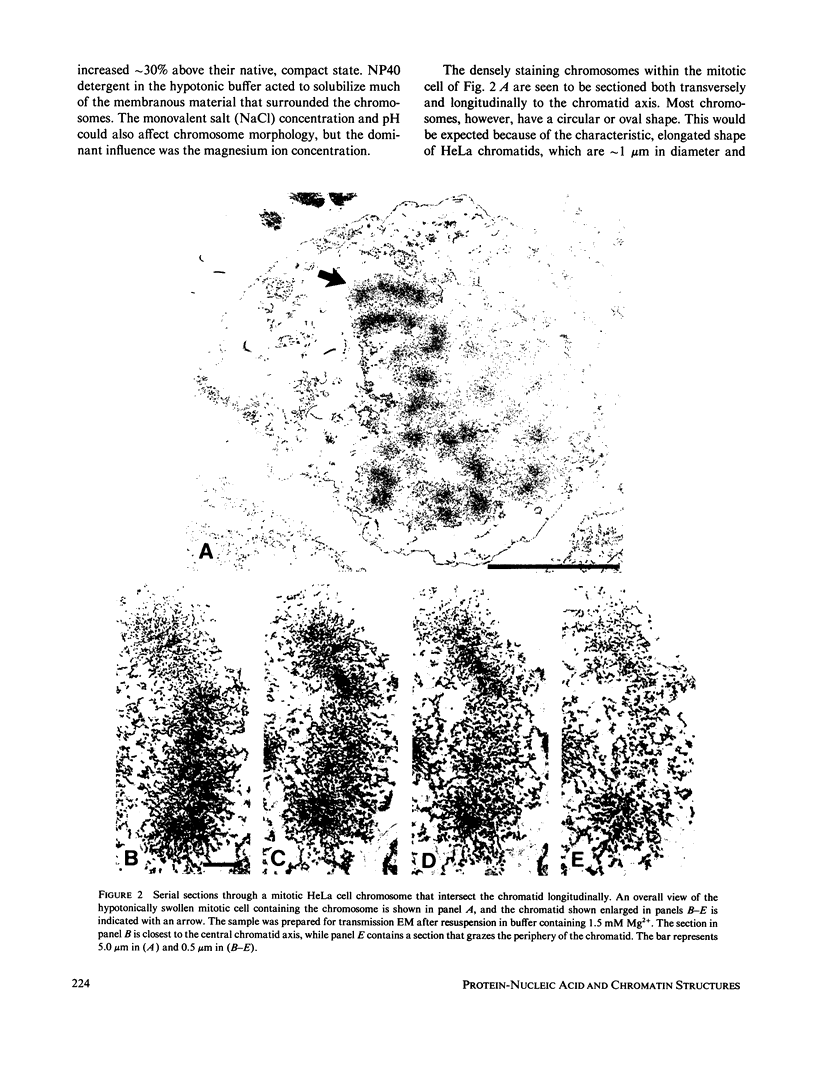
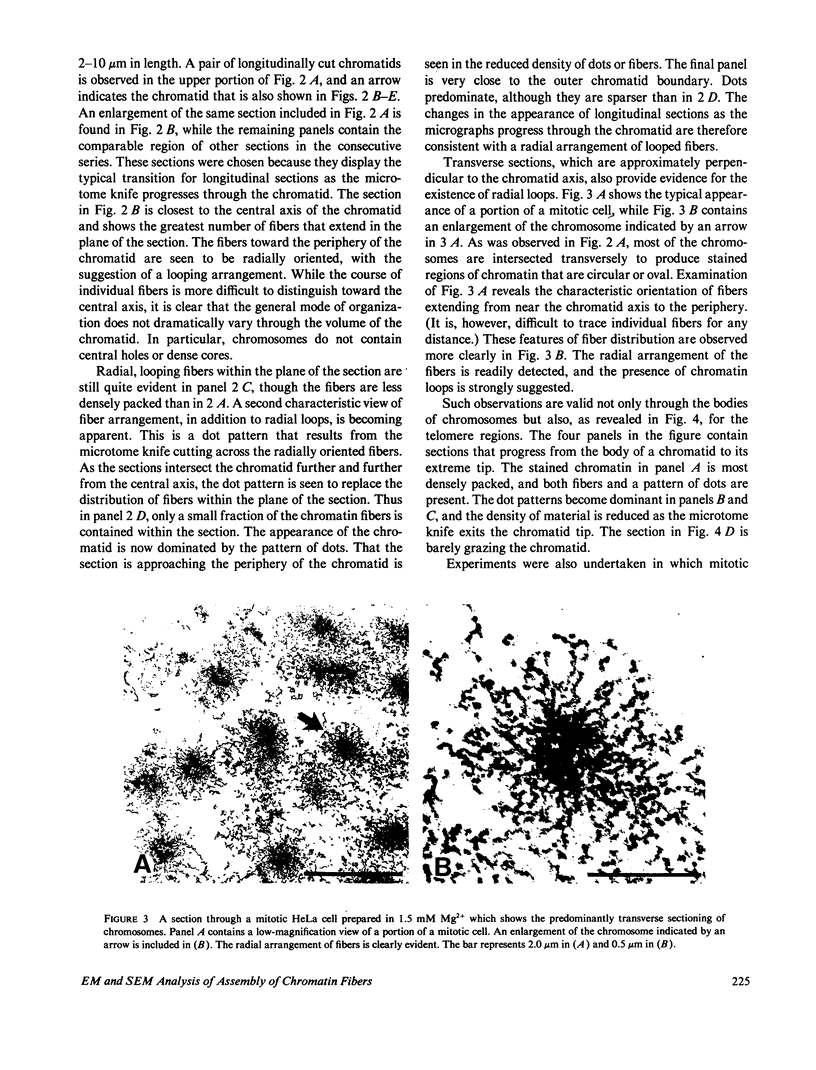
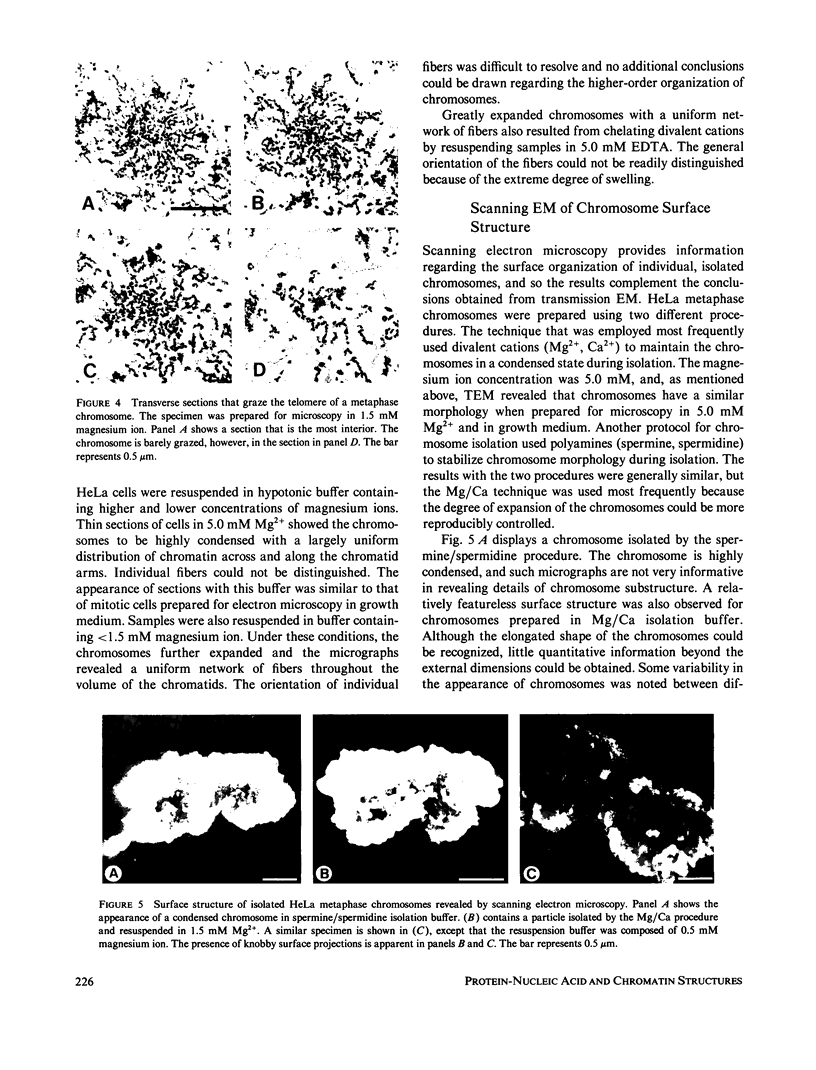
The higher-order assembly of the approximately 30 nm chromatin fibers into the characteristic morphology of HeLa mitotic chromosomes was investigated by electron microscopy. Transmission electron microscopy (TEM) of serial sections was applied to view the distribution of the DNA-histone-nonhistone fibers through the chromatid arms. Scanning electron microscopy (SEM) provided a complementary technique allowing the surface arrangement of the fibers to be observed. The approach with both procedures was to swell the chromosomes slightly, without extracting proteins, so that the densely-packed chromatin fibers were separated. The degree of expansion of the chromosomes was controlled by adjusting the concentration of divalent cations (Mg2+). With TEM, individual fibers could be resolved by decreasing the Mg2+ concentration to 1.0-1.5 mM. The predominant mode of fiber organization was seen to be radial for both longitudinal and transverse sections. Using SEM, surface protuberances with an average diameter of 69 nm became visible after the Mg2+ concentration was reduced to 1.5 mM. The knobby surface appearance was a variable feature, because the average diameter decreased when the divalent cation concentration was further reduced. The surface projections appear to represent the peripheral tips of radial chromatin loops. These TEM and SEM observations support a "radial loop" model for the organization of the chromatin fibers in metaphase chromosomes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adolph K. W., Cheng S. M., Laemmli U. K. Role of nonhistone proteins in metaphase chromosome structure. Cell. 1977 Nov;12(3):805–816. doi: 10.1016/0092-8674(77)90279-3. [DOI] [PubMed] [Google Scholar]
- Adolphs K. W., Cheng S. M., Paulson J. R., Laemmli U. K. Isolation of a protein scaffold from mitotic HeLa cell chromosomes. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4937–4941. doi: 10.1073/pnas.74.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak A. L., Zeuthen J., Crick F. H. Higher-order structure of human mitotic chromosomes. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1595–1599. doi: 10.1073/pnas.74.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D. L., Butler P. J., Pearson E. C., Thomas J. O. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur J Biochem. 1981 Oct;119(3):469–476. doi: 10.1111/j.1432-1033.1981.tb05631.x. [DOI] [PubMed] [Google Scholar]
