Abstract
Coxiella burnetii, the etiological agent of Q fever, is a gram-negative obligate intracellular bacterium. Two striking characteristics of this microorganism are its ability to thrive within a phagolysosome and its ability to persist in the environment outside a host cell. These abilities have been attributed to the existence of C. burnetii developmental cycle variants: large-cell variants (LCV), small-cell variants (SCV), and small dense cells (SDC). Variants differ in protein profiles, including differential expression of a major outer membrane protein (MOMP) of C. burnetii, designated P1. The ∼29-kDa MOMP is highly expressed in LCV, down-regulated in SCV, and not apparent in SDC. We sought to characterize P1 through purification of native protein for N-terminal analysis, cloning, and functional studies. Highly purified P1, extracted from C. burnetii membranes by using the zwitterionic detergent Empigen, allowed the determination of N-terminal and internal peptide sequences. The entire P1 coding locus was cloned by PCR amplification based upon these peptide sequences, followed by inverse PCR. Comparison of the predicted P1 amino acid sequences among the C. burnetii isolates Nine Mile, Koka, Scurry, and Kerns indicated a high degree of conservation. Structural prediction suggests that the peptide has a predominantly β-sheet conformation, consistent with bacterial porins. Typical porin characteristics were observed for native P1, including detergent solubilization properties, heat modification of purified protein, and channel formation in a planar lipid bilayer. Characterization of differentially expressed P1 as a porin increases our understanding of the function of morphological variants and their role in pathogenesis.
Coxiella burnetii is an obligately intracellular bacterium with the unique ability to survive and multiply within an acidified phagolysosome. The etiological agent of Q fever, this gram-negative member of the order Legionellales (16) has a worldwide distribution and a broad host range that includes humans, fish, birds, arthropods, rodents, and livestock (3). Though overt disease is not apparent in most of these hosts, humans may become infected and develop clinical symptoms after inhalation of contaminated aerosols. Often misdiagnosed, the disease presents in the acute form as a flu-like illness with fever, chills, malaise, and a characteristic periorbital pain (28). Epidemiologic studies suggest that some patients who contract acute Q fever progress to chronic disease years later, most commonly presenting as endocarditis or hepatitis (6). Unlike the acute disease, which is readily responsive to a short treatment with tetracycline, chronic disease patients have responded best to a prolonged regimen of combined antibiotic therapy (20).
C. burnetii survival and replication exclusively in a phagolysosome have been attributed to adaptations of an acidophilic metabolism and developmental cycle (8, 23, 24). A developmental model for C. burnetii has been proposed that is comprised of three cell variants: large-cell variants (LCV), small-cell variants (SCV), and small dense cells (SDC) (29). Cell variants differ from one another in a number of properties, including morphology, physical and chemical resistance, and metabolic activity (8, 24). The ability of C. burnetii to cause disease through persistence in the environment is likely due to an unusual extracellular stability attributed to two of the developmental-cycle variants. The LCV is approximately 0.5 to 1.0 μm in length, has diffuse chromatin, and possesses clearly distinguishable outer and cytoplasmic membranes. The LCV are fragile, probably not surviving extracellularly for extended periods. In contrast, the SCV range in size from 0.2 to 0.5 μm, have condensed chromatin, and possess an electron-dense region bounded by the outer and cytoplasmic membranes. The SDC resemble SCV in morphology but are distinct from this group in having increased physical stability (21). This most resistant form of C. burnetii is capable of withstanding pressures up to 50,000 lb/in2 (1). The SDC and SCV may represent the forms of the bacteria likely to survive as infectious particles extracellularly.
Several proteins have been shown to be differentially expressed by developmental forms, including the major outer membrane protein (MOMP) designated P1 (29). P1 was first identified by Williams and coworkers while they were comparing distinguishing antigenic determinants between phase I and phase II C. burnetii (45). McCaul and colleagues showed that P1 was not present on all variants (22). As determined by immunoelectron microscopy, monoclonal antibodies against P1 densely labeled LCV while sparsely labeling SCV and only occasionally associating with SDC. This pattern of differential expression was confirmed by Western blotting. These data frame important questions regarding the involvement of P1 in the progression between variants and its role in developmental variants.
Current vaccines against Q fever are comprised of either formalin-killed whole-cell vaccine preparations (WCV) (19) or chloroform-methanol-extracted bacterial residue (42, 46). The protective ability of WCV is well documented (14, 25, 41), but also well documented are the adverse reactions induced by WCV, including local skin reactions, fever, anorexia, and malaise in previously sensitized vaccinees (2, 13, 37, 44). New approaches to develop a safe, broad-use vaccine against Q fever are warranted. The 29-kDa MOMP P1 is a logical subunit vaccine candidate because of characteristics consistent with cell surface components, such as susceptibility to iodination reactions, resistance to detergent solubilization at low temperatures, reactivity with components of immune serum in enzyme-linked immunosorbent assays and radioimmunoprecipitation assays, and natural abundance (38; C. E. Snyder, Jr., and J. C. Williams, Abstr. Annu. Meet. Am. Soc. Microbiol. 1986, p. 193, abstr. K-1, 1986). Williams et al. found that immunity derived from exposure to a partially purified P1 protein was more efficacious in enhancing clearance of organisms from spleens of infected mice than immunity derived from other proteins or lipopolysaccharide (46).
By studying differential expression of genes and protein function, we hope to develop a better understanding of the developmental cycle model of C. burnetii. In the present study, we established a purification method for P1 that facilitated the subsequent cloning of the gene and assessment of biological function. By the generation of peptide fragments, the gene encoding P1 was cloned by using degenerate oligonucleotides followed by inverse PCR. Analysis of the resulting sequence revealed characteristics typical of bacterial porins. Planar lipid bilayer experiments with native P1 confirmed that this protein is capable of forming channels within synthetic membranes.
MATERIALS AND METHODS
Surface protein iodination of life cycle variants.
The strains and plasmids used in this study are listed in Table 1. A 2.5-mg portion of phase I C. burnetii was harvested from infected L929 cells by lysis with 1% NP-40 in 250 mM sucrose in phosphate-buffered saline (PBS) (35). After separation from host material through differential centrifugations, cell variants were separated on an isopycnic gradient (9, 34, 43). Briefly, bacteria were layered on 32% cesium chloride in 0.05 M Tris, pH 7.5, and separated by centrifugation at 208,000 × g for 16 h. Two bands were collected, pelleted out of the cesium chloride, and suspended in 500 μl of PBS. Fifty microliters of each fraction was pelleted and iodinated with Iodogen R beads (Pierce, Rockford, Ill.) by using 20 μl of carrier-free Na125I. Equal counts of labeled bacterial variants were separated by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis (PAGE).
