Abstract
Platelets share structural and functional similarities with granulocytes known to participate in antimicrobial host defense. To evaluate the potential antimicrobial activities of platelet proteins, normal human platelets were stimulated with human thrombin in vitro. Components of the stimulated-platelet supernatants were purified to homogeneity by reversed-phase high-performance liquid chromatography. Purified peptides with inhibitory activity against Escherichia coli ML35 in an agar diffusion antimicrobial assay were characterized by mass spectrometry, amino acid analysis, and sequence determination. These analyses enabled the identification of seven thrombin-releasable antimicrobial peptides from human platelets: platelet factor 4 (PF-4), RANTES, connective tissue activating peptide 3 (CTAP-3), platelet basic protein, thymosin β-4 (Tβ-4), fibrinopeptide B (FP-B), and fibrinopeptide A (FP-A). With the exception of FP-A and FP-B, all peptides were also purified from acid extracts of nonstimulated platelets. The in vitro antimicrobial activities of the seven released peptides were further tested against bacteria (E. coli and Staphylococcus aureus) and fungi (Candida albicans and Cryptococcus neoformans). Each peptide exerted activity against at least two organisms. Generally, the peptides were more potent against bacteria than fungi, activity was greater at acidic pHs, and antimicrobial activities were dose dependent. Exceptions to these observations were observed with PF-4, which displayed a bimodal dose-response relationship in microbicidal assays, and Tβ-4, which had greater activity at alkaline pHs. At concentrations at which they were individually sublethal, PF-4 and CTAP-3 exerted synergistic microbicidal activity against E. coli. Collectively, these findings suggest a direct antimicrobial role for platelets as they are activated to release peptides in response to trauma or mediators of inflammation.
The observation that rabbit serum contains bactericidal substances that act against certain gram-positive bacteria was reported over 100 years ago (13). Subsequent evidence suggested that platelets were the cellular source of this antibacterial activity (18, 41). Since these early studies, numerous investigators have sought to isolate platelet-specific antimicrobial molecules from animal and human platelets. Weksler and Nachman isolated bactericidal proteins (10, 30, and 40 kDa) from acid extracts of rabbit and human platelets (28, 41). Donaldson and Tew described a small (ca. 6 kDa) antibacterial peptide from rabbit platelets which was released from platelet granules as a result of thrombin-induced platelet aggregation (12). Carroll and Martinez reported a bactericidal peptide isolated from normal rabbit serum (4). More recently, Yeaman et al. characterized antimicrobial polypeptides from thrombin-induced released materials and acid extracts of rabbit platelets (44-46). Zaat et al. reported the presence and activities of unidentified antimicrobial substances in supernatants of thrombin-stimulated human platelets (49). In a preliminary study, we reported the isolation and tentative identification of seven antimicrobial peptides from human platelets after thrombin stimulation [Y. Q. Tang, M. R. Yeaman, and M. E. Selsted, abstracts from the American Society of Hematology 37th Annual Meeting 1995, Blood 86(Suppl. 1):556a and 910a, 1995]. Subsequently, Krijgsveld et al. reported two connective tissue activating peptide 3 (CTAP-3)-related antimicrobial peptides in supernatants of thrombin-stimulated human platelets (22). In the above reports, the platelet polypeptides exhibited relatively potent activities against pathogens that have a propensity to enter the bloodstream, including Staphylococcus aureus, Streptococcus sanguis, Escherichia coli, Candida albicans, and Cryptococcus neoformans. Collectively, these findings suggest that human platelets possess, and can be stimulated to release, several antimicrobial polypeptides.
Only limited structural and functional properties of such molecules from human platelets have been reported to date. Therefore, the present studies were undertaken to more fully define the structures and functions of antimicrobial peptides derived from human platelets. To this end, we have isolated and identified seven human platelet antimicrobial peptides (HPAPs) and evaluated their in vitro activities against common bloodborne pathogens, including bacteria and fungi.
MATERIALS AND METHODS
Thrombin stimulation and acid extraction of human platelets.
Healthy human platelet-rich plasma was obtained by low-speed centrifugation of 1 day-outdated platelets from the Blood Donor Center of the University of California, Irvine, Medical Center. The platelet-rich plasma was then dispensed into polypropylene tubes and centrifuged at 100 × g for 15 min at 25°C. The supernatants were removed and centrifuged again at 2,000 × g for 15 min at 25°C, and the sedimented platelets were washed three times with Tyrode's buffer (138 mM NaCl, 3.6 mM KCl, 10 mM NaHCO3, 0.4 mM NaH2PO4, 10 mM MgCl2, and 6 mM glucose, adjusted to pH 7.3 with phosphoric acid) (27). The washed platelets were stored at −20°C or resuspended immediately in the same buffer or Eagle's minimal essential medium (MEM; Gibco BRL). All experiments involving the use of human subjects were performed according to federal guidelines and policies of the University of California.
Preparations of freshly washed platelets, suspended to ∼5 × 108 platelets per ml in Tyrode's buffer or MEM containing 2.5 mM CaCl2, were stimulated by incubation with 1 U of human thrombin (T3010; Sigma Chemical Co., St. Louis, Mo.) per ml for 20 min at 37°C, with gentle agitation. A control suspension of platelets at the same concentration, but without thrombin stimulation, was incubated under identical conditions. Residual platelet material was removed by centrifugation as described above, and the supernatant was acidified to pH 3.0 with acetic acid.
Frozen platelet pellets (∼10 ml at 109 platelets per ml) were thawed, suspended in 6 volumes of ice-cold 30% acetic acid, and stirred in melting ice for 18 h. The resulting extract was centrifuged at 23,000 × g for 30 min at 4°C, and the supernatant was lyophilized. The lyophilized material was dissolved in 30% acetic acid, clarified by centrifugation as described above, and prepared for subsequent fractionation by size exclusion chromatography and purification by reversed-phase high-performance liquid chromatography (RP-HPLC).
Peptide purification.
Peptides contained within the acidified supernatants of thrombin-stimulated platelet released materials were purified directly by RP-HPLC. The 30% acetic acid platelet extract was fractionated by size exclusion chromatography on a 4.6- by 60-cm Bio-Gel P-60 column with an elution rate of 30 ml/h. The eluate was monitored continuously at 280 nm, and all fractions were screened for antibacterial activity (see below). Fractions containing antibacterial activity were combined and further purified by RP-HPLC. Final purification of HPAPs was achieved by use of Vydac C4 or C18 columns developed by gradient elution using water-acetonitrile containing 0.1% trifluoroacetic acid (TFA) or 0.13% heptafluorobutyric acid. Purified peptides were lyophilized, dissolved in 5% acetic acid, and stored at −20°C.
Mass spectrometry.
Masses of purified peptides were determined by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF; model Voyager DE-RP; PerSeptive Biosystems, Foster City, Calif.) or electrospray (ES; model Sciex API-1; Perkin-Elmer Corp., Boston, Mass.) mass spectrometry. Samples were introduced as a solution in 0.5% acetic acid. Typical errors of measurement were <0.1% or <0.04% of the peptide molecular weights for MALDI-TOF or ES mass spectrometry, respectively.
