Abstract
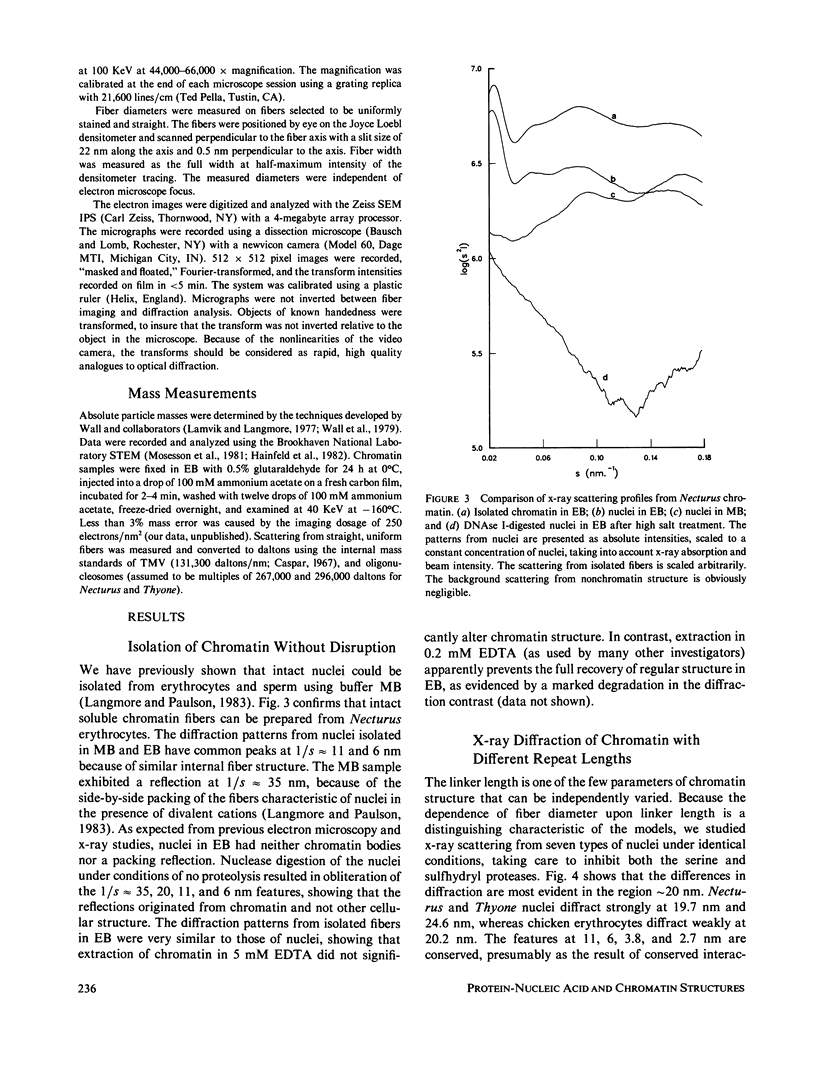
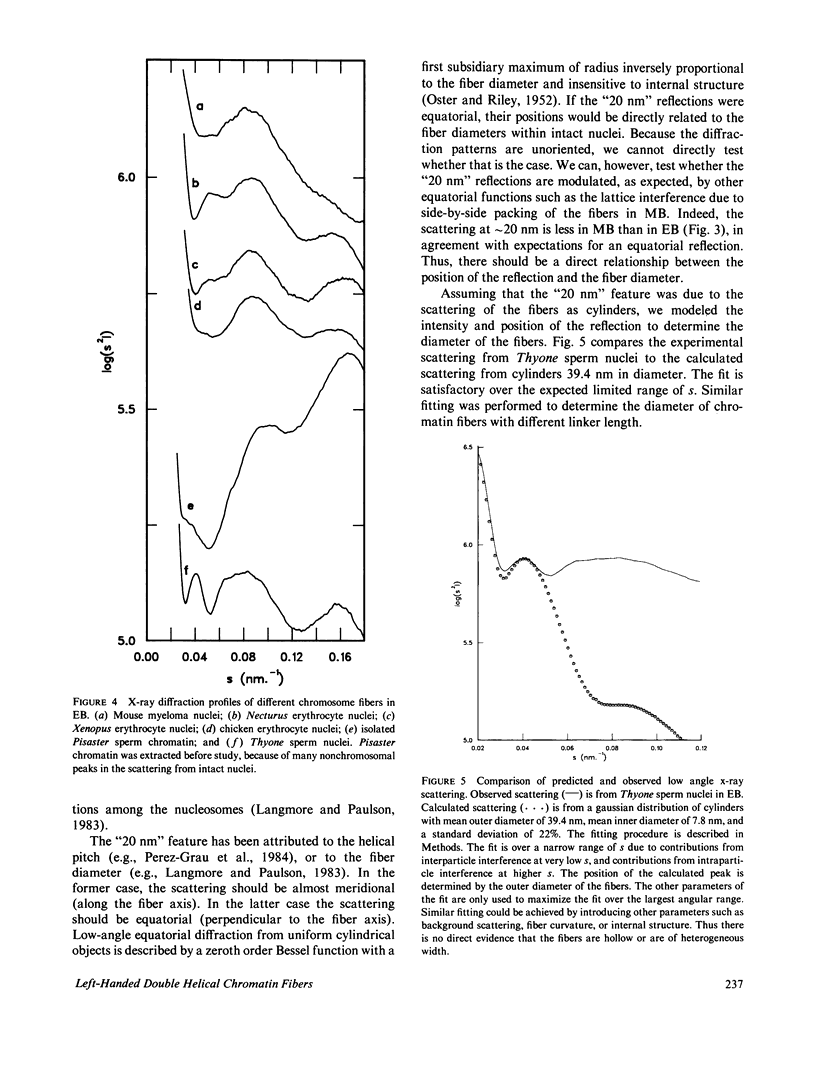
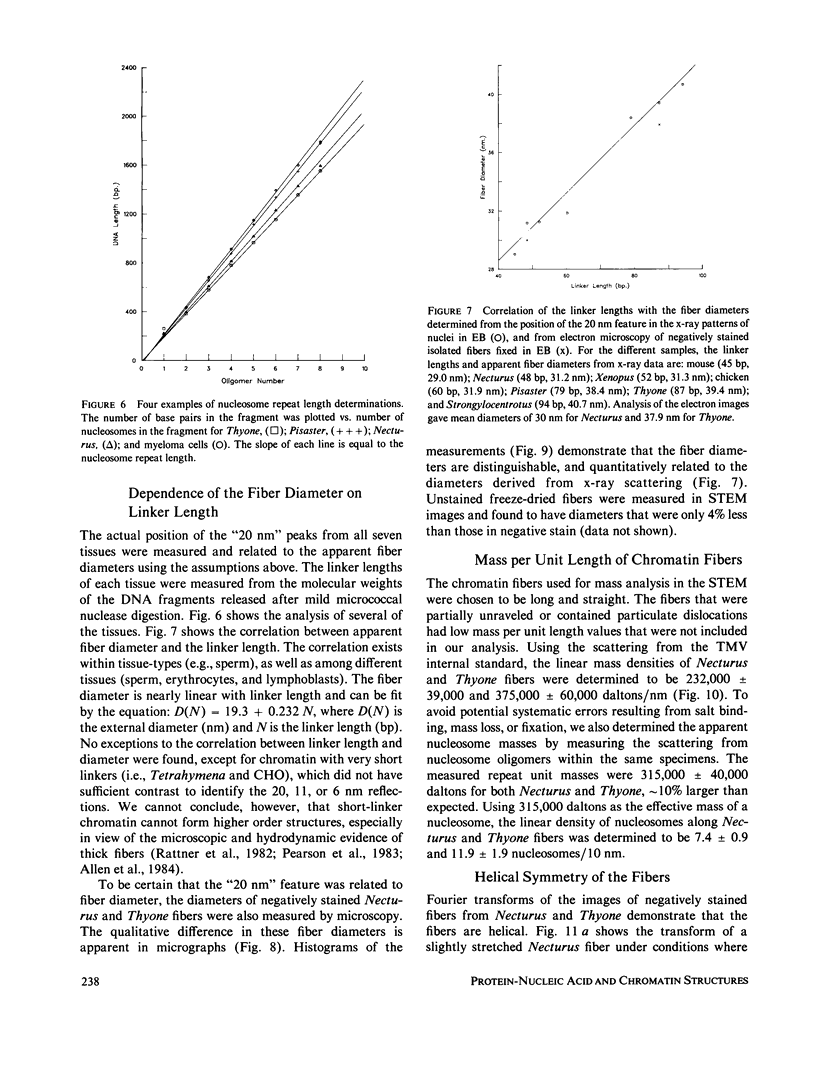
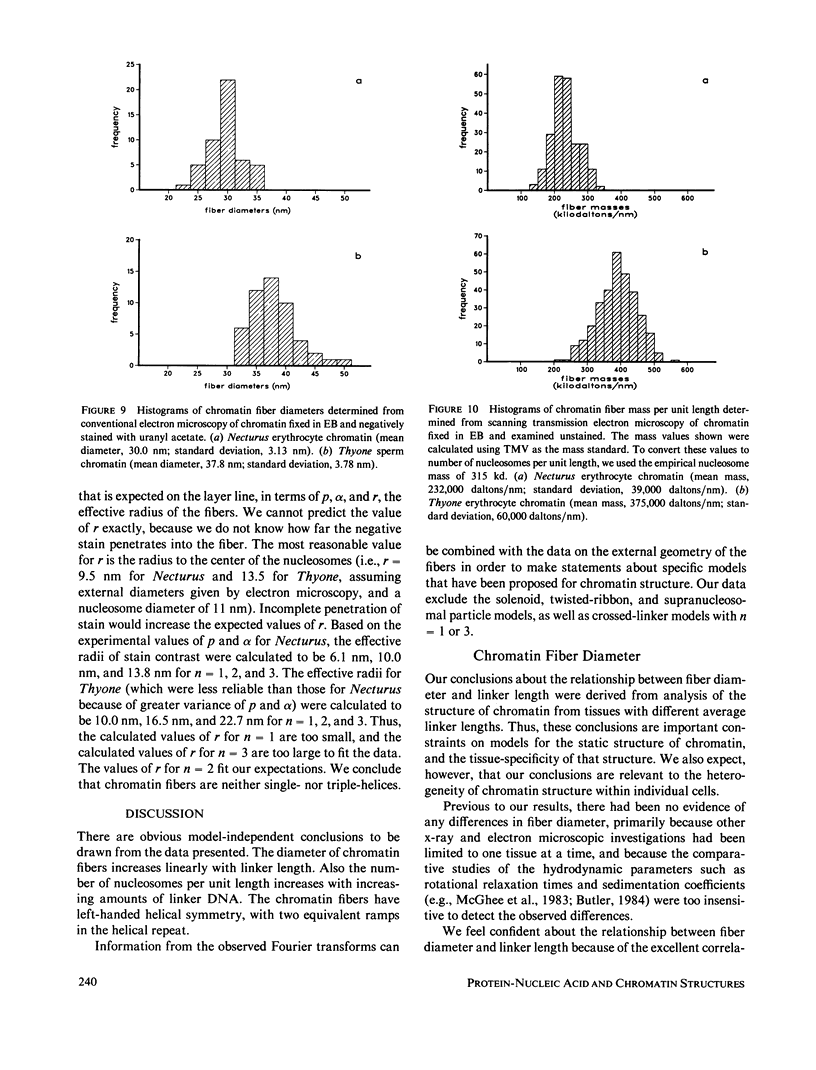
Four classes of models have been proposed for the internal structure of eukaryotic chromosome fibers--the solenoid, twisted-ribbon, crossed-linker, and superbead models. We have collected electron image and x-ray scattering data from nuclei, and isolated chromatin fibers of seven different tissues to distinguish between these models. The fiber diameters are related to the linker lengths by the equation: D(N) = 19.3 + 0.23 N, where D(N) is the external diameter (nm) and N is the linker length (base pairs). The number of nucleosomes per unit length of the fibers is also related to linker length. Detailed studies were done on the highly regular chromatin from erythrocytes of Necturus (mud puppy) and sperm of Thyone (sea cucumber). Necturus chromatin fibers (N = 48 bp) have diameters of 31 nm and have 7.5 +/- 1 nucleosomes per 10 nm along the axis. Thyone chromatin fibers (N = 87 bp) have diameters of 39 nm and have 12 +/- 2 nucleosomes per 10 nm along the axis. Fourier transforms of electron micrographs of Necturus fibers showed left-handed helical symmetry with a pitch of 25.8 +/- 0.8 nm and pitch angle of 32 +/- 3 degrees, consistent with a double helix. Comparable conclusions were drawn from the Thyone data. The data do not support the solenoid, twisted-ribbon, or supranucleosomal particle models. The data do support two crossed-linker models having left-handed double-helical symmetry and conserved nucleosome interactions.