TABLE 1.
Bacterial strains and plasmids used in this study
| Organism or plasmid | Description | Reference or source |
|---|---|---|
| C. burnetii | Nine Mile, Phase I, RSA 493 | 30 |
| Koka, Q195 | ||
| Scurry, Q217 | ||
| Kerns, Q229 | ||
| E. coli | ||
| DH5α | F− φ80d lacZΔM15Δ (lacZYA-argF) U169 deoR recA endA1 phoA relA1 | Gibco-Life BRL |
| BL21 | F−ompT hsdS (rB− mB−) gal dcm plysE | Novagen |
| XL1-MRF′ | lac[F′ proAB lacIqZΔM15 Tn10 (Tetr)] mcrA endA1 supE44 thi-1 recA1 gyrA96 relA1 | Stratagene |
| SOLR | F′ proAB lacIqZΔM15 Su− lac gyrA96 relA1 thi-1 endA1 λr sbcC recB recJ umuC::Tn5(kan) uvrC | Stratagene |
| TOP10F′ | F′ [lacIq Tn10 (tet)] lacZΔM15Δ deoR recA1 araD139 Δ(ara-leu)7679 galU galK rpsL endA1 nupG mcrA | Invitrogen |
| Plasmids | ||
| pCR 2.1-TOPO | TA cloning vector Apr | Invitrogen |
| pSV101 | pCR2.1 with 570-bp P1 Nine Mile gene insert (partial clone) | This work |
| pSV102 | pCR2.1 with 750-bp P1 Nine Mile gene insert (full clone) | This work |
| pGF-Koka | pCR2.1 with P1 Koka gene insert | This work |
| pGF-Scurry | pCR2.1 with P1 Scurry gene insert | This work |
| pGF-Kerns | pCR2.1 with P1 Kerns gene insert | This work |
| pBAD-Thio-TOPO- | Expression vector Apr | Invitrogen |
| pSV108 | pBAD-Thio-TOPO with 550-bp P1 insert | This work |
| pGF101 | pBAD-Thio-TOPO with 687-bp P1 insert | This work |
Immunoreactivity of LCV and SCV with P1 antibody.
Phase I C. burnetii (10 mg) was purified from infected eggs and used to obtain cell variants as described above. LCV, SCV, and whole bacteria (20 μg each) were separated by SDS-12% PAGE and transferred to nitrocellulose membranes (Micron Separations Inc., Westboro, Mass.). Transfers were blocked for 1 h with 10% nonfat powdered milk and 0.1% Tween-20 in PBS. Blots were then incubated with the anti-P1 monoclonal antibody 4E8 (kind gift of D. Waag, U.S. Army Medical Research Institute for Infectious Diseases) followed by goat anti-mouse immunoglobulin G horseradish peroxidase-conjugated secondary antibody (45). Western blots were developed with either a horseradish peroxidase kit (Bio-Rad, Hercules, Calif.) or an ECL Western blot detection kit (Amersham Pharmacia, Piscataway, N.J.).
Generation of N-terminal and internal sequences.
Phase I C. burnetii Nine Mile purified from yolk sacs was extracted with 1% Empigen BB (Calbiochem, La Jolla, Calif.) plus 10 mM EDTA and 50 mM NaCl at 37°C for 16 h. The soluble fraction was separated by SDS-12% PAGE and transferred onto a Problott (Applied Biosystems, Foster City, Calif.) polyvinylidene difluoride membrane. The membrane was stained with a solution of 0.1% (wt/vol) Coomassie blue, 50% (vol/vol) methanol, and 1% (vol/vol) acetic acid for 5 min and then destained with three 5-min washes of 50% methanol containing 1% acetic acid. The protein band at ∼30 kDa was excised and processed by the Protein Chemistry Sequencing Laboratory at Texas A&M University for N-terminal sequence analysis using a Hewlett Packard G1000A automated protein sequencer.
For generation of internal peptides, 5 mg of phase I C. burnetii Nine Mile was extracted with Empigen as described above. This sample was precipitated with trichloroacetic acid followed by neutralization with 0.1 N sodium hydroxide. Pellets were suspended in PBS and then incubated overnight at 4°C with an equal volume of P1 monoclonal antibody 4E8 at a concentration of 1:100. Gamma Bind Plus beads (1/10 volume; Amersham Pharmacia Biotech, Uppsala, Sweden) were added for 1 h, and bound proteins were pelleted. Pellets were suspended in sample buffer, boiled for 10 min, and separated by SDS-12% PAGE. Gels were stained with 0.1% Coomassie containing 50% methanol and 1% acetic acid. The P1 band was excised, washed, and then digested overnight at 37°C with 0.2 mg of trypsin (Promega, Madison, Wis.) in Tris, pH 9.2. Peptides were eluted with 1% trifluoroacetic acid in 60% acetonitrile. The extracted materials were submitted to the Protein Chemistry Sequencing Laboratory at Texas A&M University for sequencing. Samples, first separated by reverse-phase high-pressure liquid chromatography and analyzed by matrix-assisted laser desorption ionization mass spectroscopy, were selected for peptide sequencing based on size and relative purity.
Cloning of the P1 gene.
PCRs (100-μl mixture volumes) were performed in a DNA Thermocycler (Biometra, Tampa, Fla.) with Taq DNA polymerase (Gibco-Life BRL, Grand Island, N.Y.). Degenerate primers were purchased from Genosys Biotechnologies Inc. (Woodlands, Tex.) and used at a final concentration of 0.5 mM. Amplification consisted of an initial 5-min denaturation step at 95°C followed by 40 cycles of 30 s at 95°C, 1 min at 30°C, and 15 s at 70°C. PCR products were separated in a 1% agarose gel and purified with a gel extraction kit (Qiagen, Valencia, Calif.), ligated into pCR-2.1-TOPO (Invitrogen, Carlsbad, Calif.), and transformed into Top10F′ cells. Plasmid DNA from clones was sent to the Gene Technology Laboratory at Texas A&M University for sequencing.
