Abstract
Borrelia burgdorferi B31MI carries 18 plasmid-carried genes that form the bdr gene family. The bdr genes of B. burgdorferi encode proteins that form three distinct subfamilies, the BdrD, BdrE, and BdrF subfamilies. bdr orthologs have been demonstrated to be carried by all Borrelia species analyzed, and their widespread distribution suggests that they play an important genus-wide functional role. The biological rationale for maintaining 18 bdr alleles has not been defined. It is our hypothesis that specific paralogs function in different environments and are differentially expressed in response to environmental conditions. As a first step in testing this hypothesis, the production patterns of the Bdr proteins in spirochetes grown under a variety of conditions were assessed through immunoblot analyses. The influence of temperature, serum deprivation, tick feeding, and the mammalian environment on Bdr production was evaluated. These analyses revealed that the synthesis of some Bdr paralogs is environmentally regulated. The production of BdrF2, BdrF1, BdrE4, and BdrE5 were upregulated in host-adapted bacteria, while the production levels of other Bdr paralogs were influenced by temperature and serum starvation. These observations suggest that different Bdr paralogs function in different biological environments and provide insight into the biological basis for maintaining multiple members of this gene family.
The plasmid component of the Borrelia genome exhibits extensive genetic redundancy, with 70% of the plasmid-carried open reading frames belonging to paralogous gene families (13). The biological rationale for maintaining this redundancy, which is an energetically expensive process, is unclear. The existence of multiple copies of a given gene with variations in promoter sequences in the genome may allow for the differential regulation of individual gene family members. In view of the different environments encountered by Borrelia organisms during their natural enzootic cycles, differential gene expression may function to ensure that only genes necessary for survival in a given environment are expressed. Several borrelial genes have been shown to be temporally expressed during infection or differentially expressed in different environments (1, 10, 16, 20, 26, 27). Microarray analyses have demonstrated that a significant fraction of the Borrelia genome is differentially expressed in response to changing environmental conditions (23). However, microarray data pertaining to Borrelia gene families are difficult to interpret due to cross hybridization. Hence, less global approaches are required to assess the expression of individual members of gene families. In addition, while microarray analyses provide a snapshot of what is occurring at the transcriptional level, they do not necessarily provide an accurate picture of protein levels. The determination of production levels of individual members of protein families under different environmental conditions will provide significant insight into defining the biological niches in which individual paralogs carry out their biological functions.
In Borrelia burgdorferi B31MIpc (clone pc of strain B31MI), the bdr gene family is comprised of 18 members (13) that form three distinct subfamilies, the bdrD, bdrE, and bdrF subfamilies (5, 6, 24, 32, 33). The Bdr proteins are a highly polymorphic group of inner membrane-localized proteins. Interaction with the inner membrane occurs through a hydrophobic C-terminal membrane-spanning domain (25). While the sequence of this domain varies among the Bdr subfamilies, its overall properties are conserved. In B. burgdorferi B31MIpc, the Bdr proteins range in size from 20 to 30.6 kDa. Differences in the number of repeat motifs are largely responsible for the size differences among paralogs, and it is this feature that led to these proteins being designated Bdr proteins (for Borrelia direct repeat) (33). A revised Bdr nomenclature system has recently been developed that is applicable at the genus-wide level (6). While the function of the Bdr proteins has yet to be determined, it is important to note that the central repeat domain harbors a series of putative serine-threonine phosphorylation motifs, suggesting a possible role in cell regulation or signaling. However, it remains to be demonstrated if and under what conditions phosphorylation occurs. Database analyses have revealed that the Bdr proteins are unique to Borrelia organisms, and hybridization and immunological analyses have demonstrated that multiple bdr genes are carried and expressed by all Borrelia species and isolates (6, 7, 22, 24, 30). All bdr genes have been demonstrated to be plasmid encoded (reviewed in reference 24). In B. burgdorferi, all bdrD and bdrE genes (except bdrD5 and bdrE6) are carried by the cp32 family of plasmids, while the bdrF genes are present solely on linear plasmids (9). The bdrA, bdrB, and bdrC subfamilies of the relapsing fever spirochetes are located predominantly on linear plasmids (7). In this study, we assessed the influence of environmental conditions on Bdr production at the paralog or subfamily level and demonstrate that the synthesis of some members of the Bdr protein family is responsive to environmental changes.
MATERIALS AND METHODS
Bacterial cultivation under different environmental conditions.
In this report, we have focused our efforts on Bdr production patterns in clones or mutants of B. burgdorferi 297 and B. burgdorferi B31MIpc. The genetic composition of B. burgdorferi B31MIpc has been defined through genomic sequencing (13), and the plasmids carried by strain B31MIpc and its derivative clones has been determined through hybridization and PCR analyses (19) (Table 1). Spirochetes were cultivated in BSK-H complete medium (Sigma-Aldrich, St. Louis, Mo.) supplemented to 12% with rabbit sera (Sigma) at either 23, 33, or 37°C. In addition, cultures of B. burgdorferi 297 in which the rpoN or rpoS gene was inactivated were also employed to assess Bdr production. The influence of the presence or absence of sera on Bdr production was assessed as previously described (3). Briefly, mid-log-phase cultures were pelleted by centrifugation and gently resuspended in either RPMI (Gibco-BRL) or BSK-H incomplete media (Sigma-Aldrich) lacking rabbit sera. These cultures were maintained at 33°C for 0, 24, or 48 h, and then the cells were harvested for analysis.
TABLE 1.