Full text
PDF



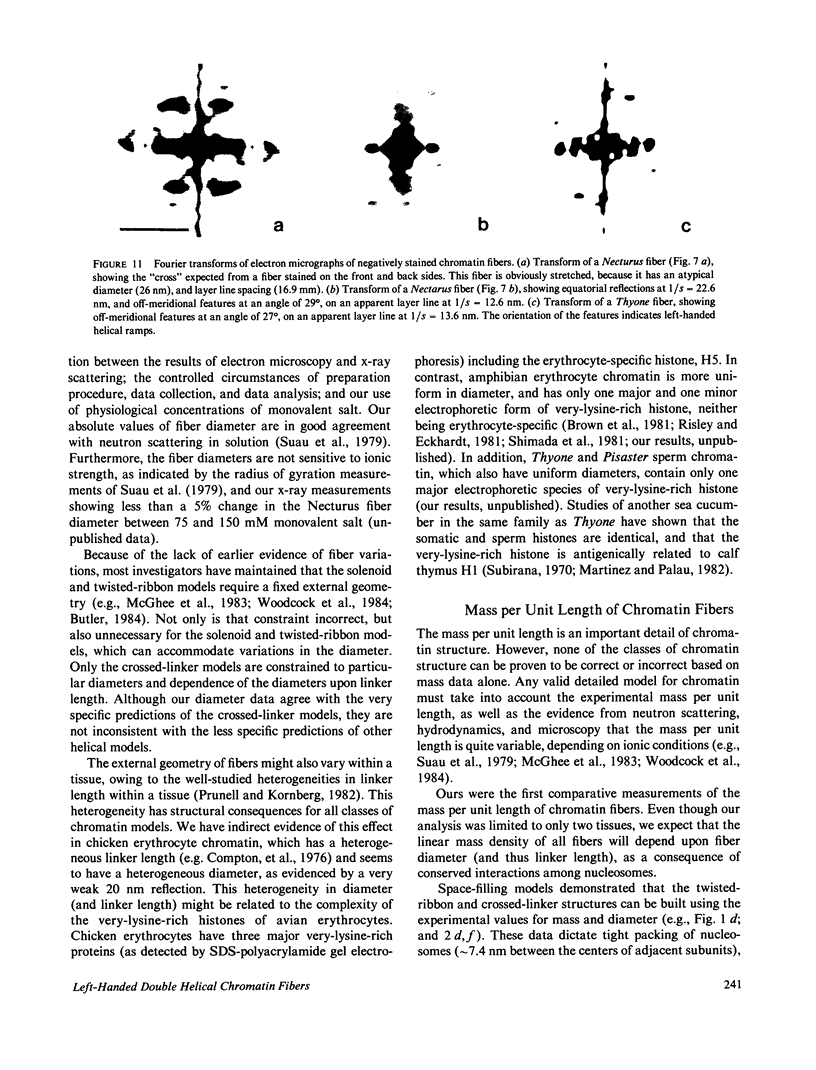




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Rau D. C., Harborne N., Gould H. Higher order structure in a short repeat length chromatin. J Cell Biol. 1984 Apr;98(4):1320–1327. doi: 10.1083/jcb.98.4.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D. L., Butler P. J., Pearson E. C., Thomas J. O. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur J Biochem. 1981 Oct;119(3):469–476. doi: 10.1111/j.1432-1033.1981.tb05631.x. [DOI] [PubMed] [Google Scholar]
- Brown G. L., Rutledge R. G., Neelin J. M. Anuran erythrocytes and liver both contain satellite histone Hls. Life Sci. 1981 Jun 29;28(26):2993–2999. doi: 10.1016/0024-3205(81)90277-0. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Skinner J. D. Avian erythrocyte chromatin degradation: the progressive exposure of the dinucleosomal repeat by bovine-pancreatic-DNAase-I-armed probes and free DNAase-I. Nucleic Acids Res. 1982 Jan 22;10(2):665–673. doi: 10.1093/nar/10.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler P. J. A defined structure of the 30 nm chromatin fibre which accommodates different nucleosomal repeat lengths. EMBO J. 1984 Nov;3(11):2599–2604. doi: 10.1002/j.1460-2075.1984.tb02180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler P. J., Thomas J. O. Changes in chromatin folding in solution. J Mol Biol. 1980 Jul 15;140(4):505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
- CASPAR D. L. ASSEMBLY AND STABILITY OF THE TOBACCO MOSAIC VIRUS PARTICLE. Adv Protein Chem. 1963;18:37–121. doi: 10.1016/s0065-3233(08)60268-5. [DOI] [PubMed] [Google Scholar]
- Campbell A. M., Cotter R. I., Pardon J. F. Light scattering measurements supporting helical structures for chromatin in solution. Nucleic Acids Res. 1978 May;5(5):1571–1580. doi: 10.1093/nar/5.5.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B. G., Baldwin J. P., Bradbury E. M., Ibel K. Organisation of subunits in chromatin. Nucleic Acids Res. 1976 Jul;3(7):1739–1746. doi: 10.1093/nar/3.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S. A., Vaughn J. P., Shaw B. R. Shortest nucleosomal repeat lengths during sea urchin development are found in two-cell embryos. Biochemistry. 1983 Nov 22;22(24):5626–5631. doi: 10.1021/bi00293a026. [DOI] [PubMed] [Google Scholar]
- Compton J. L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H. G. Electron-microscope observations on the organization of heterochromatin in certain cells. J Cell Sci. 1968 Mar;3(1):129–150. doi: 10.1242/jcs.3.1.129. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GALL J. Chromosome fibers from an interphase nucleus. Science. 1963 Jan 11;139(3550):120–121. doi: 10.1126/science.139.3550.120. [DOI] [PubMed] [Google Scholar]