Inverse PCR was used to obtain the N-terminal and C-terminal regions of the gene. C. burnetii Nine Mile total DNA (5 μg) was digested with HincII at 37°C for 3 h according to the manufacturer's protocol. The resulting fragments at a concentration of 0.5 μg/ml were ligated at 12°C for 16 h in 200 μl of ligation buffer and 1 U of T4 ligase (Gibco-Life BRL) per ml. The ligated sample was then phenol-chloroform extracted, and DNA was ethanol precipitated. The circularized DNA (0.1 μg) was used in PCR mixtures containing 50 pmol of primers (synthesized by Genosys), 500 mM deoxynucleoside triphosphates, and Taq polymerase (Gibco-Life BRL). The cycling consisted of 30 repetitions of denaturation at 94°C for 1.5 min, annealing at 48°C for 1 min, and extension at 70°C for 4 min. The amplified fragment was cloned into pCR2.1-TOPO (Invitrogen) and sent to the Gene Technology Laboratory at Texas A&M University for sequencing. Results were aligned based on the partial sequence data.
Southern blotting.
Genomic DNA from C. burnetii was digested with restriction enzyme according to the manufacturer's protocol (Boehringer Manheim, Indianapolis, Ind.). DNAs were then electrophoresed through 0.8% agarose gels and transferred to nitrocellulose membrane. Labeling of a DNA probe with [α-32P]dCTP was carried out with a random DNA labeling Decaprime II kit (Ambion, Austin, Tex.). Blots were incubated with the radiolabeled probe overnight at 65°C and then washed four times at modest stringency for 30 min each at 50°C with 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% SDS. Blots were analyzed for hybridization patterns using a PhosphorImager and appropriate software (model SF; Molecular Dynamics, Sunnyvale, Calif.).
Expression and purification of P1 fusion protein.
The pBAD-Thio-TOPO expression system (Invitrogen) was used to express recombinant P1 peptide. Briefly, a P1 gene was PCR amplified as described earlier but with amplification consisting of a 2-min denaturation step at 94°C followed by 39 cycles of 1 min at 94°C, 2 min at 45°C, and 3 min at 72°C. The purified PCR product was then ligated into the vector and transformed into LMG194 cells. Restriction digest analysis of resulting clones indicated that pSV115 had the gene in the correct orientation. Cultures of these clones were grown overnight in RM medium containing glucose and 100 μg of ampicillin per ml at 37°C with shaking. The next day 50 ml of the same medium was inoculated with 0.5 ml of the overnight culture and allowed to grow to an optical density at 600 nm of 0.5. Cultures were then induced with 0.2% l-arabinose, grown for another 4 h, and harvested by centrifugation at 3,000 × g for 5 min. The pellet was suspended in 10 ml of lysis buffer (6 M guanidine hydrochloride, 20 mM sodium phosphate, 500 mM sodium chloride; pH 7.8), incubated at room temperature for 10 min with rocking, and then sonicated. After removal of the insoluble debris by centrifugation, the lysate was applied to a pre-equilibrated ProBond resin column. The column was washed four times with denaturing buffers (8 M urea, 20 mM sodium phosphate, 500 mM sodium chloride) with decreasing pH: 7.8, 6.0, 5.3, and 4.5. Protein was subsequently eluted with 8 M urea-20 mM sodium phosphate-500 mM sodium chloride at pH 4.0. One-milliliter fractions were collected and frozen until SDS-PAGE and Western analysis.
Protein purification for porin analysis.
One milligram of C. burnetii Scurry or Nine Mile phase I was solubilized in 1% Empigen BB plus 10 mM EDTA plus 50 mM NaCl at 37°C for 4 h. The soluble proteins were separated from the insoluble fraction by centrifugation at 28,500 × g for 30 min. Soluble proteins were then precipitated with 1% trichloroacetic acid at −20°C followed by neutralization with 0.1 N NaOH. Pellets were suspended in PBS and then incubated overnight at 4°C with an equal volume of P1 monoclonal antibody 4E8. Gamma Bind Plus beads (1/10 volume; Amersham Pharmacia Biotech) were added for 1 h to immunoprecipitate protein, and bound protein was pelleted by centrifugation. P1 protein was then removed from Gamma Bind Plus beads with 100 mM glycine, pH 5.0. For channel analysis of immunoprecipitated protein, an aliquot of this sample (preparation I) was added to the cis chamber, where it was diluted 1,000-fold in 1 M NaCl plus 10 mM HEPES (pH 7.5).
For channel analysis of the trimer, the immunoprecipitated proteins were pelleted and separated by SDS-15% PAGE (preparation II). The band corresponding to the predicted P1 trimer size was excised, gel slice crushed, and eluted in water overnight at 4°C. The resulting protein was then separated from acrylamide fragments in an Ultrafree MC 0.45-μm filter unit (Millipore, Bedford, Mass.) by centrifugation at 22,000 × g for 5 min at room temperature. The bathing solution for trimer studies was 1 mM KCl-10 mM Tris-10 mM KPO4, pH 7.4.
Bilayer recordings.
Bilayer membranes were formed from a 25-mg/ml pentane suspension of 1,2-diphytanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids, Alabaster, Ala.) by the method of Montal and Mueller (26) on a 100- to 150-mm-diameter orifice in a 25-mm-thick Teflon film (Goodfellow Corp., Malvern, Pa.) separating the two chambers of a planar bilayer apparatus. Each chamber contained 2 ml of the specified solution.
Potential difference was applied across the bilayer, and currents were recorded with a model 3900A integrated patch clamp amplifier (Dagan Corp., Minneapolis, Minn.) connected to the chambers via Ag/AgCl electrodes in 3 M KCl and 1.5% agarose (Bio-Rad) bridges. The cis chamber was at virtual ground; thus, a positive potential indicates a higher potential in the trans chamber, and a positive current is one in which cations flow from the trans to the cis chamber. The built-in four-pole Bessel filter of the amplifier was set at 5 kHz. Data were acquired on a digital audiotape (DAS-75; Dagan Corp.), low-pass filtered with an eight-pole Bessel filter (model 902; Frequency Devices, Haverhill, Mass.) at 0.1 to 1 kHz (−3dB), and sampled by a computer at 5 to 20 kHz (with a Digidata 1200 A/D converter; Axon Instruments, Union City, Calif.).
Data analysis.