- Butler P. J. The folding of chromatin. CRC Crit Rev Biochem. 1983;15(1):57–91. doi: 10.3109/10409238309102801. [DOI] [PubMed] [Google Scholar]
- Butler P. J., Thomas J. O. Changes in chromatin folding in solution. J Mol Biol. 1980 Jul 15;140(4):505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A., Jost E. Characterization of nuclear structures containing superhelical DNA. J Cell Sci. 1976 Nov;22(2):303–324. doi: 10.1242/jcs.22.2.303. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Mapping sequences in loops of nuclear DNA by their progressive detachment from the nuclear cage. Nucleic Acids Res. 1980 Jul 11;8(13):2895–2906. doi: 10.1093/nar/8.13.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Halligan N., Cooke C., Rothfield N. The kinetochore is part of the metaphase chromosome scaffold. J Cell Biol. 1984 Jan;98(1):352–357. doi: 10.1083/jcb.98.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Laemmli U. K. Architecture of metaphase chromosomes and chromosome scaffolds. J Cell Biol. 1983 Jan;96(1):84–93. doi: 10.1083/jcb.96.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Laemmli U. K. Silver staining the chromosome scaffold. Chromosoma. 1984;89(3):186–192. doi: 10.1007/BF00294997. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulmer A. W., Bloomfield V. A. Higher order folding of two different classes of chromatin isolated from chicken erythrocyte nuclei. A light scattering study. Biochemistry. 1982 Mar 2;21(5):985–992. doi: 10.1021/bi00534a025. [DOI] [PubMed] [Google Scholar]
- Harrison C. J., Allen T. D., Britch M., Harris R. High-resolution scanning electron microscopy of human metaphase chromosomes. J Cell Sci. 1982 Aug;56:409–422. doi: 10.1242/jcs.56.1.409. [DOI] [PubMed] [Google Scholar]
- Harrison C. J., Allen T. D., Harris R. Scanning electron microscopy of variations in human metaphase chromosome structure revealed by Giemsa banding. Cytogenet Cell Genet. 1983;35(1):21–27. doi: 10.1159/000131831. [DOI] [PubMed] [Google Scholar]