- Gargiulo G., Worcel A. Analysis of the chromatin assembled in germinal vesicles of Xenopus oocytes. J Mol Biol. 1983 Nov 5;170(3):699–722. doi: 10.1016/s0022-2836(83)80128-4. [DOI] [PubMed] [Google Scholar]
- Hainfeld J. F., Wall J. S., Desmond E. J. A small computer system for micrograph analysis. Ultramicroscopy. 1982;8(3):263–270. doi: 10.1016/0304-3991(82)90242-x. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- KLUG A., BERGER J. E. AN OPTICAL METHOD FOR THE ANALYSIS OF PERIODICITIES IN ELECTRON MICROGRAPHS, AND SOME OBSERVATIONS ON THE MECHANISM OF NEGATIVE STAINING. J Mol Biol. 1964 Dec;10:565–569. doi: 10.1016/s0022-2836(64)80081-4. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmore J. P., Paulson J. R. Low angle x-ray diffraction studies of chromatin structure in vivo and in isolated nuclei and metaphase chromosomes. J Cell Biol. 1983 Apr;96(4):1120–1131. doi: 10.1083/jcb.96.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Organization of spacer DNA in chromatin. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6326–6330. doi: 10.1073/pnas.76.12.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov V., Dimitrov S., Smirnov V., Pashev I. A triple helix model for the structure of chromatin fiber. FEBS Lett. 1985 Feb 25;181(2):357–361. doi: 10.1016/0014-5793(85)80292-1. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Nickol J. M., Felsenfeld G., Rau D. C. Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell. 1983 Jul;33(3):831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- Mosesson M. W., Hainfeld J., Wall J., Haschemeyer R. H. Identification and mass analysis of human fibrinogen molecules and their domains by scanning transmission electron microscopy. J Mol Biol. 1981 Dec 15;153(3):695–718. doi: 10.1016/0022-2836(81)90414-9. [DOI] [PubMed] [Google Scholar]
- Pearson E. C., Butler P. J., Thomas J. O. Higher-order structure of nucleosome oligomers from short-repeat chromatin. EMBO J. 1983;2(8):1367–1372. doi: 10.1002/j.1460-2075.1983.tb01593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Grau L., Bordas J., Koch M. H. Chromatin superstructure: synchrotron radiation X-ray scattering study on solutions and gels. Nucleic Acids Res. 1984 Mar 26;12(6):2987–2996. doi: 10.1093/nar/12.6.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D. Variable center to center distance of nucleosomes in chromatin. J Mol Biol. 1982 Jan 25;154(3):515–523. doi: 10.1016/s0022-2836(82)80010-7. [DOI] [PubMed] [Google Scholar]