Data were analyzed with pCLAMP 8 software (Axon Instruments) and presented with Origin 6.0 (Microcal, Northampton, Mass.). In a few experiments, single channels in the bilayer allowed detection of discrete, usually noisy, steps, and their amplitude was measured using the cursor measurement options in the Fetchan and Clampex programs of pCLAMP 9. In most experiments, more than one channel probably existed in the bilayer, resulting in high noise and interfering with accurate measurements of the smaller discrete conductance steps. All-point amplitude histograms of the current did not always give a resolution of individual steps, and the current noise was occasionally obscured by the high channel noise.
RESULTS
Differential expression of P1 on cell variants.
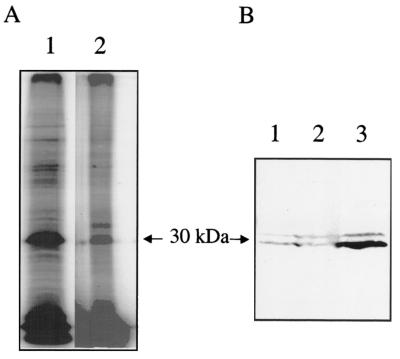
A previous study demonstrated by immunogold electron microscopy and Western blotting that developmental variants differentially expressed P1 (22). That study separated cell variants based on sensitivity to osmotic lysis and pressure lysis (French pressure cell). They reported that LCV expressed abundant P1, SCV expressed less P1 than LCV, and SDC did not express detectable P1. Our group and other investigators have employed isopycnic gradients to obtain two populations that appear to be comparable to LCV and SCV from this earlier study (7-9, 34, 35, 43). It should be noted that SCV obtained by isopycnic gradients likely contain both SCV and SDC obtained by the earlier lysis technique. To determine if P1 is differentially expressed by LCV and SCV obtained by isopycnic gradients, we employed surface iodination as well as Western blotting techniques. Surface-iodinated whole cells were applied to a cesium chloride gradient in order to separate cell variants based on respective densities (43). Equal amounts of cells by weight were then separated by SDS-PAGE. The most notable difference between the LCV and SCV was the greater intensity of an iodinated LCV protein corresponding in molecular mass to P1 (∼29 kDa) (Fig. 1A). An additional notable difference was an ∼34-kDa antigen labeled exclusively in SCV, but this difference was not consistently seen on repeated experiments. For Western analysis of the apparent differential expression, whole cells were also separated by density gradient centrifugation. Following SDS-PAGE and immunoblotting, membranes were reacted with a P1 monoclonal antibody (4E8). Two immunoreactive bands were noted, ∼29 and ∼31 kDa. Repeated experiments confirmed that there was consistently qualitatively more 29-kDa MOMP in the LCV population than in SCV (Fig. 1B, lanes 3 and 2, respectively). As noted above, the potential P1 null variant form, SDC, is likely mixed with SCV in these experiments.
FIG. 1.
SDS-PAGE and Western blot analysis of life cycle variants. (A) Iodinated LCV (lane 1) and SCV (lane 2) were separated by SDS-12.5% PAGE. (B) LCV (lane 3), SCV (lane 2), and whole cells (lane 1) were separated by SDS-12.5% PAGE, transferred to nitrocellulose, and probed with monoclonal antibody 4E8.
Purification and generation of peptide fragments.
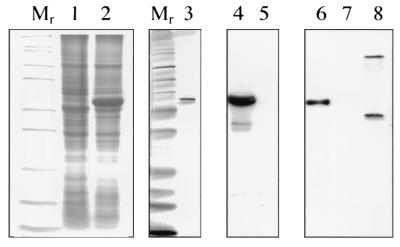
Previous attempts to identify clones encoding P1 by screening libraries with monoclonal antibodies against the MOMP resulted in the cloning of a 27-kDa lipoprotein, designated P2 (12). Use of these monoclonal antibodies to screen multiple phage C. burnetii banks also proved unsuccessful in our lab. We directed our attention to a strategy for generation of P1 peptide sequence that could be used to predict degenerate oligonucleotides to screen DNA libraries or for PCR amplification of DNA. To develop a method to purify the protein to a level acceptable for sequence analysis, a number of detergents were tested for the ability to preferentially solubilize P1. When successive extraction with Empigen BB at increasing temperatures was used, a protein corresponding in molecular weight to P1 was solubilized and highly enriched from intact C. burnetii (Fig. 2, lane 2). Immunoreactivity of the partially purified protein with P1 monoclonal antibody (4E8) was verified by Western blot (data not shown). Following SDS-PAGE, the isolated protein was transferred to a polyvinylidene difluoride membrane, stained, excised from the membrane, and sent to the Texas A&M Protein Chemistry Sequencing Laboratory for N-terminal sequencing. A 20-amino-acid sequence was obtained (Fig. 3) which showed no significant homology by BLAST analysis with proteins in the ENTREZ database.
FIG. 2.
Detergent purification of P1. Proteins solubilized in Empigen or SDS by consecutive detergent extraction at increasing temperatures were separated by SDS-PAGE on a 12.5% acrylamide gel and silver stained. Lane 1, Empigen extraction at 37°C; lane 2, Empigen extraction at 60°C; lane 3, SDS extraction at 100°C; lane 4, molecular weight marker; lane 5, starting material.
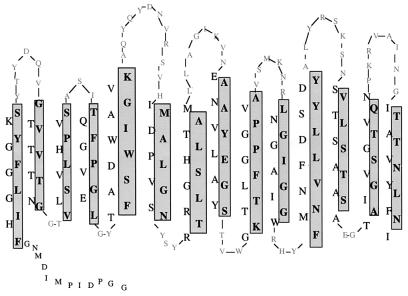
FIG. 3.
Amino acid sequence of P1 peptide. N-terminal and internal peptides derived from amino acid sequencing are indicated in bold with derived degenerate oligonucleotide primers listed beneath. The predicted amino acid sequence is based on PCR clone pSV101.
A series of low-degeneracy oligonucleotides designed from the N-terminal amino acid sequence was used to screen bacteriophage libraries for putative P1 gene clones. This technique did not identify strongly positive clones. Alternatively, we obtained internal peptides sequence data to develop a larger fragment by PCR for use in screening libraries. After purification of C. burnetii as described above, the additional step of immunoprecipitation with P1 monoclonal antibodies (4E8 and 4D6) was included to isolate P1 to near homogeneity. The resulting band in SDS-PAGE was excised and digested with trypsin overnight. Eluted P1 peptides were sent to the Texas A&M Protein Chemistry Sequencing Laboratory, and two peptides separated by reverse-phase chromatography were chosen for sequence analysis.