Summary of the plasmid profiles of postinfection clonal populations of B. burgdorferi B31MI
| Plasmida |
B. burgdorferi cloneb
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pc | c8 | c9 | c14 | c15 | c17 | c24 | c29 | c36 | c37 | |
| lp28-1 (F) | + | ND | + | + | ND | + | + | + | + | + |
| lp28-2 (G) | + | ND | + | + | ND | + | +/− | − | − | + |
| lp28-3 (H) | + | ND | − | − | ND | − | − | − | − | − |
| lp56 | + | ND | + | +/− | ND | + | + | + | + | + |
| cp32-1 (P) | + | ND | + | + | ND | + | + | + | + | + |
| cp32-3 (S) | + | ND | + | + | ND | + | + | + | + | + |
| cp32-4 (R) | + | ND | + | + | ND | + | + | + | + | + |
| cp32-6 (M) | + | ND | − | + | ND | + | − | + | + | + |
| cp32-7 (O) | + | ND | + | + | ND | + | + | − | + | + |
| cp32-8 (L) | + | ND | + | + | ND | + | + | + | + | + |
| cp32-9 (N) | + | ND | − | − | ND | − | − | + | − | − |
The letters in parentheses indicate letter designations assigned to plasmid by the Institute for Genomic Research.
The presence (+) or absence (−) of the plasmid is shown. ND, not determined; +/−, weak amplification of target sequence.
Infection of Ixodes scapularis ticks and C3H-HeJ mice with B. burgdorferi B31MIpc.
Larval-stage Ixodes scapularis ticks were infected by letting them feed on mice that had been needle inoculated with B. burgdorferi B31MIpc as previously described (28). The ticks were allowed to feed to repletion and collected. The ticks were then stored at 23°C in a desiccator containing a saturated solution of potassium sulfate to maintain 95% humidity. These ticks were then used to directly infect additional mice. The ticks were placed in a cutoff Eppendorf tube that was secured between the shoulder blades of the mice using bee's wax. The top of the tube was cut off and covered with mesh to allow airflow. All applied ticks were allowed to feed to repletion and were recovered. These ticks were used 3 days postrepletion as described below. Infection of the mice was confirmed by cultivation of ear punch biopsies.
Cultivation of B. burgdorferi within implanted dialysis membrane chambers.
To obtain B. burgdorferi in a mammalian host-adapted state, a modified version of the dialysis membrane chamber implant model developed by Akins et al. (2) was employed. Briefly, Spectra/Por 6 membrane (molecular size cutoff, 5 kDa; Fisher Scientific) was sterilized by boiling in 1 mM EDTA and then soaked in purified water (20 min). The dialysis bags were submerged in BSK-H medium containing Borrelia antibiotic cocktail (Sigma-Aldrich). To prepare the dialysis membrane chambers, a section of dialysis tubing was tied at one end and filled with 5 ml of an actively growing culture of B. burgdorferi B31MIpc or B. burgdorferi 297 (103 spirochetes ml−1) in BSK-H medium. Prior to the procedure, the Borrelia cultures were maintained in BSK-H medium at 23°C. The tubing was then tied, and excess membrane was removed from both ends. Two dialysis membrane chambers (DMCs) were implanted into the peritoneal cavity of a rat using strict aseptic technique. The chambers were explanted 8 to 12 days later and rinsed with BSK-H medium, and their contents were removed using a syringe (18-gauge needle). The bacteria were collected by centrifugation (8,000 × g for 20 min), washed twice with phosphate-buffered saline (PBS) (pH 7.4), and resuspended in PBS.
Generation of Bdr peptides and antipeptide antisera.
Bdr subfamily- or paralog-specific peptides were designed on the basis of earlier analyses of Bdr sequences that demonstrated that these subfamilies can be differentiated on the basis of the sequences of their N-terminal domains. The peptides were designed on the basis of sequences that were either absolutely unique to a given paralog or subfamily. In addition, a scan of the B. burgdorferi genome sequence revealed that the peptide sequences were unique to the Bdr proteins. Prior to their synthesis, each potential peptide sequence was analyzed using the PepTool program to assess its predictive antigenicity. The peptides (Table 2) were synthesized using a new method for peptide synthesis (A. Holm, patent pending) and were coupled to the OVA323-339 T-cell epitope. Sequences homologous to this T-cell epitope sequence are not present in the Borrelia genome. To generate antisera to the peptides, each peptide (50 μg) was injected into C3H-HeJ mice (three mice per set) in combination with either incomplete Freund's adjuvant and alum or montanide. Two booster doses were administered at 2-week intervals. Sera were collected by snipping the tail, and antibody (Ab) titers were assessed by an enzyme-linked immunosorbent assay.
TABLE 2.
Bdr peptide sequences used to generate specific antisera
| Peptide | Peptide sequence | Notes |
|---|---|---|
| BdrD | CSSEEAIDFVFLHNDN | The peptide sequence represents a conserved segment shared by BdrD subfamily members. |
| BdrE | CMETVSTNIASVTQE | This peptide sequence represents a conserved segment shared by BdrE subfamily members. |
| BdrF3 | AVLATTNITEDQIYR | This peptide represents a unique sequence within the BdrF3 protein. |
| BdrF1 | CSMEQLIAQDLSKRYYY | This peptide represents a unique sequence within the BdrF1 protein |
| OVA323-339 | ISQAVHAAHAEINEAGR | T-cell epitope conjugated to each peptide. |
Electrophoresis and immunoblot analyses.
Cells were resuspended in sodium dodecyl sulfate (SDS) solubilizing buffer and boiled. The samples (2 × 106 cells per lane as determined by spectrophotometry) were then resolved in 15% polyacrylamide gels using SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The fractionated proteins were transferred onto Immobilon P membranes by electroblotting, and immunoblot analyses were performed as previously described (24). The antipeptide antisera were used at dilutions of 1:200, and the anti-Bdr antiserum was used at a dilution of 1:1,000. An immuno-pure goat anti-mouse immunoglobulin G (IgG) secondary Ab conjugated to peroxidase was used at a dilution of 1:40,000 to 1:60,000. Detection was by chemiluminescence using the Supersignal West Pico stable peroxide solution and the Supersignal West Pico Luminol-enhancer (Pierce). In some cases, the immunoblot signals were quantitated using a Alpha Innotech Imager (Alpha Innotech Corp.). To facilitate interpretation of the immunoblot data, the properties and other relevant features of the Bdr proteins of B. burgdorferi B31MIpc are described in Table 3.