Structural determination.
Amino acid compositions of the peptides were determined after hydrolysis in 6.0 N HCl (2 h, 150°C). Samples (0.3 to 1.0 nmol of peptide) were prepared for automated Edman sequence analysis after reduction with mercaptoethanol and alkylation with 4-vinylpyridine. Peptides found to be blocked at the amino terminus were deblocked using pyroglutamate aminopeptidase (Boehringer Mannheim, GmbH, Mannheim, Germany). Derivatized peptides were purified by RP-HPLC prior to sequence analysis. Proteolytic digestion of selected peptides was carried out using α-chymotrypsin (Boehringer Mannheim, GmbH). Two nanomoles of peptide was dissolved in 50 μl of 0.125 M ammonium bicarbonate buffer and incubated with enzyme (1.0 μg) for 60 min at 37°C. Digested fragments were subsequently purified by RP-HPLC, characterized by amino acid analysis, and in some cases, sequenced. Carboxyl-terminal amino acid sequences were determined by analysis of residues released following digestion with carboxypeptidase Y (Boehringer Mannheim, GmbH).
Antimicrobial assays.
The antimicrobial properties of purified peptides were evaluated in vitro against E. coli ML-35, S. aureus 502A, C. albicans 16820, and C. neoformans 271A. Each organism has been used in prior studies to evaluate antimicrobial peptide activities (23, 33, 44, 46). Two assays were used for these purposes: a radial diffusion agarose plate assay (23) and a solution-phase microbicidal assay (32). For either assay, organisms were grown to mid-log phase in Trypticase soy broth (TSB; Difco Laboratories, Detroit, Mich.) for bacteria or Sabouraud dextrose broth (SDB; Difco) for fungi. For the diffusion assay, organisms were diluted to a concentration of 106 CFU/ml in 10 ml of 1% agarose (molecular biology grade; melted and then held at 42°C) containing 3 mg of glucose. In initial bioassay screens of crude or semipurified samples, the agarose was buffered with 10 mM phosphate to a pH of 7.4. For purified peptide samples, the agarose was buffered to pH values of 5.5 or 6.5 using 10 mM MES [2-(N-morpholino)ethanesulfonic acid; Sigma] or to pH 7.5 using PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); Sigma]. The buffered agarose seeded with a microorganism was used as the underlayer in the radial diffusion assay. Purified peptides stored in 5% acetic acid were relyophilized, dissolved in 0.01% acetic acid (5 μl, representing a range of concentrations from 50 to 200 nmol/ml or 0.25 to 1 nmol/well), pipetted into wells of the underlay agar, and incubated at 37°C for 3 h. The agar plates were then overlaid with buffered agarose containing TSB (for bacteria) or SDB (for fungi). After incubation at 37°C overnight (for bacteria and C. albicans) or for 48 h (for C. neoformans), the diameter of the zone of inhibition surrounding each well was measured.
For the solution-phase microbicidal assay, washed organisms were inoculated into one of the following buffer systems: MES buffer (2 mM, pH 5.5) for microbicidal assays of platelet factor 4 (PF-4), platelet basic protein (PBP), and CTAP-3 against the four test organisms; PIPES buffer (10 mM, pH 7.4) for microbicidal assays of thymosin β-4 (Tβ-4) against the four test organisms; or sodium acetate buffer (2 mM, pH 5.5) for the microbicidal assay of PF-4 and CTAP-3 in combination against E. coli. Buffers were inoculated with an appropriate organism at 106 CFU/ml and incubated with a peptide(s) at concentrations ranging from 0 to 20 nmol/ml for 1 h at 37°C. After incubation, the suspensions were quantitatively cultured in duplicate onto nutrient agar. Colonies were enumerated after incubation for a minimum of 24 h at 37°C. All antimicrobial assays were performed a minimum of two times, and representative data are presented.
RESULTS
Purification of HPAPs.
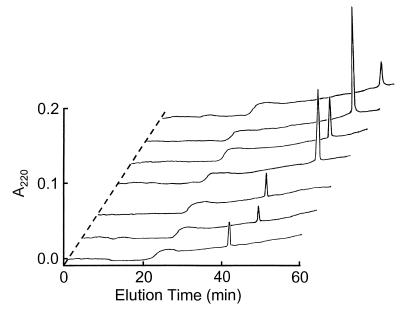
Isolation and purification of antimicrobial peptides from acid-extracted human platelets proceeded in a two-step manner. First, acetic acid extracts of human platelets were subjected to size exclusion chromatography (Fig. 1A). The resulting fractions were screened for antimicrobial activity by a diffusion assay. Activity against bacteria was observed in the initial screening of P-60 chromatography fractions 17 to 48 (data not shown). Fractions exhibiting such activity were combined, lyophilized, and subjected to RP-HPLC purification (see below). Acidified supernatants of nonstimulated human platelets and thrombin-stimulated human platelets were purified by RP-HPLC (Fig. 1B, panels I and II). In addition, fractions 17 to 48 eluted from the P-60 column were purified under identical RP-HPLC conditions (Fig. 1B, panel III). By comparing RP-HPLC profiles of control (Fig. 1B, panel I) and thrombin-stimulated (Fig. 1B, panel II) platelets, it was evident that seven predominant proteins (peaks 1 to 7 in the latter sample) were released from platelets as a result of thrombin stimulation. Similarly, peaks with corresponding antimicrobial activity were likewise identified in acid-extracted platelet samples. These peaks were absent from control RP-HPLC chromatograms of nonstimulated platelets (Fig. 1B, panel I) or the thrombin reagent alone (data not shown). Each of the above peptides was collected and purified to homogeneity (Fig. 2) by RP-HPLC using one or both buffer systems (water-acetonitrile containing 0.1% TFA or 0.13% heptafluorobutyric acid). Based on their increasing retention times on RP-HPLC, these peptides were numbered 1 to 7 as shown in Fig. 2 and Table 1. Peptides 3 to 7 were detected in thrombin-induced released materials (Fig. 1B, panel II) and in acid extracts of human platelets (Fig. 1B, panel III). The respective molecules coeluted under at least two RP-HPLC conditions and exhibited identical migration characteristics upon sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown).
FIG. 1.
Purification of human platelet antimicrobial peptides. (A) Bio-Gel P-60 size exclusion chromatography. An acetic acid extract from human platelets was loaded onto a 4.8- by 60-cm Bio-Gel P-60 column and eluted with 30% acetic acid at a flow rate of 30 ml/h. Several fractions, encompassed by fractions 17 to 48, demonstrated antibacterial activity upon initial screening assays. These fractions were further purified by RP-HPLC. (B) RP-HPLC purification. RP-HPLC conditions were a 1.0- by 25-cm Vydac C4 column that was equilibrated in 0.1% TFA-water at a flow rate of 2.5 ml/min. Linear gradients of acetonitrile containing 0.1% TFA were applied over 60 min (0 to 40%) and 40 min (40 to 100%). The following samples were injected onto the column: supernatant from nonstimulated human platelets (I), supernatant from thrombin-stimulated human platelets (II), and fractions 17 to 48 from the Bio-Gel P-60 column (III). Seven active peptides were obtained from peaks 1 to 7 in panel II and identified as shown in Fig. 2. Corresponding peaks 3 to 7 in panels II and III were identified to be identical. Peak A (panels I and II) and peak D (panel III) were identified as human serum albumin and dioctylphthalate, respectively.