Cloning and expression of P1.
Degenerate primers were designed based on the amino acid sequence obtained from N-terminal analysis of complete mature P1 and one internal peptide fragment (Fig. 3). PCR amplification of genomic C. burnetii Nine Mile DNA with primers from the N-terminal and internal peptide sequences resulted in the generation of a 570-bp product, which was cloned into pSV101. Sequencing indicated that the PCR product contained codons corresponding to the amino acids encoded by the primers as well as the 12 amino acids from the N-terminal analysis not incorporated in primer design (Fig. 3). This partial clone of P1 represented approximately two-thirds of the predicted mature gene.
Attempts to screen gene libraries for putative clones with this gene fragment were not successful, although this fragment did hybridize to a single restriction enzyme (EcoRI)-digested DNA fragment on Southern blots with C. burnetii chromosomal DNA. We employed an inverse PCR technique to obtain the remaining C-terminal portion of the gene, as well as the N-terminal signal sequence and potential promoter regions. HincII-digested C. burnetii fragments were self-ligated and then used in inverse PCRs. Sequence analysis of the resultant 1.1-kb fragment indicated that both regions were cloned (AY082614).
Based on sequence analysis of this cloned region, the P1 gene contained an open reading frame of 756 bp encoding 252 amino acids. The G+C content was 44%, consistent with the 43- to 45-mol% G+C content of the C. burnetii genome (27). BLAST comparison identified no sequences with striking similarity. Based on the TESS program (Baylor College of Medicine Search Launcher [http://searchlauncher.bcm.tmc.edu]), a putative ribosome binding site with the sequence GGAGA was located between 7 and 11 nucleotides upstream of the initiating methionine codon. Additionally, a 23-amino-acid signal peptide sequence, which was consistent with N-terminal sequence analysis of the MOMP, was also predicted. From these results we predict the mature P1 protein to have a molecular mass of 24,515 Da and a theoretical isoelectric point of 8.7.
To determine whether P1 sequence is conserved among different strains of C. burnetii, Southern blot analysis of total DNA from various C. burnetii isolates was carried out. This analysis demonstrated a single hybridizing band under modestly stringent conditions (approximately 15% mismatch allowed) in all isolates (data not shown). To understand the level of variation among isolate groups, we cloned and sequenced the mature genes from the Koka, Kerns, and Scurry isolates and compared these to the sequence from the Nine Mile isolate (18). We expected minor differences, with greatest variation occurring between isolates from chronic disease patients (Kerns and Scurry) and isolates that have been grouped with acute-disease-causing organisms (Nine Mile and Koka). The P1 genes from Koka and Nine Mile were highly conserved, with only one base pair difference (Fig. 4). The Kerns isolate contained the most differences, with 31 base pair changes altering 19 amino acids, and the Scurry isolate contained some of the variations found in the Kerns isolate.
FIG. 4.
Comparison of the predicted amino acid sequence by Clustal W for the cloned P1 gene from four strains (Nine Mile, Koka, Scurry, and Kerns) of C. burnetii. The bottom sequence is the consensus.
Efforts to express recombinant P1 from the native promoter were unsuccessful. P1 peptide was not expressed as a fusion protein in pBAD-TOPO but was expressed in pBAD-Thio-TOPO, which uses the fusion peptide partner thioredoxin to facilitate folding of membrane proteins (pSV108 and pGF101). A fusion peptide containing the entire mature peptide was purified under denaturing conditions with a nickel bond column to absorb C-terminal polyhistidine residues. By Western blot analysis, the P1-thioredoxin fusion peptide reacted with P1-specific monoclonal antibodies (4E8 and 4D6) (Fig. 5, lanes 3, 4, and 6).
FIG. 5.
Expression of pBAD-Thio-TOPO clone fusion protein. The open reading frame encoding the entire mature P1 protein was cloned into the fusion vector to generate pGF101 and transformed into E. coli Top10F′. Expression of fusion protein was monitored by Coomassie blue staining of induced (lane 2) and uninduced (lane 1) cultures and by Western blotting with P1-specific monoclonal antibody (4E8) (induced [lane 4] and uninduced [lane 5]) in SDS-12% PAGE. Fusion protein was purified by His-tagged nickel affinity chromatography and analyzed for purity by silver staining (lane 3) and Western blotting with P1-specific monoclonal antibody (4E8) (lane 6) compared with an irrelevant thioredoxin fusion protein (lane 7) and C. burnetii whole cells (lane 8). Note that P1 was detected as a monomer (∼29 kDa) and a trimer (∼90 kDa).
Predicted topology of P1.
Analysis of the obtained P1 sequence using the Protein Sequence Analysis server (http://bmirc-www.bu.edu/psa) indicated that mature P1 was predicted to contain over 95% β-sheet confirmation (data not shown). This finding was in agreement with original speculations of the possible biological role of P1 as a porin (4). A model of how P1 may reside in the outer membrane (Fig. 6) was developed based on the following assumptions of known characteristics of bacterial porins (15, 33): (i) transmembrane β strands were primarily amphipathic, (ii) periplasmic turns were short and contained turn promoters such as D, N, E, G. P, and S, and (iii) external loops were generally long and associated with sequences likely to be antigenic. Our model predicts a higher proportion of charged residues within the third antigenic loop than any other extracellular domain, as is consistent with other porins whose primary antigenic determinants reside within this same region (32, 36). We found a large number of charged amino acids residing within the predicted pore of the protein. Because of the ratio of positive to negative amino acids within this channel, we predict that P1 may have anion selectivity and plan to experimentally test this possibility.
FIG. 6.
Predicted membrane topology of P1. Large letters indicate amino acids embedded within the membrane, with boxed regions indicating amino acids directly in contact with the hydrophobic membrane and unboxed regions indicating those lining the pore itself.
Planar lipid bilayer reconstitution.