TABLE 3.
Description of the Bdr proteins of B. burgdorferi B31MI and summary of production data
| Bdr paraloga | Massb | Plasmid | Bdr protein productionc under various conditions
|
|||||
|---|---|---|---|---|---|---|---|---|
| Temp (°C)
|
HA | Serum deprivation for:
|
||||||
| 23 | 37 | 0 h | 24 h | 48 h | ||||
| BdrD1 (BBL35) | 22.1 | cp32-8 | + | + | + | + | + | + |
| BdrD2 (BBM34) | 25.4 | cp32-6 | + | + | + | + | + | + |
| BdrD3 (BBO34) | 22.0 | cp32-7 | + | + | + | + | + | + |
| BdrD4 (BBP34) | 24.1 | cp32-1 | + | + | + | + | + | + |
| BdrD5 (BBQ42) | 20.6 | lp56 | + | + | + | + | + | + |
| BdrD6 (BBS37) | 22.7 | cp32-3 | + | + | + | + | + | + |
| BdrD10 (BBN34) | 20.7 | cp32-9 | + | + | + | + | + | + |
| BdrE1 (BBL27) | 22.4 | cp32-8 | + | + | + | + | ↓1.5[0] | ↓1.5[0] |
| BdrE2 (BBN27) | 22.3 | cp32-9 | − | − | + | − | + | ++ |
| BdrE3 (BBO27) | 23.7 | cp32-7 | + | ↓1.5(23) | ↑2.5(37) | + | ↓1.5[0] | ↓2.0[0] |
| BdrE4 (BBR27) | 20.3 | cp32-4 | + | + | ↑4.4(37) | + | ↓1.5[0] | ↓1.5[0] |
| BdrE5 (BBS29) | 24.1 | cp32-3 | + | ↓4.8(23) | ↑6.4(37) | + | ↓1.5[0] | ↓1.5[0] |
| BdrE6 (BBQ34) | 27.3 | lp56 | + | ↓1.9(23) | ↑1.6(37) | + | ↓2.0[0] | ↓2.0[0] |
| BdrF1 (BBF03) | 20.0 | lp28-1 | − | − | + | − | + | ++ |
| BdrF2 (BBG33) | 30.6 | lp28-2 | + | + | ↑3.4(37) | + | + | + |
| BdrF3 (BBH13) | 25.8 | lp28-3 | + | + | + | + | + | + |
The ORF designation assigned by the Institute for Genomic Research is shown in parentheses after the Bdr paralog.
Molecular mass (in kilodaltons).
A plus sign (+) indicates production of the protein, while a minus sign (−) indicates that production was not detected by immunoblotting. For those proteins that are not expressed during cultivation under normal conditions, two plus signs indicate that production increased with time during serum deprivation. The numerical values listed indicate fold changes in production level (↑ indicating fold increase and ↓ indicating fold decrease) relative to the production level at a certain temperature or time (i.e., 0 h, or not serum starved) (indicated in parentheses or brackets, respectively, in superscript). All values, except those for BdrE3 were determined by scanning densitometry. The values for BdrE3 could not be determined by this approach due to the close proximity of other Bdr bands and instead were visually estimated.
HA, host-adapted bacteria.
IFA.
Slides for indirect immunofluorescence assay (IFA) were prepared by immersion in a solution of 70% ethanol and 1% HCl for 30 min, followed by air drying. Slides were then immersed in a 0.01% poly-l-lysine solution (Sigma-Aldrich) for 5 min and air dried. To analyze Bdr production in ticks, infected nymphal-stage I. scapularis ticks were used in the IFAs either 2 days or 1.5 months postrepletion. The ticks were dissected on a slide, and the midgut material was mixed with 10 μl of 1× PBS. The suspension was distributed over a 15-mm diameter. The samples were outlined using a hydrophobic marking pen and allowed to air dry overnight, and the spirochetes were fixed to the slides by immersion in acetone for 10 min, followed by air drying. The slides were blocked by flooding with 80 μl of 10% fetal goat serum (FGS) and incubation in a 37°C humidified chamber for 30 min. The blocking agent was removed, and the samples were flooded with 80 μl of either (i) rabbit anti-Bdr Ab at a 1:100 dilution in 10% FGS, (ii) monoclonal mouse anti-Fla Ab at a 1:10 dilution in 10% FGS, or (iii) both rabbit anti-Bdr Ab (1:100 dilution) and monoclonal mouse anti-Fla Ab (1:10 dilution) in 10% FGS. The slides were incubated in a humidified chamber at 37°C for 30 min and washed three times for 1 min each time by immersion in 1× PBS, and then 80 μl of secondary fluorescent-labeled Ab was added to each slide (1:1,000 dilution in 10% FGS). For a control, additional slides were blocked as described above and screened with either prebleed serum or screened only with a secondary Ab. The Abs used were goat anti-rabbit IgG (heavy and light chains) conjugated with fluorescein (Pierce) and goat anti-mouse IgG (heavy and light chains) conjugated with rhodamine (Pierce). The secondary Abs were incubated for 30 min in a humidified chamber at 37°C, and the slides were washed three times in 1× PBS, rinsed with deionized water, and allowed to dry. Ten microliters of Fluoromount G (ethyl methanesulfonate) was placed onto each sample, and a coverslip was placed on the slide. The coverslips were sealed to the slide using clear nail polish around all the edges and allowed to air dry. The samples were analyzed using an Olympus BX51 fluorescence microscope with Fluor pan objectives (oil immersion). The images were captured using a MagnaFire camera.