FIG. 2.
Analytical RP-HPLC of HPAPs. Samples (0.5 nmol) of each of the peaks, 1 to 7, isolated for Fig. 1B were injected onto a 0.4- by 25-cm Vydac C18 column and eluted at a flow rate of 1.0 ml/min in the same solvent system as that described for Fig. 1B. A linear gradient of 0 to 40% acetonitrile containing 0.1% TFA was applied over 60 min. Peak identification (also see Table 1): 1, FP-A; 2, FP-B; 3, Tβ-4; 4, PBP; 5, CTAP-3; 6, RANTES; 7, PF-4. The methods for identification of these peptides are shown in Table 1.
TABLE 1.
Comparative biochemical characteristics of HPAPs
| Peptide | HPLC peak no.a | Molecular mass (Da)b | N-terminal or partial sequencec | C-terminal sequenced | Net charge at pH:e
|
Concn in platelets (nmol/1010 platelets)f | |
|---|---|---|---|---|---|---|---|
| 7.5 | 5.5 | ||||||
| FP-A | 1 | NDg | ADSGEGDFLAEGGGVR | ND | −3.00 | −2.86 | <1.0 |
| FP-B | 2 | 1,551.6 (1,551.6) | G2VNDNEEGFFSA13 | ND | −3.00 | −2.88 | <1.0 |
| Tβ-4 | 3 | 4,962.2 (4,963.5) | K18KTETQEKNPLPSKE32 | G-E-S (0.1-0.1-0.8) | −3.00 | −2.52 | 2.0 |
| PBP | 4 | 10,260.6 (10,261.9) | S1STKGQTKRNLAKGK15 | ND | +3.55 | +5.95 | 2.0 |
| CTAP-3 | 5 | 9,287.0 (9,287.8) | N1LAKGKEESLDSDLY15 | S-A-D (0.1-0.3-0.4) | +0.53 | +2.95 | 23 |
| RANTES | 6 | 7,850.5 (7,847.1) | S1PYSSDTTPCCFAYI15 | ND | +4.52 | +5.99 | 0.2 |
| PF-4 | 7 | 7,765.0 (7,766.2) | E1AEEDGDLQCLCVKT15 | L-L-E-S (2.2-0.8-2.8) | +2.57 | +4.84 | 15 |
See Fig. 2.
Molecular masses determined by mass spectrometry compared with those (in parentheses) calculated from amino acid sequences of corresponding peptides (see references in the text).
Sequences of the peptides are shown in single-letter code. The numbering of residues is matched to the sequences of the corresponding peptides.
Released amounts (nanomoles) of C-terminal amino acids upon digestion with carboxypeptidase Y are shown in parentheses. The C-terminal sequences of peaks 3, 5, and 7 were identical with those of the respective peptides.
Net charges were calculated by using the pI Graph Viewprogram in the GeneWorks (version 2.5.1) suite.
Approximate content of each peptide in the released material of platelets based on quantity of recovery as described in Results.
not determined.
Thrombin-induced release of HPAPs.
Amino acid analysis was used to estimate the quantity of HPAPs 1 to 7 released from thrombin-stimulated human platelets. The quantity of each HPAP recovered from 1010 platelets is summarized in Table 1. The most abundant HPAPs in the thrombin-stimulated released material were HPAP 5 (52% of the total HPAP mass recovered) and HPAP 7 (34% of the total HPAP mass recovered); the relative quantities of HPAPs 1, 2, 3, 4, and 6 were ∼2, ∼2, 4, 4, and ∼0.4%, respectively. As noted above, HPAPs 3 to 7 were present in both the thrombin-stimulated and acid-extracted human platelet preparations, whereas HPAPs 1 and 2 were found only in the former preparation. Recovery of HPAP 3 was ∼10 times higher from the acid extracts than from the thrombin-induced released material; however, the relative abundance of HPAPs 4 to 7 was equivalent in the two preparations (Fig. 1B, panels II and III). Peaks not numbered in Fig. 1B were either inactive in antimicrobial assays or appeared as a complex of broad minor peaks upon repurification; these peaks were not characterized further.
Identification of HPAPs.
The identities of purified HPAPs were determined by analysis of primary sequences, molecular masses, and amino acid compositions. The molecular masses of HPAPs 2 to 7, as determined by ES or MALDI-TOF mass spectrometry, are shown in Table 1. N-terminal or internal amino acid sequences (≥15 residues each) were obtained for HPAPs 1, 4, 5, 6, and 7 (Table 1). Sequence analysis of HPAP 1 (∼0.4 nmol) yielded no amino acids beyond Arg16. HPAPs 2 and 3 were found to be N-terminally blocked and were subjected to deblocking with pyroglutamate aminopeptidase. Twelve residues were sequenced from the N terminus of deblocked HPAP 2, beginning with amino acid residue number 2 (Table 1). However, HPAP 3 remained refractory to sequencing following deblocking and was therefore subjected to proteolysis using α-chymotrypsin. Three fragments of the resulting HPAP 3 digest were purified by RP-HPLC (data not shown), and a 15-amino-acid sequence was obtained from one fragment (Table 1).
A sequence similarity search using the BLAST algorithm (1) revealed the partial sequences of HPAPs 1 to 7 to be identical to those of seven known peptides (see below). The amino acid compositions of HPAPs 1 to 7 were consistent with those calculated from the corresponding sequences of the respective known peptides (data not shown). Thus, the amino acid sequence data, molecular masses, and amino acid compositions enabled the identification of HPAPs 1 to 7 as the following: fibrinopeptide A (FP-A) (15), fibrinopeptide B (FP-B) (15, 34), Tβ-4 (8), PBP (42), CTAP-3 (9), regulated upon activation-normal T-cell expressed and secreted protein (RANTES) (31), and PF-4 (10). Additionally, analysis of carboxypeptidase Y-released amino acids of the C termini of HPAPs 3, 5, and 7 corroborated that the C-terminal sequences are identical to those of Tβ-4, CTAP-3, and PF-4, respectively (Table 1). However, the C-terminal residue Arg14 of HPAP 2, which was anticipated on comparison of amino acid composition and partial sequence of HPAP 2 with FP-B, was not detected by sequence analysis.
Human serum albumin (Fig. 1B, panels I and II, peak A) was identified based on coelution with a human serum albumin standard (Sigma) in RP-HPLC (data not shown) and 10 N-terminal sequence (D-A-H-K-S-E-V-A-H-R) identity with the albumin standard (24). Dioctylphthalate (Fig. 1B, panel III, peak D) was active against bacteria and was identified by fast-atom bombardment mass spectrometry and coelution with a known standard in RP-HPLC (data not shown). This compound is used as a plasticizer in plastic bags for storing human platelets; similar results were reported by Racz et al. (30).
Antimicrobial activities of purified HPAPs.
