Abstract
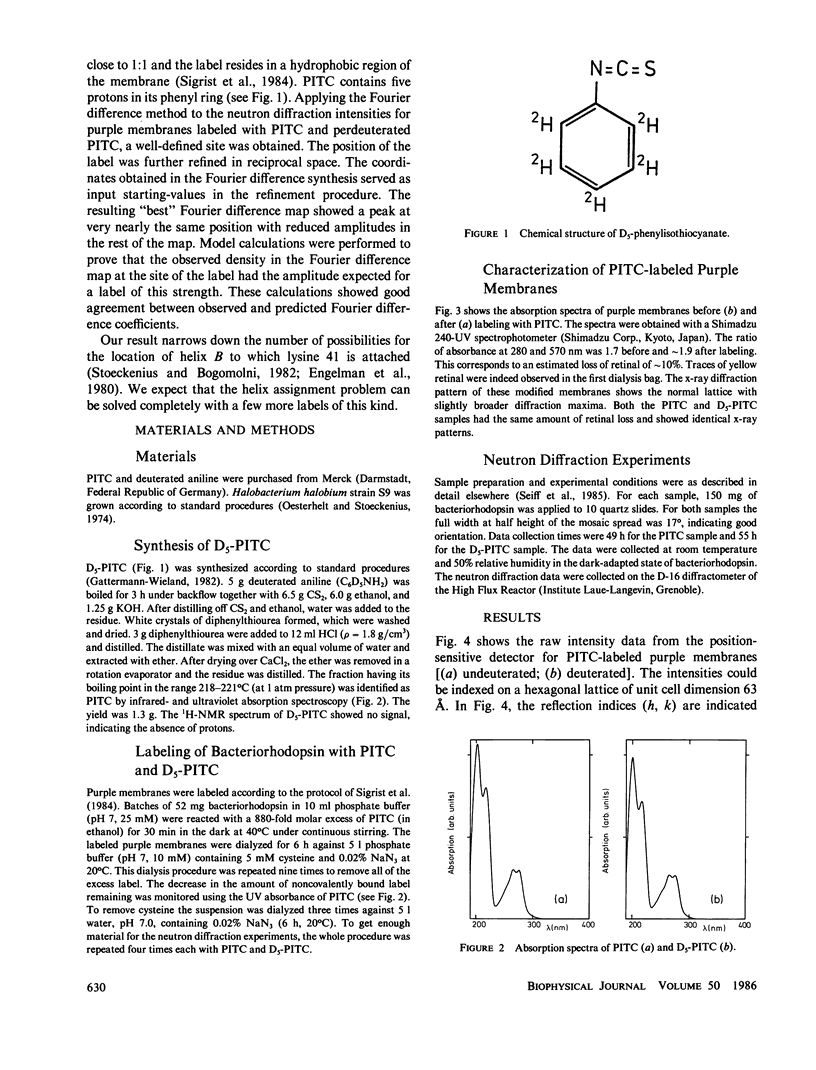
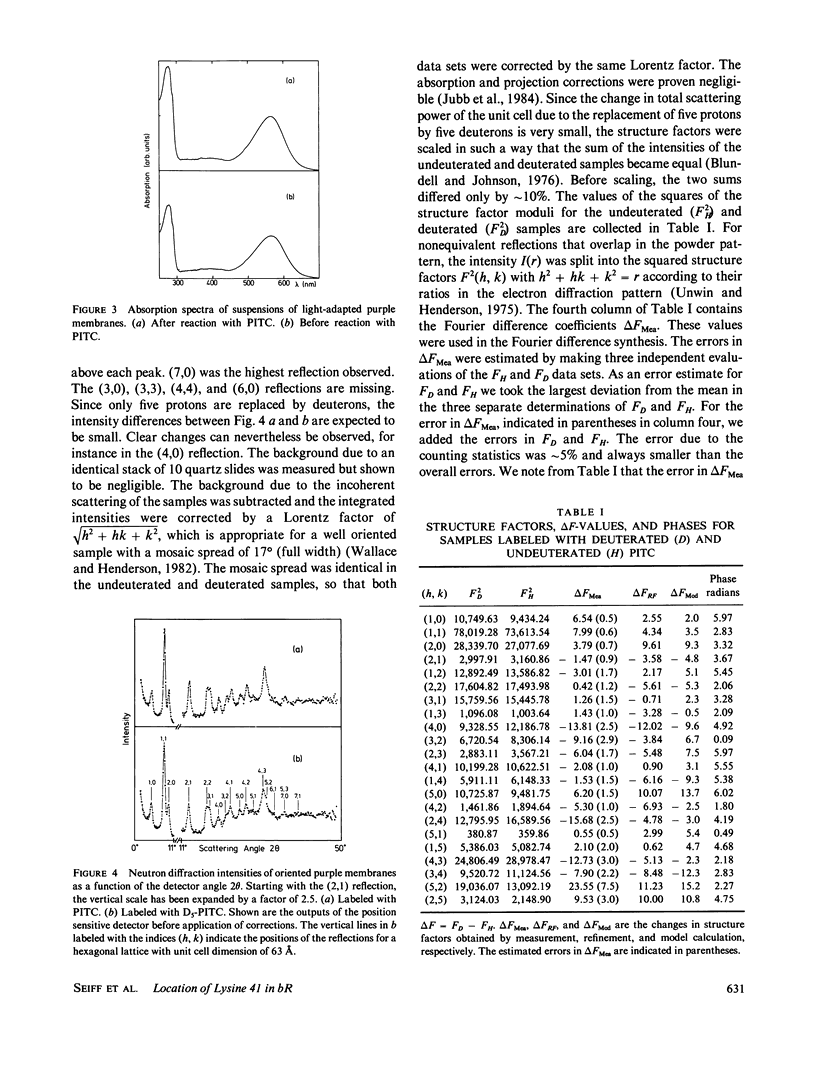
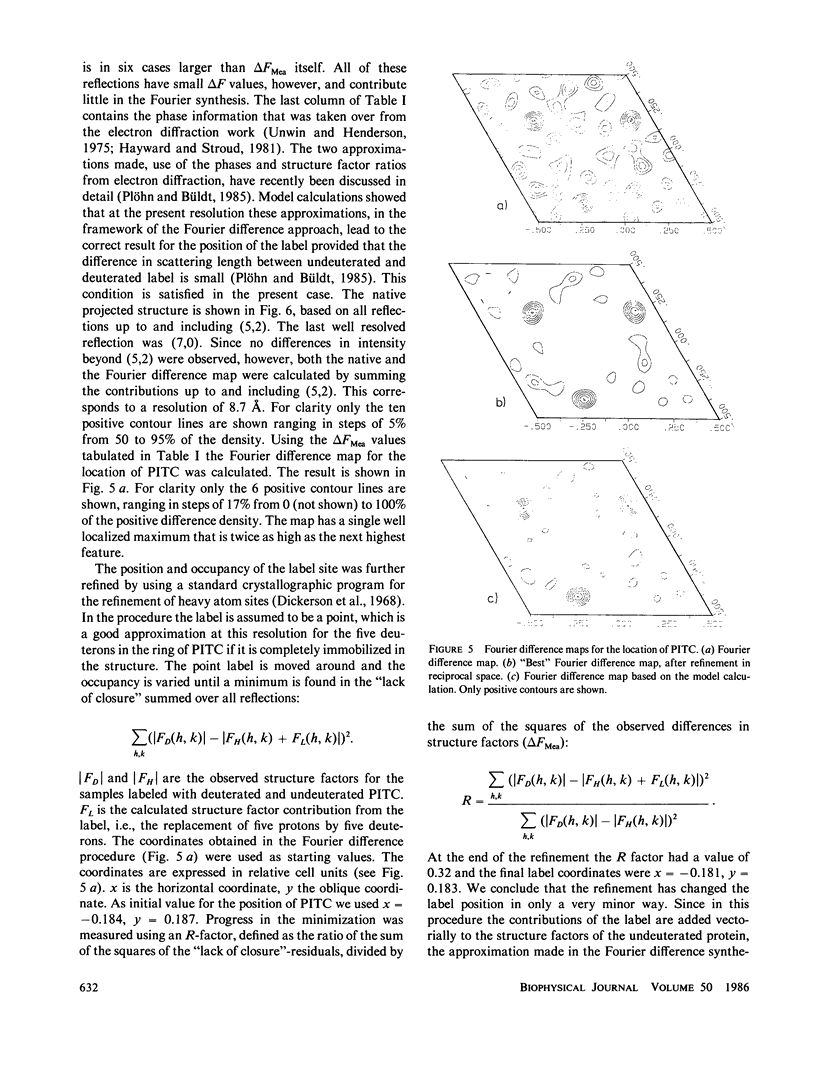
Purple membranes were prepared in which bacteriorhodopsin was labeled at lysine 41 with phenylisothiocyanate (PITC) and with perdeuterated PITC. The in-plane position of this small label containing only five deuterons was determined from the differences between the neutron diffraction intensities of the two samples. At 8.7-Å resolution the Fourier difference map revealed a well-defined site between helices 3 and 4. This position was confirmed by a refinement procedure in reciprocal space. Model calculations showed that the observed difference density had the right amplitude for the label. Thus it is possible to locate a small group in a large protein structure by replacing as few as five hydrogens by deuterium. The observed location of PITC restricts the number of possibilities for the assignment of helix B in the sequence (to which lysine 41 is attached) to one of the seven helices of the structure. Taking into account the size of the label and the length of the lysine side chain our result excludes helices 1, 2, and 7 as candidates for B.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agard D. A., Stroud R. M. Linking regions between helices in bacteriorhodopsin revealed. Biophys J. 1982 Mar;37(3):589–602. [PMC free article] [PubMed] [Google Scholar]
- Allegrini P. R., Sigrist H., Schaller J., Zahler P. Site-directed fluorogenic modification of bacteriorhodopsin by 7-chloro-4-nitrobenz-2-oxa-1,3-diazole. Eur J Biochem. 1983 May 16;132(3):603–608. doi: 10.1111/j.1432-1033.1983.tb07406.x. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Henderson R., McLachlan A. D., Wallace B. A. Path of the polypeptide in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2023–2027. doi: 10.1073/pnas.77.