RESULTS
Generation of Bdr subfamily- and paralog-specific antisera.
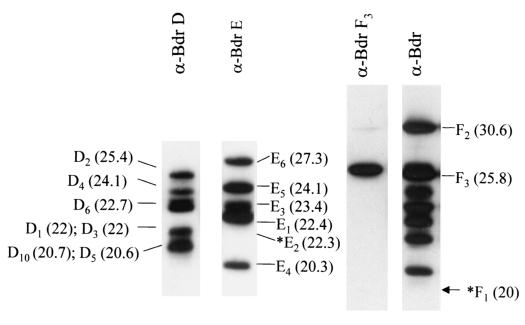
To allow for the identification of individual Bdr proteins, peptides were designed on the basis of unique N-terminal domain sequences and used to generate Bdr subfamily- or paralog-specific antisera. Immunoblot analyses of whole-cell lysates of B. burgdorferi B31MIpc revealed each antisera to be completely specific. Briefly, identical immunoblot strips were screened with either anti-BdrD, anti-BdrE, anti-BdrF3, or anti-Bdr antiserum (Fig. 1). The anti-Bdr antiserum recognizes, in part, a conserved domain of the Bdr proteins and thus recognizes most Bdr paralogs and orthologs, regardless of subfamily affiliation (24). The anti-BdrD and anti-BdrE antisera recognized proteins with molecular masses consistent with those of known members of the corresponding subfamily. However, the BdrD paralogs BdrD5 and BdrD10 were not immunoreactive with the anti-Bdr antiserum. This is likely due to sequence variation within the repeat motif region of BdrD5 and BdrD10. Due to sequence variation in the BdrF proteins, antisera that would selectively recognize all BdrF proteins could not be generated. However, it was possible to generate anti-BdrF3 antiserum. This antiserum reacted with a single protein of 25.8 kDa, which is consistent with the mass of BdrF3. While specific antisera were not generated against BdrF1 and BdrF2, these proteins are recognized by the anti-Bdr family-wide antisera, and upon immunoblotting, they can be differentiated from other paralogs based on their molecular mass. Note that BdrF1 was not detected in spirochetes cultivated under standard conditions (Fig. 1), because as described below, the production of this paralog is environmentally regulated. For reference, the predicted migration position of BdrF1 is indicated by an asterisk (Fig. 1).
FIG. 1.
Demonstration of the specificity of Bdr subfamily- or paralog-specific antisera. A cell lysate of B. burgdorferi B31MI served as the antigenic substrate to test the specificity of the various antisera. The lysates were fractionated in large-format (16 by 20 cm) SDS-polyacrylamide gels (12.5% polyacrylamide) and then immunoblotted (as described in Materials and Methods). The immunoblot strips were generated from gels run under identical conditions. Individual Bdr proteins were identified on the basis of their molecular mass (in kilodaltons [indicated in parentheses to the sides of the gels]). The migration positions of Bdr proteins are shown in the figure without the Bdr prefix. The asterisks indicate the expected migration positions of BdrE2 and BdrF1 . Production of these proteins was not detected under the growth conditions employed. The antisera used (anti-BdrD [α- BdrD]) is indicated above each panel.
Influence of temperature and host environment on Bdr production patterns.
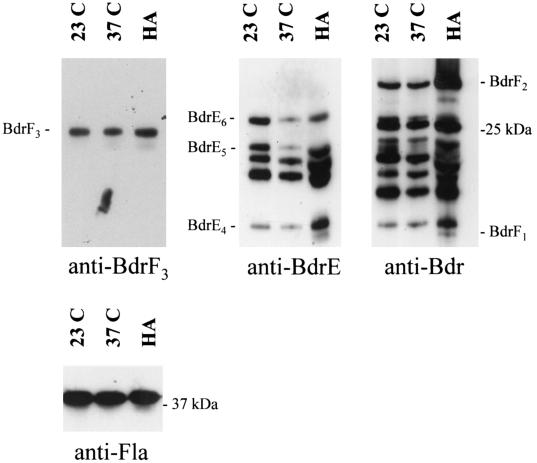
To assess the influence of temperature on Bdr production, B. burgdorferi B31MIpc bacteria grown at either 23, 33, or 37°C were harvested for immunoblot analysis. In all analyses, the same number of bacteria was loaded in each lane. Ensuring that the bacterial loads were equal was a critically important aspect of interpreting the data presented here. To verify that the gel bacterial loads were equal in all experiments, Western blot analyses of the Fla protein were performed. Because the transcriptional expression and production of this protein are not influenced by environmental conditions, it has been widely used as a control in differential expression analyses (2-4, 8, 11, 12, 14-16, 18, 21, 23, 30, 31). The Fla immunoblot analyses confirmed that the gel bacterial loads were equal in all cases. Identical immunoblots were then screened with either anti-Bdr, anti-BdrD, anti-BdrE, or anti-BdrF3 antiserum. Bacteria grown at 37°C produced lower levels of several BdrE paralogs than bacteria grown at 23°C (Fig. 2; data summarized in Table 3). The decreases in production of BdrE6 and BdrE5 in bacteria cultivated at 37°C versus 23°C were measured by scanning densitometry and determined to be 1.9- and 4.8-fold, respectively.
FIG. 2.