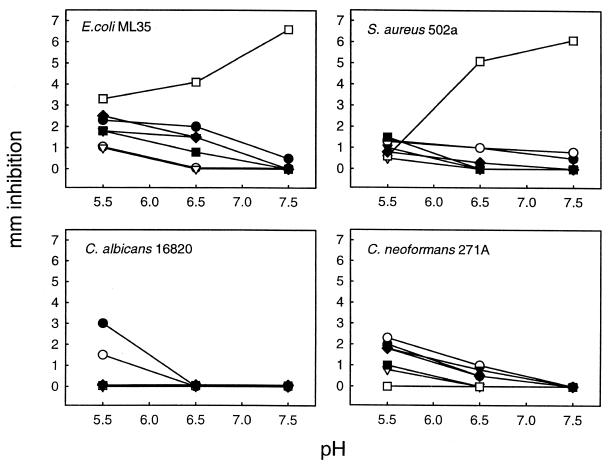
The antimicrobial activity of each HPAP was evaluated against E. coli ML35, S. aureus 502A, C. albicans 16820, and C. neoformans 271A by use of a radial diffusion agarose plate assay (23). This assay was used to analyze the effect of pH and peptide concentration on antimicrobial activities. The amount of peptide applied to each well was 100 nmol/ml (150 μg to 1.0 mg per ml, depending on the molecular weight of each peptide) and pH values tested were 5.5, 6.5, and 7.5. As shown in Fig. 3, the majority of HPAPs were more active against the test organisms in slightly acidic media (e.g., pH 5.5); however, Tβ-4 exhibited more antibacterial activity under slightly alkaline conditions (e.g., pH 7.5). Compared to the other three test organisms, C. albicans was generally less sensitive to most HPAPs; only RANTES (HPAP 6) and PF-4 (HPAP 7) exhibited anticandidal activity in this assay at pH 5.5.
FIG. 3.
Antimicrobial activities of HPAPs as influenced by pH. Purified peptides (0.5 nmol) were pipetted into wells produced in agarose-glucose plates preseeded with corresponding organisms. The plates were buffered with MES (pH 5.5 or 6.5) or PIPES (pH 7.5). After incubation, antimicrobial activity was assessed by measurement of the diameters of the clear zones of inhibition (in millimeters). ▵, FP-A; ▪, FP-B; □, Tβ-4; ▾, PBP; ▿, CTAP-3; •, RANTES; ○, PF-4.
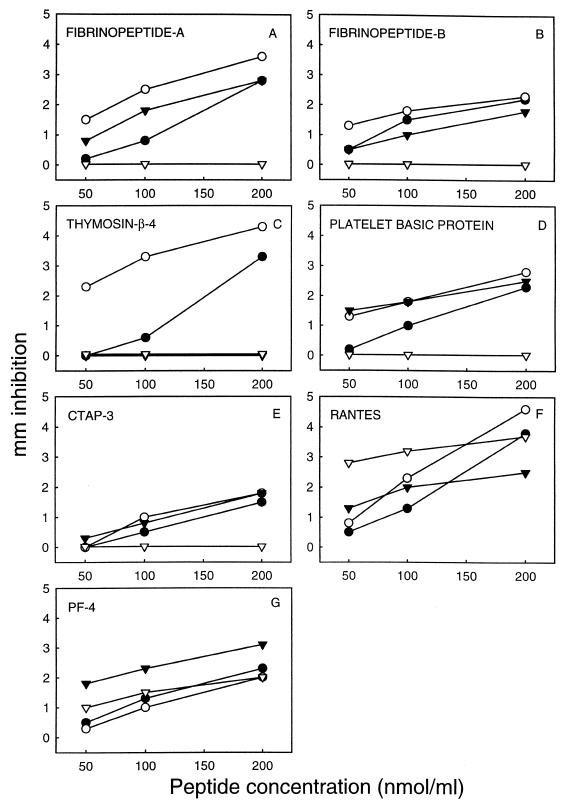
Similar experiments were conducted with various peptide concentrations from 50 to 200 nmol/ml at pH 5.5. In these studies, FP-A, FP-B, and CTAP-3 exhibited a direct relationship between increasing concentration and inhibition of E. coli, S. aureus, and C. neoformans (Fig. 4A, B, and E). However, these HPAPs failed to demonstrate any detectable inhibitory effect against C. albicans under these conditions. Similarly, Tβ-4 exerted a dose-related pattern of inhibition of bacterial pathogens (E. coli and S. aureus) but lacked detectable activity against either fungal pathogen (Fig. 4C). In contrast, RANTES and PF-4 exhibited dose-related activities against all organisms tested (Fig. 4F and G). The antibacterial activities of most of the HPAPs were greater against E. coli than S. aureus under these assay conditions; the converse was true for PF-4. Alternatively, the antifungal activities of most HPAPs were greater against C. neoformans than C. albicans; however, RANTES was more active against the latter than the former.
FIG. 4.
Dose-dependent antimicrobial activities of HPAPs. Purified peptides FP-A (A), FP-B (B), Tβ-4 (C), PBP (D), CTAP-3 (E), RANTES (F), and PF-4 (G) in the concentration ranges indicated were loaded into wells of MES-buffered (pH 5.5) glucose-agarose plates preseeded with specific organisms. ○, E. coli ML35; •, S. aureus 502A; ▿, C. albicans 16820; ▾, C. neoformans 271A. Other conditions were identical to those described for Fig. 3.
Sodium phosphate buffer (10 mM, pH 7.4) has been used widely in radial diffusion antimicrobial assays (23). The crude platelet acid extract, and Bio-Gel P-60 column fractions thereof, exhibited activity against bacteria tested in this system (data not shown). However, after purification, PF-4 and CTAP-3 exhibited very weak activities against bacteria under these conditions, and a precipitate was observed around wells containing these peptides in the assay plates. These results suggested that the solubility of the purified peptides may negatively influence their antimicrobial activity in the phosphate-buffered TSB-agarose system used for our preliminary bioassays (data not shown). In contrast, all HPAPs were highly soluble in the biological buffers MES or PIPES. Peptides resuspended in these buffers exhibited correspondingly greater activities against bacteria (in MES at pH 5.5 or in PIPES at pH 7.5) (Fig. 3 and 4) than when suspended in phosphate buffer. Furthermore, we observed that TSB or SDB inhibited the antimicrobial activity of purified PF-4 (data not shown), and others have found a similar inhibition of various cationic peptides (16). Thus, our studies employed glucose rather than TSB or SDB as the nutrient source in microbe-containing underlay agars.