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M., Zaccai G. Bacteriorhodopsin is an inside-out protein. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5894–5898. doi: 10.1073/pnas.77.10.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S. B., Stroud R. M. Projected structure of purple membrane determined to 3.7 A resolution by low temperature electron microscopy. J Mol Biol. 1981 Sep 25;151(3):491–517. doi: 10.1016/0022-2836(81)90007-3. [DOI] [PubMed] [Google Scholar]
- Henderson R. The structure of the purple membrane from Halobacterium hallobium: analysis of the X-ray diffraction pattern. J Mol Biol. 1975 Apr 5;93(2):123–138. doi: 10.1016/0022-2836(75)90123-0. [DOI] [PubMed] [Google Scholar]
- Jubb J. S., Worcester D. L., Crespi H. L., Zaccaï G. Retinal location in purple membrane of Halobacterium halobium: a neutron diffraction study of membranes labelled in vivo with deuterated retinal. EMBO J. 1984 Jul;3(7):1455–1461. doi: 10.1002/j.1460-2075.1984.tb01996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katre N. V., Finer-Moore J., Stroud R. M., Hayward S. B. Location of an extrinsic label in the primary and tertiary structure of bacteriorhodopsin. Biophys J. 1984 Aug;46(2):195–203. doi: 10.1016/S0006-3495(84)84013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorana H. G., Gerber G. E., Herlihy W. C., Gray C. P., Anderegg R. J., Nihei K., Biemann K. Amino acid sequence of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5046–5050. doi: 10.1073/pnas.76.10.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D., Stoeckenius W. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Abdulaev N. G., Feigina M. Y., Kiselev A. V., Lobanov N. A. The structural basis of the functioning of bacteriorhodopsin: an overview. FEBS Lett. 1979 Apr 15;100(2):219–224. doi: 10.1016/0014-5793(79)80338-5. [DOI] [PubMed] [Google Scholar]