Influence of cultivation temperature and the mammalian host environment on B. burgdorferi Bdr synthesis profiles. Cell lysates of B. burgdorferi B31MIpc cultivated at either 23 or 37°C (indicated above each lane) were analyzed by SDS-PAGE in 15% polyacrylamide gels (16 by 20 cm) and immunoblotted. In addition, host-adapted bacteria (HA) obtained from DMCs that had been implanted in rats for 96 h were also analyzed. The immunoblots were screened with either anti-BdrD (data not shown), anti-Bdr, anti-BdrE, anti-BdrF3, or anti-Fla Ab as indicated below each panel. For reference, the identities of a subset of the expressed proteins are indicated. The Fla immunoblot demonstrates that protein loadings were equal in all lanes.
To determine if host-specific factors influence Bdr production, host-adapted bacteria were generated using the dialysis membrane chamber model (2). Immunoblot analyses of these bacteria revealed that the production of BdrF1, BdrF2, and several BdrE proteins were upregulated relative to their production during cultivation at 37°C (Fig. 2; data summarized in Table 3), while the production of other Bdr proteins and Fla remained unchanged. Using four independently generated batches of host-adapted bacteria, the average increase in BdrF2 production (in comparison to bacteria cultivated at 37°C) was 3.43-fold. The fold increases in BdrE6, BdrE5, and BdrE4 were 1.6, 6.4, and 4.4, respectively. BdrF1 could be detected only in host-adapted and serum-starved bacteria. Hence, the increased production of BdrF1, BdrF2, and the BdrE proteins in the host environment indicates that undefined host specific factors or environmental parameters differentially influence Bdr production.
Analysis of Bdr production profiles in rpoN and rpoS mutants of B. burgdorferi 297.
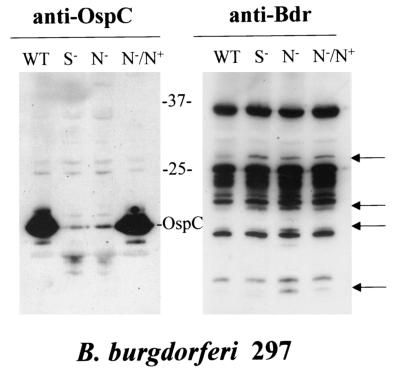
The RpoN and RpoS alternative sigma factors form a regulatory system that is thought to be part of B. burgdorferi's environmental sensing system (17). To determine if Bdr expression is either directly or indirectly regulated by RpoN or RpoS, immunoblot analyses were conducted using the wild-type 297 and rpoN and rpoS mutants of strain 297 (17). Isolate 297 was used for these analyses, because an rpoN or rpoS knockout of strain B31MIpc was not available. For a control, an immunoblot was screened with anti-OspC antiserum, since ospC expression is regulated through RpoN or RpoS (17). As expected, OspC production was completely repressed in both the rpoS and rpoN knockout and restored by rpoN complementation (Fig. 3). While significant changes in Bdr production levels were not observed for most paralogs, four immunoreactive proteins were detected in the rpoN mutant that were not observed in wild-type B. burgdorferi 297 (Fig. 3). Two of these four proteins were also observed in the rpoS mutant. Upon complementation of the rpoN mutant with RpoN expressed from a plasmid, the level of two of the four upregulated proteins returned to that observed in the wild type.
FIG. 3.
Bdr production profiles in B. burgdorferi 297 rpoN and rpoS knockout mutants. B. burgdorferi 297 mutants (rpoS or rpoN mutants) (17) were cultivated in the presence of erythromycin and harvested, and the cells were lysed by sonication. The lysates were analyzed by SDS-PAGE (15% polyacrylamide gels [16 by 20 cm]) and immunoblotted. The immunoblots were screened with the anti-Bdr antiserum. The migration positions of paralogs whose production was influenced by the rpo mutations are indicated by the arrows. The migration positions of OspC and of molecular mass markers (in kilodaltons) are shown between the two gels. The antiserum employed is indicated above each panel. Abbreviations are as follows: WT, wild-type B. burgdorferi 297; S−, rpoS mutant; N−, rpoN mutant.
Analysis of the influence of plasmid composition on Bdr production patterns.
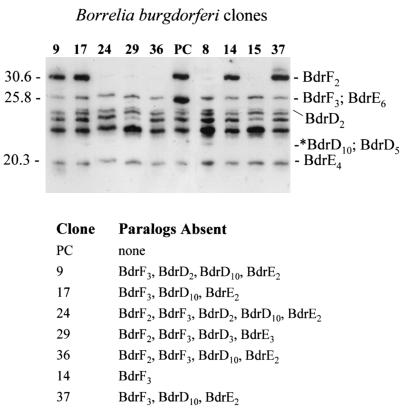
To determine if the loss of certain bdr genes results in a compensatory increase in the production of other Bdr paralogs, production patterns in isolates lacking one or more bdr-carrying plasmids were assessed. The plasmid profiles for these clones (Table 1 and Fig. 4) were determined in a previous analysis (19). Compensating increases in Bdr production were not observed in clones that lost one or more bdr-carrying plasmids. This suggests that during in vitro cultivation B. burgdorferi can tolerate some degree of variation in its overall Bdr profile and that not all Bdr paralogs are required for growth in vitro.
FIG. 4.
Bdr production patterns in B. burgdorferi clones carrying different plasmids. A series of clones derived from B. burgdorferi B31MIpc were obtained by subsurface plating of a culture derived from an ear punch biopsy of an infected mouse. The plasmid profiles of the clones had been previously determined using PCR and hybridization approaches (19). Cell lysates of the clones were fractionated by SDS-PAGE (18% polyacrylamide gel) and immunoblotted. The clones are shown over the lanes without the c prefix. The blot was screened with anti-Bdr antiserum. The migration positions of a few Bdr paralogs are indicated for reference to the right of the gel. The expected migration positions of the BdrD10 and BdrD5 paralogs, which are not recognized by anti-Bdr antiserum, are indicated by an asterisk. The migration positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel. Note that clone pc (PC) is the only one that carries the lp28-3 plasmid, which encodes BdrF3. Hence, the strong band seen in this clone at 25.8 kDa is due to the production of both BdrF3 and BdrD2. The Bdr paralog that are not carried on the clones are shown under the gel.