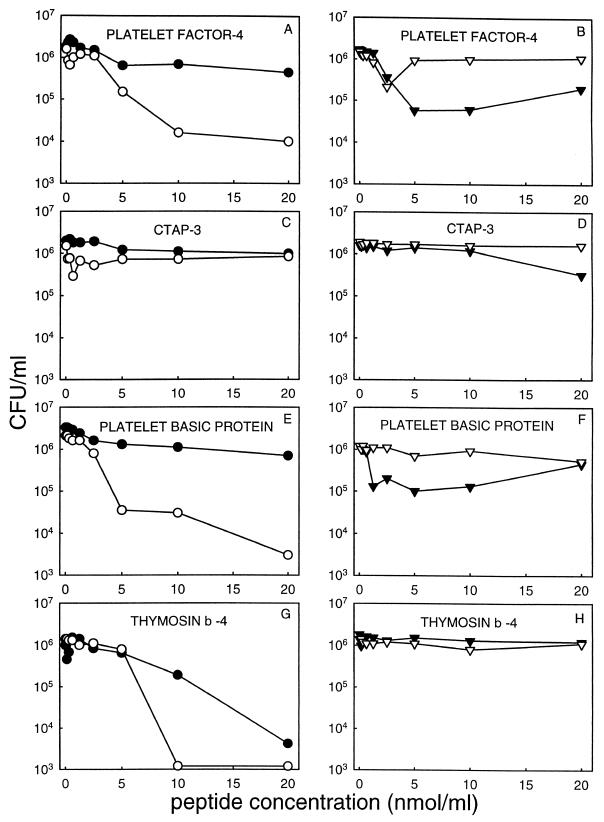
A solution-phase assay was also used to evaluate the effect of peptide concentration on microbicidal activities against four organisms. Peptide concentrations ranged from 0 to 20 nmol per ml, in sodium acetate (pH 5.5), MES (pH 5.5), or PIPES (pH 7.5) buffers. As shown in Fig. 5, it is notable that PF-4, PBP, and Tβ-4 had considerably greater activity against E. coli than against S. aureus and generally greater activity against bacteria than fungi under these test conditions. Moreover, these peptides exhibited substantially greater antimicrobial activities than those of CTAP-3 under these conditions. In PIPES buffer (pH 7.5), Tβ-4 caused a reduction of 2 to 3 log10 CFU/ml in S. aureus or E. coli, respectively (Fig. 5G). However, there was no killing activity against C. albicans or C. neoformans observed for this peptide under this assay condition (Fig. 5H). The acidic buffer MES was also used as described above to examine microbicidal activities of Tβ-4 against these selected organisms. Our results indicated that there was no significant killing of any organism by this peptide under such test conditions (data not shown). These results are consistent with our observation that Tβ-4 is more active in nonacidic buffer in the radial diffusion assay (Fig. 3).
FIG. 5.
Microbicidal activity of PF-4, CTAP-3, PBP, and Tβ-4. Two million CFU of the indicated organism per milliliter was incubated with 0 to 20 nmol of peptide per ml in 50 μl (final volume) of MES buffer (2 mM, pH 5.5) for PF-4 (A and B), CTAP-3 (C and D), and PBP (E and F) or PIPES buffer (10 mM, pH 7.50) for Tβ-4 (G and H). After 1.0 h of incubation at 37°C, samples of each incubation were serially diluted in 10 mM sodium phosphate buffer (pH 7.4) and plated in duplicate on nutrient agar plates. Surviving CFU were enumerated following incubation (24 to 48 h) at 37°C. ○, E. coli ML35; •, S. aureus 502A; ▿, C. albicans 16820; ▾, C. neoformans 271A.
Interestingly, our results revealed that PF-4 exhibited a bimodal antifungal effect against C. albicans (Fig. 5B). At concentrations of less than 5 nmol/ml, PF-4 caused a reduction of ∼0.8 log10 CFU/ml. However, at concentrations exceeding 5 nmol/ml, PF-4 failed to exert significant anticandidal activity. This dose-response relationship was highly reproducible (n = 3) but was observed only for the bioassays of PF-4 against C. albicans.
As shown in Fig. 4 and 5, purified HPAPs were generally more potent in the microbicidal assay than in the agar diffusion assay. For example, in the solution-phase microbicidal assay, we detected a decrease of >1 log10 CFU/ml of E. coli, C. albicans, and C. neoformans at concentrations of ≤5 nmol of PF-4 per ml (Fig. 5A and B). However, in the agar diffusion assay, we first observed antimicrobial activities of PF-4 at a concentration of 50 nmol/ml (Fig. 4G), which was 10 times higher than in the microbicidal assay. Moreover, the antimicrobial activities of HPAPs were generally more active in 2 mM than in 10 mM MES or sodium acetate buffer (data not shown).
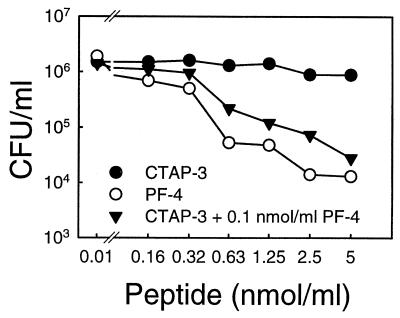
In addition to testing HPAPs for individual antimicrobial potencies and spectra, we examined the potential for combined activities of sublethal concentrations of PF-4 and CTAP-3 against E. coli ML-35. As shown in Fig. 6, the addition of increasing amounts of CTAP-3 to a constant concentration of PF-4 (0.1 nmol/ml) resulted in progressively increased bactericidal activity over a 60-min period. The resulting colicidal activity of this peptide combination (≤0.3 nmol/ml and 0.1nmol/ml for CTAP-3 and PF-4, respectively) occurred despite a lack of such activity for either peptide alone at these concentrations (Fig. 6). However, this synergistic interaction may be pH and buffer dependent, since the crude thrombin-induced platelet supernatant (equivalent to ∼107 platelets) had no detectable killing activity against E. coli or S. aureus at pH 7.3 in the solution-phase microbicidal assay. Moreover, no synergistic effect of these two peptides against E. coli was observed in MES buffer (2 mM, pH 5.5; data not shown).
FIG. 6.
Microbicidal activity of PF-4 and CTAP-3 in combination. Assay conditions were identical to those described for Fig. 5A, except that sodium acetate buffer (2 mM, pH 5.5) was used. Dose-dependent killing of E. coli by PF-4 (○) and CTAP-3 (•) and the synergistic effect of CTAP-3 and PF-4 (▾) are shown. Synergy was demonstrated in incubations containing increasing concentrations of CTAP-3 supplemented with 0.1 nmol of PF-4 per ml. Each data point represents the mean of two independent experiments.
DISCUSSION
Platelets have multiple functional attributes suggestive of an integral role in antimicrobial host defense (47, 48). These functions include navigation toward the inflammatory chemoattractant N-f-MetLeuPhe, expression of Fc and complement C3a/C5a receptors, and the capacity to generate antimicrobial oxygen metabolites including superoxide, hydrogen peroxide, and hydroxyl free radicals. Moreover, platelets interact directly with microorganisms, contribute to clearance of pathogens from the bloodstream, and significantly participate in antibody-dependent cell cytotoxicity against microbial pathogens (47, 48). Yet, the molecules and mechanisms contributing to the antimicrobial effects of platelets have not been fully defined.
In the present study, we identified seven HPAPs as FP-A, FP-B, Tβ-4, PBP, CTAP-3, RANTES, and PF-4 isolated from the released material of human platelets following thrombin stimulation. Each of these proteins has been structurally characterized previously. However, our prior studies [Tang et al., Blood 86(Suppl. 1):556a and 910a, 1995] and present findings demonstrated the direct antimicrobial properties of this group of human platelet proteins. Additionally, our data indicate that two of the predominant HPAPs, PF-4 and CTAP-3, interact synergistically to effect microbicidal activity in vitro.