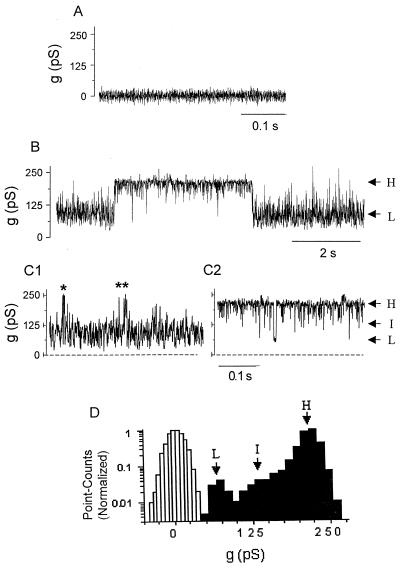
The predicted secondary structure from the P1 sequence provided indirect evidence that P1 may function as a porin. Porin activity was tested by electrophysiological measurements in a planar lipid bilayer. Before addition of P1, the bilayer exhibits zero conductance. No conductance change occurred upon addition of the “carrier” buffer containing Empigen (final concentration, 0.01%) and antibody to the cis chamber of the bilayer apparatus (Fig. 7A). In contrast, increase in conductance, indicating incorporation into the bilayer was seen several minutes after the addition of immunoprecipitated P1 (diluted 1:1,000, with a final Empigen concentration of 0.001%). Conductance through the pore was noisy and fluctuated rapidly but primarily existed as one of two predominant substates of either low conductance (65 to 75 pS) or high conductance (210 to 230 pS) (Fig. 7B). Closer inspection of the conductance traces at faster time sweep (Fig. 7C) revealed an intermediate conductance step of ∼130 to 140 pS. When the low conductance predominated (Fig. 1), fast transitions to the intermediate level contributed to the large noise of this level. Occasional transitions to the high level can be resolved. The noise in the stretches of the high conductance level (Fig. 2) appear to arise from fast transitions to the intermediate level, with occasional transitions to the low level. Figure 7D shows an all-point amplitude histogram of the predominant high conductance stretch (from Fig. 2), with a major peak at ∼225 pS, and smaller, less resolved peaks at ∼130 pS and 70 pS, and a histogram, with a peak at zero, obtained from control experiments (Fig. 7A).
FIG. 7.
Porin activity of P1 in the planar lipid bilayer. (A) The addition of the carrier solution, containing Empigen plus antibody without P1, did not result in increase conductance. (B) Representative current traces recorded after application of immunoprecipitated P1 to the cis chamber of an artificial bilayer. The currents fluctuate between two main conductances, a low conductance of ∼70 pS (L) and a high conductance of 210 pS (H). (C) Parts of the trace from panel B on an expanded time scale, with the low conductance state predominating (C1) and with the high conductance state predominating (C2). The noise in the current trace in C1 appears as fast transitions between the low conductance level (L) and an intermediate level (I) of ∼120 pS. In C2, fast transitions from the main high conductance to the intermediate level with a few transitions to the low level can be observed. Occasional transitions to the high level are indicated with asterisks. (D) Conductance distribution. An all-point amplitude histogram of the trace shown in panel B is presented (dark bars). The peak representing the high conductance level (H) predominates. The smaller peaks represent the less abundant intermediate (I) and low (L) conductance levels. The open histogram is of the trace shown in panel A with no P1, with a peak around 0 pS. Conditions were as follows: 1 M NaCl-10 mM HEPES, pH 7.5, with a holding potential of +80 mV.
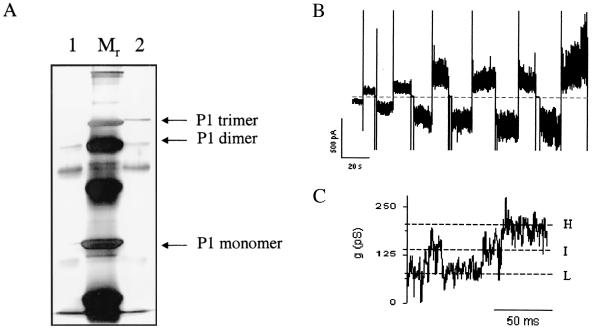
To specifically test whether the trimer had pore-forming abilities, the immunoprecipitated P1 still bound to Gamma Bind Plus beads was separated by SDS-15% PAGE (Fig. 8A). The band corresponding to the predicted P1 trimer size was eluted and applied to the planar lipid bilayer apparatus. Figure 8B depicts current traces from a bilayer containing multiple channels at various voltages. Figure 8C shows current traces from a different experiment (using the same trimer extraction method) with fewer channels in the bilayer (presumably a single channel). At applied potential of +40 mV, three current levels can be detected. The highest level may correspond to the main conductance level (−200 pS), while the lower levels represent the flow through one (∼60 to 70 pS) or two (∼120 to 130 pS) monomers. Both of these values are comparable to what was obtained with the immunoprecipitated protein that was not eluted from the gel.
FIG. 8.
Activity of a gel-purified P1 trimer. (A) Immunoprecipitated P1 was separated by SDS-PAGE on a 15% gel. Lane 1, protein boiled in sample buffer for 10 min; lane 2, protein solubilized in sample buffer at room temperature. The band representing the P1 trimer was excised and eluted in water overnight at 40°C. (B and C) P1 trimer was added to the cis chamber of a bilayer setup, resulting in increase conductance. (B) Bilayer containing multiple channels. The current is in response to alternating negative and positive potentials (from 10 to 60 mV in 10-mV increments). (C) Currents recorded from a bilayer with presumably a single trimer at an applied potential of +40 mV. Three conductance levels can be detected (dashed lines) representing the conductances of one monomer in the trimer (L), two monomers (I), and three monomers (H). The recording solution was 1 M KCl-10 mM KPO4, pH 7.4.
DISCUSSION
Previous attempts to clone P1 by screening with monoclonal antibodies were not successful (D. Waag, personal communication). Therefore, we employed a reverse genetic approach to obtain a protein sequence that would allow screening of DNA libraries with degenerate oligonucleotides for putative clones. Earlier MOMP purification strategies employed techniques to solubilize membranes in SDS at high temperatures and separate the soluble material by size chromatography (4; Snyder and Williams, Abstr. Annu. Meet. Am. Soc. Microbiol.). This method required large amounts of bacteria and resulted in only a partially purified P1. A study conducted by Lukacova et al. showed purification of two membrane proteins of 29 and 40 kDa, using the zwitterionic detergent Empigen (17). Although these proteins were subsequently found to be protective in mouse vaccine studies, researchers did not identify the lower-molecular-mass protein as P1. Using this detergent, we developed a purification protocol that enabled us to generate a peptide sequence that was ultimately used to clone the full gene. The mature protein encoded by the P1 gene is predicted to migrate with a mass that is approximately 5 kDa smaller than what was calculated by SDS-PAGE. The increased apparent size may be explained by a tight association between peptidoglycan and P1, as was noted for the MOMP of Legionella pneumophila (10, 11). Originally, the L. pneumophila MOMP was thought to be a hetero-oligomeric protein, composed of 28- and 31-kDa subunits, but protein mapping revealed that the two peptides were identical with the addition of peptidoglycan to the larger form. An alternative explanation for the size discrepancy of P1 could be that the protein is not fully denatured, a phenomenon similar to that observed with the N-terminal and C-terminal regions of OmpA of E. coli. Only by heating OmpA in SDS at 100°C will both domains be denatured, resulting in a slower gel migration, presumably because of the expanded conformation (39). Finally, it has been determined that pretreatment of purified P1 protein with formic aid prior to PAGE analysis results in an immunoreactive P1 protein migrating at approximately 24 kDa (C. Snyder, personal communication).