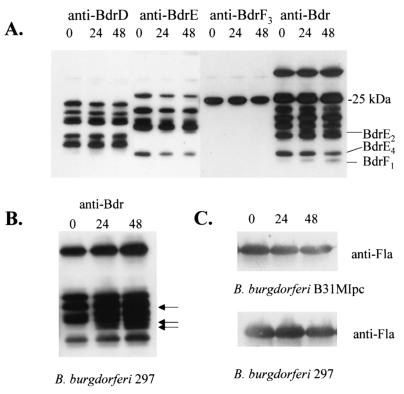
Influence of serum starvation on Bdr production.
To determine if serum influences Bdr production, immunoblot analyses were conducted on B. burgdorferi B31MIpc and B. burgdorferi 297 that were deprived of rabbit serum for either 0, 24, or 48 h (at 33°C). The production of some Bdr paralogs proved to be responsive to serum starvation (Fig. 5; data summarized in Table 3). In B. burgdorferi B31MIpc, the production of BdrE2 and BdrF1 were specifically upregulated in response to serum deprivation. Neither of these proteins were produced by spirochetes grown under typical culture conditions (i.e., BSK-H medium with 12% rabbit sera at 33°C). Since the complete composition of the bdr gene family is undefined in isolate 297, it was not possible to determine the identities of the Bdr paralogs upregulated by serum starvation in this isolate. However, the production of at least three Bdr proteins (Fig. 5B) was significantly upregulated in 297. As expected, the production levels of the constitutively expressed Fla protein were not affected by serum deprivation (Fig. 5C).
FIG. 5.
Influence of serum deprivation on Bdr production patterns. B. burgdorferi B31MI (A) or 297 (B) were cultivated in the absence of added rabbit serum for either 0, 24, or 48 h (indicated above each lane). Identical immunoblots of B. burgdorferi B31MI were screened with the anti-BdrD, anti-BdrE, anti-BdrF3, or anti-Bdr antiserum as indicated above the lanes. The arrows in panel B indicate the migration positions of Bdr paralogs that were upregulated by serum starvation in isolate 297. (C) Immunoblot analyses of the Fla protein in isolates B31MIpc and 297 that were subjected to serum deprivation for the times (in hours) indicated over the lanes. This control demonstrates that the production of the normally constitutively expressed Fla protein is not affected by serum deprivation.
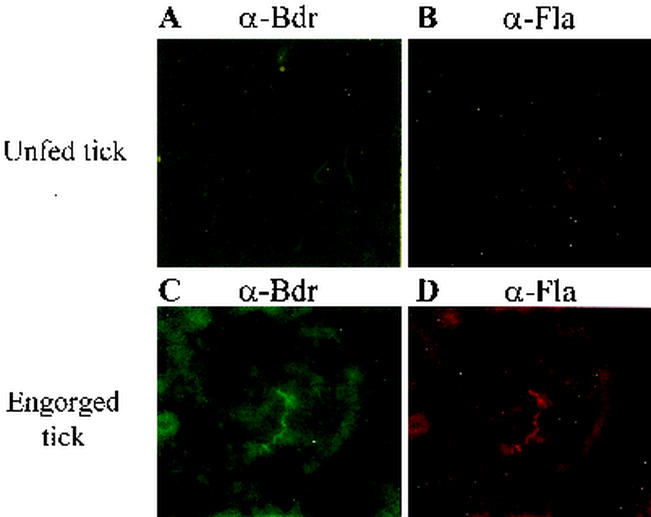
Analysis of Bdr production in fed and unfed Ixodes scapularis ticks using IFAs.
Bdr production in fed and unfed ticks was assessed through IFAs. Ixodes scapularis nymphal ticks that were fed on mice and then maintained for 1.5 months in a humidified chamber served as the unfed ticks. These ticks, which were no longer engorged, were viable and highly active and were demonstrated to be capable of transmitting infection to uninfected C3H-HeJ mice (data not shown). Since the Bdr proteins are located in the inner membrane, the macerated tick midgut material was treated with acetone to permeabilize the cells. While the spirochetal burden was relatively low in the unfed ticks, all spirochetes detected by dark-field microscopy were immunoreactive when the anti-Bdr antiserum was used as the primary Ab (Fig. 6). Detection of Ab binding was through a fluorescein-labeled secondary Ab. For the IFAs, detection of flagellum-Ab binding using a rhodamine red secondary Ab for detection served as the positive control. To assess expression in fed ticks, several of the infected Ixodes scapularis ticks described above were fed on uninfected mice to repletion. The engorged ticks were analyzed at 3 days postrepletion. Bdr expression was readily detected in all spirochetes. In both the fed and unfed ticks, the identities of the specific paralogs that were expressed could not be determined, because we were unable to detect Bdr proteins using the subfamily- or paralog-specific antisera in the IFAs. This is presumably due to the inaccessibility of the N-terminal domain, the region to which these antisera were generated, in the native Bdr proteins as they exist in the spirochetes.
FIG. 6.
Demonstration of Bdr production in fed and unfed ticks through IFAs. IFAs were performed as described in Materials and Methods. IFAs were performed using unfed ticks (A and B) and fed ticks (C and D). In panels A and C, anti-Bdr antiserum (α-Bdr) was used as the primary Ab and goat anti-rabbit Ab conjugated with fluorescein was the secondary Ab, and in panels C and D, anti-Fla antiserum (α-Fla) was used as the primary Ab and goat anti-mouse Ab conjugated with rhodamine was the secondary Ab. As a negative control, IFAs were performed using preimmune sera (data not shown).