The data presented demonstrate that thrombin stimulates human platelets to release HPAPs. Human PF-4, RANTES, PBP, and CTAP-3 have previously been shown to be released from platelets upon thrombin stimulation (20, 21, 26, 39). Holt et al. (20) showed that 1 mg of CTAP-3 (also termed low-affinity PF-4) and 0.1 mg of PBP were recovered from 1,600 ml of human blood, respectively. However, we are unaware of studies that have quantified thrombin-induced release of PF-4 and RANTES from human platelets. Moreover, while Tβ-4 has been isolated from total extracts of human platelets (17), we believe that this is the first study to show that Tβ-4 is released from human platelets upon thrombin stimulation. Two HPAPs, FP-A and FP-B, were unrecoverable from total protein extracts of platelets, suggesting that these HPAPs are generated as a result of thrombin stimulation of platelets. It is conceivable that these HPAPs may be produced by thrombin-mediated proteolysis of fibrinogen which is secreted by human platelets (15, 19). Consistent with this hypothesis is the fact that both the α- and β-chains of human fibrinogen have thrombin-cleavable Arg-Gly motifs at their N termini (15).
The above findings are relevant to the potential roles of platelets in antimicrobial host defense (47, 48). Platelets respond chemotactically to soluble signals generated by complement fixation or to microorganisms themselves. Additionally, thrombin stimulation increases platelet adherence to injured or infected vascular endothelial cells, promoting platelet accumulation at these sites (3). At sites of damaged or infected vascular endothelium, adherent platelets may be induced to release antimicrobial polypeptides where they may contribute to host defense against microbial colonization and/or invasion.
Although the physiologic relevance of thrombin-mediated HPAP release in host defense is not yet proven, several studies suggest that thrombin and platelets play a role in preventing and/or limiting infection. For example, thrombocytopenia has been shown to put experimental animals (37) and humans (5, 38) at significantly increased risk of infection. Furthermore, thrombin participates in key cellular events that regulate inflammatory responses (28, 40, 47, 48). For example, platelets and neutrophils, both involved in wound healing, are stimulated by thrombin and other soluble mediators generated in response to tissue injury or infection. In turn, platelets recognize and interact with vascular endothelial cells injured due to infecting pathogens and recruit and facilitate adhesion of neutrophils and other leukocytes to these sites (11, 21, 28, 47, 48). Frohm et al. (14) reported that Tβ-4 and other antimicrobial peptides are detectable within human wound and blister fluid.
The varying abundance, antimicrobial potencies and spectra, and conditional optima for antimicrobial activity suggest complementary roles for HPAPs in antimicrobial host defense. PF-4 exerted the broadest spectrum of activity against pathogens studied, with antimicrobial activity against all organisms tested (Fig. 4). Similarly, RANTES was active against the four organisms tested but was much less abundant than PF-4. CTAP-3 was the most abundant HPAP, but its antimicrobial potency was substantially less than those of PF-4 or RANTES. The relative amounts of the other HPAPs in either the thrombin-induced released material or acid extract were substantially less than CTAP-3 or PF-4.
Many of the HPAPs identified appear to have different conditional optima for antimicrobial activity. For example, we observed that Tβ-4, an anionic peptide, exhibited an antimicrobial optimum at slightly alkaline pH. This differs from results obtained for the cationic HPAPs, or anionic FP-A and FP-B, each of which exerted greater antimicrobial activities and spectra under slightly acidic conditions. Paralleling these findings, CTAP-3 has been reported to be significantly more active in tissue-remodeling functions than its precursor, PBP (40). In contrast, PBP exerts significantly greater antimicrobial activity than its derivative, CTAP-3. Interestingly, Krijgsfeld et al. did not detect any bactericidal activity of purified PF-4 or CTAP-3 against E. coli or S. aureus in a phosphate-buffered system (pH 7) (22). Our studies, however, demonstrate that both of these peptides exhibit significant or potent antimicrobial activities under slightly acidic buffer conditions. We speculate that our results differ from those of the previous study (22) due to the distinct buffer and pH conditions employed. In this regard, we have observed that sodium phosphate markedly inhibits the antimicrobial properties of several antimicrobial peptides (unpublished data).
We found that PF-4 exhibited a unique concentration-microbicidal activity relationship. For example, PF-4 appeared to have a concentration optimum of ≤5 nmol/ml for direct microbicidal activity (Fig. 5B). Mayo and Chen (25) demonstrated that human PF-4 exists in a monomer-dimer-trimer equilibrium in solution. Decreases in peptide concentration, pH, or ionic strength in the solution shift the equilibrium to favor a monomeric form of PF-4, while the opposite conditions strongly favor a tetrameric configuration. We hypothesize that monomeric PF-4 exerts greatest microbicidal activity, with diminishing potency as the peptide concentration increases to favor a multimeric state(s) in solution.
The inflammatory microenvironment, containing adherent, activated platelets, may be ideal for HPAP-mediated antimicrobial action in vivo. For example, the pH of leukocyte phagolysosomes descends to 4.5 to 6.0 subsequent to phagocytosis of microorganisms (36). Likewise, abscess exudate, serum, and interstitial fluid in the setting of an inflammatory response may reach acidic pH values following oxidative burst responses of leukocytes (35, 36, 39, 43). All but one of the HPAPs exert greatest activity at slightly acidic pH compared to alkaline conditions. Similarly, the in vitro antibacterial activities of 37K CAP, a peptide isolated from human neutrophil granules, are enhanced under acidic (pH 5.5) compared with alkaline conditions (35). Likewise, magainin-2, an antimicrobial peptide found in frog skin secretions, exerts greater in vitro bactericidal activity at acidic pH than in neutral or alkaline environments (2). For comparison, the antibacterial effects of rabbit neutrophil α-defensins are also significantly influenced by pH conditions. However, in contrast to most HPAPs, the α-defensins exhibit optimal bactericidal activities at pHs ranging from 7 to 8 and relatively little antimicrobial activity at pH 5.8 (32). Bessalle et al. (2) recently proposed that pH influences the mode of peptide-target cell interaction, presumably by influencing conformational properties of the peptide and/or target ligand.
Shafer et al. (35) and Yeaman et al. (47, 48) have suggested that certain antimicrobial peptides, including microbicidal proteins from platelets, potentiate the antimicrobial mechanisms of leukocytes. For example, peptides that are active under mildly acidic conditions may enhance the ability of leukocytes to kill microbial pathogens through nonoxidative mechanisms, particularly as the maturing phagolysosome becomes acidified. Thus, acidic pH may amplify the direct microbicidal potencies of HPAPs and/or potentiate the antimicrobial mechanisms of leukocytes following phagocytosis of organisms previously exposed to these peptides.