P1 protein demonstrated characteristics common to many porins of gram-negative bacteria. These shared features include a typical signal peptide sequence that is cleaved from the mature protein, amphipathic antiparallel β sheets, an abundance of polar residues, and a C-terminal phenylalanine (47). The natural abundance, surface localization, detergent solubility, and trimer stability in SDS of P1 provide additional indications that the biological function of P1 is as a porin. Studies conducted by Banerjee-Bhatnagar et al. that tested the ability of a partially purified sample of P1 to induce liposomal swelling indicated the presence of a pore-forming protein (4). Our studies indicate that immunoprecipitated MOMP is capable of forming a pore within a planar lipid bilayer, thus confirming its role as a porin. Similar to other bacterial porins, P1 exhibits a trimer conformation that is also incorporated into a bilayer.
These porin studies were conducted with native protein. The difficulty of working with this intracellular pathogen and the multiple steps involved in the extraction of the protein placed a limit on the amount of protein generated. Expression of the full recombinant protein may provide a more readily accessible supply for porin analysis. Work with other bacterial porins has indicated that toxicity can result from foreign porin expression in E. coli. Although several cloning vectors were unable to confer detectable P1 expression, the tightly regulated, folding assisting fusion vector, pBAD-Thio-TOPO enabled adequate expression of the fusion protein for purification using denaturing conditions. The adaptation of an efficient nondenaturing purification protocol for this fusion protein will allow testing of the porin activity from a recombinant source.
Additional characterization of porin activity for P1 will aid in understanding the role of differential expression by C. burnetii cell variants. Immunoprecipitated P1 exhibited several conductance levels, with a maximum of 225 pS (Fig. 7A). Expanded analysis of the conductance suggested that porin activity switched between three states, each approximately 70 pS, which would indicate a relatively small channel. We speculated that this represents states of opening for a monomer (70 pS), dimer (∼145 pS), and trimer (225 pS). The conductance levels could also have been affected by the presence of a monoclonal antibody that partially blocked pore activity. Therefore, we isolated trimeric P1 after separation by SDS-PAGE from monomers, dimers, and precipitating antibody (Fig. 8A). P1 trimer prepared in this manner also exhibited porin activity with a similar three-level activity. This suggests that monoclonal antibodies 4E8 and 4D6 do not interfere with porin activity and that each monomer of the trimeric P1 had the ability to open and close independently, although potentially in a cooperative manner.
Ionic and size selectivities of the channel, as well as the effect pH may have on the porin activity of P1, are important issues to address given the acidified but nutrient-rich phagolysosomal environment within which C. burnetii resides. Several porins of Escherichia coli, OmpF, OmpC, and PhoE, have been studied under conditions of extreme pH. Benz et al. saw a correlation with increased channel size and low pH (5), while Schindler and Rosenbusch correlated increased conductance value with increased pH (31). Specifically looking at low versus high pH, Todt et al. observed that small channels were stabilized in this acidic environment whereas basic environments stabilized larger channels (40). One may speculate that since the “metabolic home” of C. burnetii is in the acidified phagolysosome, P1 channel activity may be opposite that of E. coli, with the porin developing a more open state under acidic conditions.
The cloning of the differentially expressed MOMP provides an exciting opportunity to more clearly define the roles of developmental cycle variants. It will be interesting to determine why one variant (LCV) requires higher expression of P1 porin activity and why SDC appear not to require this porin. It is logical to surmise that SDC are metabolically down-regulated and do not require the substrates that enter via this porin (23). The general diffusion porins OmpF and OmpC of E. coli primarily use the growth environment as the source for a signal to regulate expression. Since general vacuole nutrients are available to both variants, it seems unlikely that sampling of a specific nutrient(s) would provide the signal to differentially regulate P1 expression in C. burnetii. One attractive alternative signal might be by-products of an active metabolism, since LCV are more metabolically active than the SCV, signaling the need for more substrate to enter via this porin.
Cloning and expression of P1 recombinant protein will also allow evaluation of the value pf P1 as a subunit vaccine and diagnostic target using enzyme-linked immunosorbent assay or alternate serological monitoring of infection. Current diagnostic tools employ inactivated phase I and phase II organisms, requiring biosafety level 3 containment to prepare antigen, to monitor specific immune responses during infection. Our preliminary studies with infected mice suggest that animals recognize P1 early in infection as an immunodominant antigen.
Acknowledgments
This work was supported by Public Health Service grants AI37744 and AI448191 from the National Institute of Allergy and Infectious Diseases.
We thank David Waag, USAMRIID, for providing monoclonal antibodies against P1, Larry Harris-Haller, Gene Technology Laboratory, Texas A&M University, for sequence analysis, and Jon Skare for advice with P1 structural prediction.
Editor: B. B. Finlay
REFERENCES
- 1.Amano, K., and J. C. Williams. 1984. Sensitivity of Coxiella burnetii peptidoglycan to lysosome hydrolysis and correlation of sacculus rigidity with peptidoglycan-associated proteins. J. Bacteriol. 160:989-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ascher, M. S., M. A. Berman, and R. Ruppanner. 1983. Initial clinical and immunologic evaluation of a new phase I Q fever vaccine and skin test in humans. J. Infect. Dis. 148:214-222. [DOI] [PubMed] [Google Scholar]
- 3.Baca, O. G., and D. Paretsky. 1983. Q fever and Coxiella burnetii: a model for host-parasite interactions. Microbiol. Rev. 47:127-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee-Bhatnagar, N., C. R. Bolt, and J. C. Williams. 1996. Pore-forming activity of Coxiella burnetii outer membrane protein oligomer comprised of 29.5- and 31-kDa polypeptides. Ann. N. Y. Acad. Sci. 791:378-401. [DOI] [PubMed] [Google Scholar]
- 5.Benz, R., R. E. W. Hancock, and T. Nakae. 1982. Outer membranes. In R. Antolini (ed.), Transport in biomembranes: model systems and reconstitution. Raven Press, New York, N.Y.