DISCUSSION
Previous analyses of the 18-member Bdr protein family of B. burgdorferi B31MI have demonstrated that these proteins form three distinct subfamilies (6), are located in the inner membrane (25), and possess putative serine-threonine phosphorylation motifs (29). It has been postulated that the Bdr proteins play a role in sensing and/or transducing environmental signals (25). The goals of this study were to determine if Bdr production is responsive to environmental conditions. Bdr production under different environmental conditions was assessed through immunoblot or IFAs. These analyses revealed that the production of some Bdr paralogs were influenced to various degrees by environmental conditions (Table 3). The data presented here regarding BdrF2 production are consistent with the results of microarray analyses that have demonstrated at least a threefold increase in BdrF2 transcript in host-adapted bacteria in comparison to that observed in in vitro-cultivated bacteria (23). Using microarrays, Akins and colleagues (personal communication) also observed a 3.7-fold increase in BdrF2 transcript in host-adapted bacteria. While temperature alone influenced the production of some Bdr paralogs, increased BdrF2 production clearly requires host-specific factors. The DMC model employed here to generate host-adapted bacteria has some limitations in that the spirochetes are protected from direct contact with host cells and from many host factors that would be encountered during natural infection. Only small polypeptides or other soluble factors can diffuse into the DMCs which have a molecular size cutoff of 5 kDa. Hence, it is possible that the production of other Bdr paralogs may also be influenced by host-specific factors that could not transverse the DMCs.
Since the Lyme disease spirochetes cycle between ticks and mammals, Bdr production in ticks was also assessed. The tick environment is highly variable and is influenced by external temperature, physiological changes associated with tick feeding, and molting. Spirochetes in fed ticks are exposed to an ample supply of sera that decreases with time postrepletion, while sera is absent from the midgut of an unfed tick. IFAs revealed that the Bdr proteins are expressed in both fed and unfed ticks. The presence or absence of serum in the growth medium is thought to partially mimic the environments encountered by the spirochetes in fed and unfed ticks, respectively. Serum deprivation has been demonstrated to induce production of at least 20 proteins (3), some of which have molecular masses and pIs similar to those of Bdr proteins. Here we demonstrate that serum deprivation induces the production of specific Bdr proteins in both B31MI and isolate 297. While serum deprivation may trigger a general stress response, it is important to note that its effects on Bdr production were selective and affected only a subset of paralogs. In summary, it can be concluded that the production of some Bdr proteins is differentially influenced by environmental conditions.
To determine if the loss of individual bdr-carrying plasmids results in an alteration in the production levels of the remaining Bdr proteins, Bdr synthesis patterns were determined for clones with different plasmids. The loss of one or more bdr-encoding plasmids did not alter the production levels of other bdr genes. From this, it can be concluded that variation in Bdr production profiles can be tolerated by the spirochetes and that not all paralogs are required for survival during in vitro cultivation. Clearly, there must be some degree of functional redundancy in the Bdr proteins, and it is possible that one function of plasmid redundancy overall is to protect the cell against the potential adverse effects of loss of one or more plasmids. The loss of a paralog encoded by one plasmid may be complemented by a paralog encoded by a different plasmid.
Alternative sigma factors have been demonstrated to regulate the expression of some Borrelia genes such as OspC and DbpA (17). It has been postulated that the RpoN-RpoS system may regulate several genes that are involved in stress responses (17). Although the production of some Bdr proteins is clearly responsive to environment and stress, Bdr production profiles in isolate 297 were not dramatically altered in rpoN and rpoS mutants. Consistent with this, a strict RpoN consensus site was not detected upstream of the bdr genes. Analysis of the sequences upstream of these genes revealed that there is extensive variation which could influence the transcription of these genes and serve as the basis for their differential regulation. Future analyses will seek to determine if bdr expression is regulated at the transcriptional level.
The data presented here demonstrate that the production of the Bdr protein family is complex and environmentally regulated. With the identification of paralogs whose production is environmentally influenced, it will now be possible to target specific alleles for gene inactivation and then test the contribution of those paralogs to the biology of the spirochetes in different environments.
Acknowledgments
We thank Michael Norgard and Xiaofen Yang for supplying the rpo mutants. We thank the Molecular Pathogenesis group at Virginia Commonwealth University for helpful discussions.
This work was supported in part by grants from National Institutes of Health National Institute of Allergy and Infectious Diseases grant AI51586 to R.T.M and D.M.R. and grant AI-29735 to J.R. and M.C. and National Institute of Neurological Disorders and Stroke grant NS43088 to J.M.