Consistent with this hypothesis is the fact that many of the HPAPs identified are members of the intercrine family of chemokines (29). Thus, HPAPs and related antimicrobial peptides from platelets may recruit leukocytes to sites of infection and potentiate the antimicrobial mechanisms of these cells (11, 21, 28, 47, 48). Consistent with this concept, Cocchi et al. (6) recently reported that RANTES, a β-chemokine, plays a role in suppression of human immunodeficiency virus proliferation or pathogenesis via modulation of T-cell function and/or direct antiviral effects. Similarly, Cole et al. recently reported that some ELR-CXC chemokines are directly antibacterial (7). Thus, considering that they are released from platelets by thrombin, it is possible that HPAPs contribute two key mechanisms to antimicrobial host defense, by (i) directly inhibiting or killing pathogens where platelets accumulate at sites of microbial colonization or infection and (ii) recruiting and amplifying leukocyte and/or lymphocyte antimicrobial responses in these settings.
Acknowledgments
This research was supported in part by funds from Large Scale Biology (M.E.S.) and by National Institutes of Health grants AI22931 to M.E.S. and AI39001 and AI48031 to M.R.Y.
We thank Patti Tran and Dat Tran, Jack Edwards, and Arnie Bayer for their assistance with various aspects of this project. We are also grateful to Agnes H. Henschen and Aftab Ahmed for expert assistance in peptide sequence analysis. We thank John Greaves for identifying dioctylphthalate and Kym Faull (UCLA) for mass spectroscopic analysis.
Editor: V. J. DiRita
Footnotes
For a commentary on this article, see page 6515 in this issue.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bessalle, R., H. Haas, A. Goria, I. Shalit, and M. Fridkin. 1992. Augmentation of the antibacterial activity of magainin by positive-charge chain extension. Antimicrob. Agents Chemother. 36:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carney, D. H. 1992. Postclotting cellular effects of thrombin mediated by interaction with high-affinity thrombin receptors, p. 351-370. In L. J. Berliner (ed.), Thrombin: structure and function. Plenum Press, New York, N.Y.
- 4.Carroll, S. F., and R. J. Martinez. 1981. Antibacterial peptide from normal rabbit serum. 1. Isolation from whole serum, activity, and microbicidal spectrum. Biochemistry 20:5973-5981. [DOI] [PubMed] [Google Scholar]
- 5.Chang, F. Y., N. Singh, T. Gayowski, M. M. Wagener, S. M. Mietzner, J. E. Stout, and I. G. Marino. 2000. Thrombocytopenia in liver transplant recipients: predictors, impact on fungal infections, and role of endogenous thrombopoietin. Transplantation 69:70-75. [DOI] [PubMed] [Google Scholar]
- 6.Cocchi, F., A. L. DeVico, A. Garzino-Demo, S. K. Arya, R. C. Gallo, and P. Lusso. 1995. Identification of RANTES, MIP-1α, and MIP-1β as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811-1815. [DOI] [PubMed] [Google Scholar]
- 7.Cole, A. M., T. Ganz, A. M. Liese, M. D. Burdick, L. Liu, and R. M. Strieter. 2001. IFN-inducible ELR-CXC chemokines display defensin-like antimicrobial activity. J. Immunol. 167:623-627. [DOI] [PubMed] [Google Scholar]
- 8.Conlon, J. M., L. Grimelius, G. Wallin, and L. Thim. 1988. Isolation and structural characterization of thymosin-β4 from a human medullary thyroid carcinoma. J. Endocrinol. 118:155-159. [DOI] [PubMed] [Google Scholar]
- 9.Costor, C. W., E. M. Smith, M. C. Bignall, P. A. Hossler, and T. H. Sisson. 1991. Preparation and bioassay of connective tissue activating peptide III and its isoforms. Methods Enzymol. 198:405-416. [DOI] [PubMed] [Google Scholar]
- 10.Deuel, T. F., P. S. Keim, M. Farmer, and R. L. Heinrikson. 1977. Amino acid sequence of platelet factor 4. Proc. Natl. Acad. Sci. USA 74:2256-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diacovo, T. G., K. D. Puri, R. A. Warnock, T. A. Springer, and U. H. von Andrian. 1996. Platelet-mediated lymphocyte delivery to high endothelial venules. Science 273:252-255. [DOI] [PubMed] [Google Scholar]
- 12.Donaldson, D. M., and J. G. Tew. 1977. Beta-lysin of platelet origin. Bacteriol. Rev. 41:501-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fodor, J. 1887. Die Fahigkeit des Blutes. Bakterien zu vernichten. Dtsch. Med. Wochenschr. 13:745-747. [Google Scholar]
- 14.Frohm, M., H. Gunne, A.-C. Bergman, B. Agerberth, T. Bergman, A. Boman, S. Lidén, H. Jörnvall, and H. G. Boman. 1996. Biochemical and antibacterial analysis of human wound and blister fluid. Eur. J. Biochem. 237:86-92. [DOI] [PubMed] [Google Scholar]
- 15.Furlan, M. 1988. Structure of fibrinogen and fibrin, p. 17-64. In J. L. Francis (ed.), Fibrinogen, fibrin stabilisation, and fibrinolysis. VCH, New York, N.Y.
- 16.Gabay, J. E., R. W. Scott, D. Campanelli, J. Griffith, C. Wilde, M. N. Marra, M. Seeger, and C. F. Nathan. 1989. Antibiotic proteins of human polymorphonuclear leukocytes. Proc. Natl. Acad. Sci. USA 86:5610-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannappel, E., and M. V. Kampen. 1987. Determination of thymosin β4 in human blood cells and serum. J. Chromatogr. 397:279-285. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch, J. G. 1960. Comparative bactericidal activities of blood serum and plasma serum. J. Exp. Med. 112:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmsen, H. 1994. Platelet secretion and energy metabolism, p. 524-545. In R. W. Colman, J. Hirsh, V. J. Marder, and E. V. Salzman (ed.), Hemostasis and thrombosis: basic principles and clinical practice, 3rd ed. J. B. Lippincott Co., Philadelphia, Pa.