- Harrison C. J., Britch M., Allen T. D., Harris R. Scanning electron microscopy of the G-banded human karyotype. Exp Cell Res. 1981 Jul;134(1):141–153. doi: 10.1016/0014-4827(81)90471-7. [DOI] [PubMed] [Google Scholar]
- Langmore J. P., Paulson J. R. Low angle x-ray diffraction studies of chromatin structure in vivo and in isolated nuclei and metaphase chromosomes. J Cell Biol. 1983 Apr;96(4):1120–1131. doi: 10.1083/jcb.96.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. S., Mandelkern M., Crothers D. M. Solution structural studies of chromatin fibers. Biochemistry. 1981 Mar 17;20(6):1438–1445. doi: 10.1021/bi00509a006. [DOI] [PubMed] [Google Scholar]
- Lewis C. D., Laemmli U. K. Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell. 1982 May;29(1):171–181. doi: 10.1016/0092-8674(82)90101-5. [DOI] [PubMed] [Google Scholar]
- Marsden M. P., Laemmli U. K. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979 Aug;17(4):849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- McCready S. J., Godwin J., Mason D. W., Brazell I. A., Cook P. R. DNA is replicated at the nuclear cage. J Cell Sci. 1980 Dec;46:365–386. doi: 10.1242/jcs.46.1.365. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Langmore J. P. Low angle x-ray diffraction studies of HeLa metaphase chromosomes: effects of histone phosphorylation and chromosome isolation procedure. J Cell Biol. 1983 Apr;96(4):1132–1137. doi: 10.1083/jcb.96.4.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedat J., Manuelidis L. A direct approach to the structure of eukaryotic chromosomes. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):331–350. doi: 10.1101/sqb.1978.042.01.035. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T. Unravelled nucleosomes, nucleosome beads and higher order structures of chromatin: influence of non-histone components and histone H1. J Mol Biol. 1981 Jul 15;149(4):709–733. doi: 10.1016/0022-2836(81)90354-5. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Butler P. J. Size-dependence of a stable higher-order structure of chromatin. J Mol Biol. 1980 Nov 25;144(1):89–93. doi: 10.1016/0022-2836(80)90215-6. [DOI] [PubMed] [Google Scholar]