- Seiff F., Wallat I., Ermann P., Heyn M. P. A neutron diffraction study on the location of the polyene chain of retinal in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1985 May;82(10):3227–3231. doi: 10.1073/pnas.82.10.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist H., Allegrini P. R., Stauffer K., Schaller J., Abdulaev N. G., Rickli E. E., Zahler P. Group-directed modification of bacteriorhodopsin by arylisothiocyanates. Labeling, identification of the binding site and topology. J Mol Biol. 1984 Feb 15;173(1):93–108. doi: 10.1016/0022-2836(84)90405-4. [DOI] [PubMed] [Google Scholar]
- Stoeckenius W., Bogomolni R. A. Bacteriorhodopsin and related pigments of halobacteria. Annu Rev Biochem. 1982;51:587–616. doi: 10.1146/annurev.bi.51.070182.003103. [DOI] [PubMed] [Google Scholar]
- Trewhella J., Anderson S., Fox R., Gogol E., Khan S., Engelman D., Zaccai G. Assignment of segments of the bacteriorhodopsin sequence to positions in the structural map. Biophys J. 1983 Jun;42(3):233–241. doi: 10.1016/S0006-3495(83)84391-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin P. N., Henderson R. Molecular structure determination by electron microscopy of unstained crystalline specimens. J Mol Biol. 1975 May 25;94(3):425–440. doi: 10.1016/0022-2836(75)90212-0. [DOI] [PubMed] [Google Scholar]
- Wallace B. A., Henderson R. Location of the carboxyl terminus of bacteriorhodopsin in purple membrane. Biophys J. 1982 Sep;39(3):233–239. doi: 10.1016/S0006-3495(82)84513-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolber P. K., Stoeckenius W. Retinal migration during dark reduction of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1984 Apr;81:2303–2307. doi: 10.1073/pnas.81.8.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]