- 20.Holt, J. C., M. E. Harris, A. M. Holt, E. Lange, A. Henschen, and S. Niewiarowski. 1986. Characterization of human platelet basic protein, a precursor form of low-affinity platelet factor 4 and β-thromboglobulin. Biochemistry 25:1988-1996. [DOI] [PubMed] [Google Scholar]
- 21.Kameyoshi, Y., A. Dörschner, A. I. Mallet, E. Christophers, and J.-M. Schröder. 1992. Cytokine RANTES released by thrombin-stimulated platelets is a potent attractant for human eosinophils. J. Exp. Med. 176:587-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krijgsveld, J., S. A. J. Zaat, J. Meeldijk, P. A. van Veelen, G. Fang, B. Poolman, E. Brandt, J. E. Ehlert, A. J. Juijpers, G. H. M. Engbers, J. Feijen, and J. Dankert. 2000. Thrombocidins, microbicidal proteins from human blood platelets, are C-terminal deletion products of CXC chemokines. J. Biol. Chem. 275:20374-20381. [DOI] [PubMed] [Google Scholar]
- 23.Lehrer, R. I., M. Rosenman, S. S. L. Harwig, R. Jackson, and P. Eisenhauer. 1991. Ultrasensitive assays for endogenous antimicrobial polypeptides. J. Immunol. Methods 137:167-173. [DOI] [PubMed] [Google Scholar]
- 24.Lichenstein, H. S., D. E. Lyons, M. M. Wurfel, D. A. Johnson, M. D. McGinley, J. C. Leidli, D. B. Trollinger, J. P. Mayer, S. D. Wright, and M. M. Zukowski. 1994. Afamin is a new member of the albumin, alpha-fetoprotein, and vitamin D-binding protein gene family. J. Biol. Chem. 269:18149-18154. [PubMed] [Google Scholar]
- 25.Mayo, K. H., and M. J. Chen. 1989. Human platelet factor 4 monomer-dimer-tetramer equilibria investigated by 1H NMR spectroscopy. Biochemistry 28:9469-9478. [DOI] [PubMed] [Google Scholar]
- 26.Moore, S., D. S. Pepper, and J. D. Cash. 1975. Platelet antiheparin activity. The isolation and characterization of platelet factor 4 released from thrombin-aggregated washed human platelets and its dissociation into subunits and the isolation of membrane-bound antiheparin activity. Biochim. Biophys. Acta 379:370-384. [PubMed] [Google Scholar]
- 27.Mustard, J. F., R. L. Kinlough-Rathbone, and M. A. Packham. 1989. Isolation of human platelets from plasma by centrifugation and washing. Methods Enzymol. 169:3-11. [DOI] [PubMed] [Google Scholar]
- 28.Nachman, R. L., and B. B. Wecksler. 1972. The platelet as an inflammatory cell. Ann. N. Y. Acad. Sci. 201:131-137. [DOI] [PubMed] [Google Scholar]
- 29.Oppenheim, J. J., C. O. C. Zachariae, N. Mukaida, and K. Matsushima. 1991. Properties of the novel proinflammatory supergene “intercrine” cytokine family. Annu. Rev. Immunol. 9:617-648. [DOI] [PubMed] [Google Scholar]
- 30.Racz, Z., J. Pick, K. Baroti, J. Pinter, and J. Szabo. 1993. Blood products stored in plastic bags: release of plasticizers from the bag material. Orv. Hetil. 134:1581-1586. (In Hungarian.) [PubMed] [Google Scholar]
- 31.Schall, T. J., J. Jongstra, B. J. Dyer, J. Jorgensen, C. Clayberger, M. M. Davis, and A. M. Krensky. 1988. A human T cell-specific molecule is a member of a new gene family. J. Immunol. 141:1018-1025. [PubMed] [Google Scholar]
- 32.Selsted, M. E., D. Szklarek, and R. I. Leher. 1984. Purification and antibacterial activity of antimicrobial peptides of rabbit granulocytes. Infect. Immun. 45:150-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selsted, M. E., Y.-Q. Tang, W. L. Morris, P. A. McGuire, M. J. Novotny, W. Smith, A. H. Henschen, and J. S. Cullor. 1993. Purification, primary structures, and antibacterial activities of β-defensins, a new family of peptides from bovine neutrophils. J. Biol. Chem. 268:6641-6648. [PubMed] [Google Scholar]
- 34.Senior, R. M., W. F. Skogen, G. L. Griffin, and G. D. Wilner. 1986. Effects of fibrinogen derivatives upon the inflammatory response. J. Clin. Investig. 77:1014-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shafer, W. M., L. E. Martin, and J. K. Spitznagel. 1986. Late intraphagosomal hydrogen ion concentration favors the in vitro antimicrobial capacity of a 37-kilodalton cationic granule protein of human neutrophil granulocytes. Infect. Immun. 53:651-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spitznagel, J. K. 1984. Non-oxidative antimicrobial reactions of leukocytes. Contemp. Top. Immunobiol. 14:283-343. [DOI] [PubMed] [Google Scholar]
- 37.Sullam, P. M., U. Frank, M. R. Yeaman, M. G. Tauber, A. S. Bayer, and H. F. Chambers. 1993. Effect of thrombocytopenia on the early course of streptococcal endocarditis. J. Infect. Dis. 168:910-914. [DOI] [PubMed] [Google Scholar]
- 38.Viscoli, C., P. Bruzzi, E. Castagnola, L. Boni, T. Calandra, H. Gaya, F. Meunier, R. Feld, S. Zinner, J. Klastersky, M. Glauser, and the International Antimicrobial Therapy Group (IATCG) of the European Organization for Research and Treatment of Cancer (EORTC). 1994. Factors associated with bacteraemia in febrile, granulocytopenic cancer patients. Eur. J. Cancer 30:430-437. [DOI] [PubMed] [Google Scholar]
- 39.Walz, A., B. Dewald, V. von Tscharner, and M. Baggiolini. 1989. Effects of neutrophil-activating peptide NAP-2, platelet basic protein, connective tissue-activating peptide III, and platelet factor 4 on human neutrophils. J. Exp. Med. 170:1745-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wecksler, B. B. 1992. Platelets, p. 727-746. In J. I. Gallin, I. M. Goldstein, and R. Snyderman (ed.), Inflammation: basic principles and clinical correlation, 2nd ed. Reven Press, Ltd., New York, N.Y.
- 41.Wecksler, B. B., and R. L. Nachman. 1971. Rabbit platelet bactericidal protein. J. Exp. Med. 134:1114-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wenger, R. H., A. N. Wicki, A. Walz, N. Kieffer, and K. J. Clemetson. 1989. Cloning of cDNA coding for connective tissue activating peptide III from a human platelet-derived λgt11 expression library. Blood 73:1498-1503. [PubMed] [Google Scholar]
- 43.Wright, J., J. H. Schwartz, R. Olson, J. M. Kosowsky, and A. I. Tauber. 1986. Proton secretion by the sodium/hydrogen ion antiporte in the human neutrophil. J. Clin. Investig. 77:782-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeaman, M. R., A. S. Ibrahim, J. E. Edwards, Jr., A. S. Bayer, and M. A. Ghannoum. 1993. Thrombin-induced rabbit platelet microbicidal protein is fungicidal in vitro. Antimicrob. Agents Chemother. 37:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeaman, M. R., S. M. Puentes, D. C. Norman, and A. S. Bayer. 1992. Partial characterization and staphylocidal activity of thrombin-induced platelet protein. Infect. Immun. 60:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeaman, M. R., Y.-Q. Tang, A. J. Shen, A. S. Bayer, and M. E. Selsted. 1997. Purification and in vitro activities of rabbit platelet microbicidal proteins. Infect. Immun. 65:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeaman, M. R. 1997. The role of platelets in antimicrobial host defense. Clin. Infect. Dis. 25:951-970. [DOI] [PubMed] [Google Scholar]
- 48.Yeaman, M. R., and A. S. Bayer. 1999. Antimicrobial peptides from platelets. Drug Resist. Updates 2:116-126. [DOI] [PubMed] [Google Scholar]
- 49.Zaat, S. A. J., P. S. Hiemstra, and J. Dankert. 1994. Initial characterization of antibacterial proteins from thrombin-stimulated platelets involved in clearance of Streptococcus sanguis from cardiac vegetation in experimental endocarditis, p. 473-475. In A. Totolian (ed.), Pathogenic streptococci, present and future. Lancer Publication, St. Petersburg, Russia.