Editor: V. J. DiRita
REFERENCES
- 1.Akins, D., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 18:507-520. [DOI] [PubMed] [Google Scholar]
- 2.Akins, D. R., K. W. Bourell, M. J. Caimano, M. V. Norgard, and J. R. Radolf. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Invest. 101:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alban, P. S., P. W. Johnson, and D. R. Nelson. 2000. Serum-starvation-induced changes in protein synthesis and morphology of Borrelia burgdorferi. Microbiology 146:119-127. [DOI] [PubMed] [Google Scholar]
- 4.Anguita, J., S. Samanta, B. Revilla, K. Suk, S. Das, S. W. Barthold, and E. Fikrig. 2000. Borrelia burgdorferi gene expression in vivo and spirochete pathogenicity. Infect. Immun. 68:1222-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlyon, J. A., and R. T. Marconi. 1998. Cloning and molecular characterization of a multicopy, linear plasmid-carried, repeat motif-containing gene from Borrelia turicatae, a causative agent of relapsing fever. J. Bacteriol. 180:4974-4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlyon, J. A., D. M. Roberts, and R. T. Marconi. 2000. Evolutionary and molecular analyses of the Borrelia bdr super gene family: delineation of distinct subfamilies and demonstration of the genus wide conservation of putative functional domains, structural properties and repeat motifs. Microb. Pathog. 28:89-105. [DOI] [PubMed] [Google Scholar]
- 7.Carlyon, J. A., D. M. Roberts, M. Theisen, C. Sadler, and R. T. Marconi. 2000. Molecular and immunological analyses of the Borrelia turicatae Bdr protein family. Infect. Immun. 68:2369-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, J. A., N. El-Hage, J. C. Miller, K. Babb, and B. Stevenson. 2001. Borrelia burgdorferi RevA is a surface-exposed outer membrane proteins whose expression is regulated in response to environmental temperature and pH. Infect. Immun. 69:5286-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 10.de Silva, A. M., and E. Fikrig. 1997. Borrelia burgdorferi genes selectively expressed in ticks and mammals. Parasitol. Today 13:267-270. [DOI] [PubMed] [Google Scholar]
- 11.El-Hage, N., K. Babb, J. A. Carroll, N. Lindstrom, E. R. Fischer, J. C. Miller, R. D. Gilmore, Jr., M. L. Mbow, and B. Stevenson. 2001. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147:821-830. [DOI] [PubMed] [Google Scholar]
- 12.Elias, A. F., J. L. Bono, J. A. Carroll, P. Stewart, K. Tilly, and P. Rosa. 2000. Altered stationary-phase response in a Borrelia burgdorferi rpoS mutant. J. Bacteriol. 182:2909-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser, C., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischman, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 14.Golde, W. T., B. Robinson-Dunn, M. G. Stobierski, D. Dykhuzien, I.-N. Wang, V. Carlson, H. Stiefel, S. Shiflett, and G. L. Campbell. 1998. Culture-confirmed reinfection of a person with different strains of Borrelia burgdorferi sensu stricto. J. Clin. Microbiol. 36:1015-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagman, K. E., P. Lahdenne, T. G. Popova, S. F. Porcella, D. R. Akins, J. D. Radolf, and M. V. Norgard. 1998. Decorin binding protein of Borrelia burgdorferi is encoded within a two-gene operon and is protective in the murine model of Lyme borreliosis. Infect. Immun. 66:2674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and J. R. Akins. 2001. Regulation of the OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubner, A., X. Yang, D. M. Nolen, T. G. Popova, F. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 98:12724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Indest, K. J., J. K. Howell, M. B. Jacobs, D. School-Meeker, S. J. Norris, and M. T. Philipp. 2001. Analysis of Borrelia burgdorferi vlsE gene expression and recombination in the tick vector. Infect. Immun. 69:7083-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDowell, J. V., J. T. Skare, S.-Y. Sung, and R. T. Marconi. 2001. Analysis of mechanisms associated with the loss of infectivity of clonal populations of Borrelia burgdorferi B31MI. Infect. Immun. 69:3670-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDowell, J. V., S.-Y. Sung, G. Price, and R. T. Marconi. 2001. Demonstration of the genetic stability and temporal expression of select members of the Lyme disease spirochete OspF protein family during infection in mice. Infect. Immun. 69:4831-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porcella, S. F., C. A. Fitzpatrick, and J. L. Bono. 2000. Expression and immunological analysis of the plasmid-borne mlp genes of Borrelia burgdorferi strain B31. Infect. Immun. 68:4992-5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porcella, S. F., T. G. Popova, D. R. Akins, M. Li, J. R. Radolf, and M. V. Norgard. 1996. Borrelia burgdorferi supercoiled plasmids encode multicopy open reading frames and a lipoprotein gene family. J. Bacteriol. 178:3293-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts, D. M., J. A. Carlyon, M. Theisen, and R. T. Marconi. 2000. The bdr gene families of the Lyme disease and relapsing fever spirochetes: possible influence on biology, pathogenesis and evolution. Emerg. Infect. Dis. 6:110-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts, D. M., M. Theisen, and R. T. Marconi. 2000. Analysis of the cellular localization of Bdr paralogs in Borrelia burgdorferi, a causative agent of Lyme disease: evidence for functional diversity. J. Bacteriol. 182:4222-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suk, K., S. Das, W. Sun, B. Jwang, S. W. Barthold, R. A. Flavell, and E. Fikrig. 1995. Borrelia burgdorferi genes selectively expressed in the infected host. Proc. Natl. Acad. Sci. USA 92:4269-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sung, S. Y., J. McDowell, J. A. Carlyon, and R. T. Marconi. 2000. Mutation and recombination in the upstream homology box-flanked ospE-related genes of the Lyme disease spirochetes results in the development of new antigenic variants during infection. Infect. Immun. 68:1319-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theisen, M. 1996. Molecular cloning and characterization of nlpH, encoding a novel surface-exposed, polymorphic, plasmid-encoded 33-kilodalton lipoprotein of Borrelia afzelii. J. Bacteriol. 178:6435-6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, X., T. G. Popova, K. E. Hagman, S. K. Wikel, G. B. Schoeler, M. J. Caimano, J. D. Radolf, and M. Norgard. 1999. Identification, characterization, and expression of three new members of the Borrelia burgdorferi Mlp (2.9) lipoprotein gene family. Infect. Immun. 67:6008-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, J.-R., J. M. Hardham, A. G. Barbour, and A. G. Norris. 1997. Antigenic variation in Lyme disease Borreliae by promiscuous recombination of vmp like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]
- 32.Zuckert, W. R., and J. Meyer. 1996. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J. Bacteriol. 178:2287-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuckert, W. R., J. Meyer, and A. G. Barbour. 1999. Comparative analysis and immunological characterization of the Borrelia Bdr protein family. Infect. Immun. 67:3257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]