- Rattner J. B., Saunders C., Davie J. R., Hamkalo B. A. Ultrastructural organization of yeast chromatin. J Cell Biol. 1982 Apr;93(1):217–222. doi: 10.1083/jcb.93.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz M., Nehls P., Hozier J. Involvement of histone H1 in the organization of the chromosome fiber. Proc Natl Acad Sci U S A. 1977 May;74(5):1879–1883. doi: 10.1073/pnas.74.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Risley M. S., Eckhardt R. A. H1 histone variants in Xenopus laevis. Dev Biol. 1981 May;84(1):79–87. doi: 10.1016/0012-1606(81)90372-9. [DOI] [PubMed] [Google Scholar]
- Shimada T., Okihama Y., Murata C., Shukuya R. Occurrence of H1o-like protein and protein A24 in the chromatin of bullfrog erythrocytes lacking histone 5. J Biol Chem. 1981 Oct 25;256(20):10577–10582. [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Strauss F., Prunell A. Organization of internucleosomal DNA in rat liver chromatin. EMBO J. 1983;2(1):51–56. doi: 10.1002/j.1460-2075.1983.tb01379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suau P., Bradbury E. M., Baldwin J. P. Higher-order structures of chromatin in solution. Eur J Biochem. 1979 Jul;97(2):593–602. doi: 10.1111/j.1432-1033.1979.tb13148.x. [DOI] [PubMed] [Google Scholar]
- Subirana J. A. Nuclear proteins from a somatic and a germinal tissue of the echinoderm Holothuria tubulosa. Exp Cell Res. 1970 Dec;63(2):253–260. doi: 10.1016/0014-4827(70)90211-9. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L. G. The polymerization of actin. II. How nonfilamentous actin becomes nonrandomly distributed in sperm: evidence for the association of this actin with membranes. J Cell Biol. 1976 Apr;69(1):51–72. doi: 10.1083/jcb.69.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., Frado L. L., Rattner J. B. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol. 1984 Jul;99(1 Pt 1):42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Strogatz S., Riley D. Structure of chromatin and the linking number of DNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1461–1465. doi: 10.1073/pnas.78.3.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentgraf H., Franke W. W. Differences of supranucleosomal organization in different kinds of chromatin: cell type-specific globular subunits containing different numbers of nucleosomes. J Cell Biol. 1984 Jul;99(1 Pt 1):272–286. doi: 10.1083/jcb.99.1.272. [DOI] [PMC free article] [PubMed] [Google Scholar]