- 6.Brouqui, P. 1993. Chronic Q fever. Arch. Intern. Med. 153:642-648. [DOI] [PubMed] [Google Scholar]
- 7.Heinzen, R. A., and T. Hackstadt. 1996. A developmental stage-specific histone H1 homolog of Coxiella burnetii. J. Bacteriol. 178:5049-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinzen, R. A., T. Hackstadt, and J. E. Samuel. 1999. Developmental biology of Coxiella burnetii. Trends Microbiol. 7:149-154. [DOI] [PubMed] [Google Scholar]
- 9.Heinzen, R. A., D. Howe, L. P. Mallavia, D. D. Rockey, and T. Hackstadt. 1996. Developmentally regulated synthesis of an unusually small, basic peptide by Coxiella burnetii. Mol. Microbiol. 22:9-19. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman, P., J. Seyer, and C. Butler. 1992. Molecular characterization of the 28- and 31-kilodalton subunits of the Legionella pneumophila major outer membrane protein. J. Bacteriol. 174:908-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffman, P. S., M. Ripley, and W. Risini. 1992. Cloning and nucleotide sequence of a gene (ompS) encoding the major outer membrane protein of Legionella pneumophila. J. Bacteriol. 174:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoover, T. A., and J. C. Williams. 1990. A Coxiella burnetii gene encodes a 29-kD antigen with a bacterial lipoprotein signal sequence. Vaccines 90:447-450. [Google Scholar]
- 13.Kazar, J., R. Brezina, A. Palanova, B. Turda, and S. Schramek. 1982. Immunogenicity and reactogenicity of Q fever chemovaccine in persons professionally exposed to Q fever in Czechoslovakia. Bull. W. H. O. 60:389-394. [PMC free article] [PubMed] [Google Scholar]
- 14.Kishimoto, R. A., J. W. Johnson, R. H. Kenyon, M. S. Ascher, E. W. Larson, and C. E. Pederson, Jr. 1978. Cell-mediated immune responses of guinea pigs to an inactivated phase I Coxiella burnetii vaccine. Infect. Immun. 19:194-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koebnik, R., K. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 16.Labrenz, M., and P. Hirsch. The genus Coxiella. In G. Garrity, D. R. Boone, and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., in press. Springer-Verlag, New York, N.Y.
- 17.Lukacova, M., E. Gajdosova, L. Skultety, E. Kovacova, and J. Kazar. 1994. Characterization of protective effects of a 29 kDa protein isolated from Coxiella burnetii by detergent Empigen BB. Eur. J. Epidemiol. 10:227-230. [DOI] [PubMed] [Google Scholar]
- 18.Mallavia, L. P. 1991. Genetics of rickettsiae. Eur. J. Epidemiol. 7:213-221. [DOI] [PubMed] [Google Scholar]
- 19.Marmion, B. P., R. A. Ormsbee, M. Kyrkou, J. Wright, D. A. Worswick, A. A. Izzo, A. Esterman, B. Feery, and R. A. Shapiro. 1990. Vaccine prophylaxis of abattoir-associated Q fever: eight years' experience in Australian abattoirs. Epidemiol. Infect. 104:275-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurin, M., and D. Raoult. 1996. Optimum treatment of intracellular infection. Drugs 2:45-55. [DOI] [PubMed] [Google Scholar]
- 21.McCaul, T. F. 1991. The developmental cycle of Coxiella burnetii, p. 223-258. In J. C. Williams and H. A. Thompson (ed.), Q fever: the biology of Coxiella burnetii. CRC Press, Boca Raton, Fla.
- 22.McCaul, T. F., N. Banerjee-Bhatnagar, and J. C. Williams. 1991. Antigenic differences between Coxiella burnetii cells revealed by postembedding immunoelectron microscopy and immunoblotting. Infect. Immun. 59:3243-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCaul, T. F., T. Hackstadt, and J. C. Williams. 1981. Ultrastructural and biological aspects of Coxiella burnetii under physical disruptions, p. 267. In W. Burgdorfer and R. L. Anacker (ed.), Rickettsiae and rickettsial diseases. Academic Press, New York, N.Y.
- 24.McCaul, T. F., and J. C. Williams. 1981. Developmental cycle of Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. J. Bacteriol. 147:1063-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meiklejohn, G., and E. H. Lennette. 1950. Q fever in California. I. Observation on vaccine of human beings. Am. J. Hyg. 52:54-61. [PubMed] [Google Scholar]
- 26.Montal, M., and P. Mueller. 1972. Formation of bimolecular membranes from lipid bilayers and study of their electrical properties. Proc. Natl. Acad. Sci. USA 69:3561-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers, W. F., O. G. Baca, and C. L. Wisseman. 1980. Genome size of the rickettsia Coxiella burnetii. J. Bacteriol. 144:460-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raoult, D., and T. Marrie. 1995. Q fever. Clin. Infect. Dis. 20:489-496. [DOI] [PubMed] [Google Scholar]
- 29.Samuel, J. E. 2000. Developmental cycle of Coxiella, p. 427-440. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C..
- 30.Samuel, J. E., M. E. Frazier, and L. P. Mallavia. 1985. Correlation of plasmid type and disease caused by Coxiella burnetii. Infect. Immun. 49:775-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindler, H., and J. P. Rosenbusch. 1978. Matrix protein from Escherichia coli outer membranes forms voltage-controlled channels in lipid bilayers. Proc. Natl. Acad. Sci. USA 75:3751-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schirmer, T. 1998. General and specific porins from bacterial outer membranes. J. Struct. Biol. 121:101-109. [DOI] [PubMed] [Google Scholar]
- 33.Schulz, G. E. 2000. β-barrel membrane proteins. Curr. Opin. Struct. Biol. 10:443-447. [DOI] [PubMed] [Google Scholar]
- 34.Seshadri, R., L. R. Hendrix, and J. E. Samuel. 1999. Differential expression of translational elements by life cycle variants of Coxiella burnetii. Infect. Immun. 67:6026-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seshadri, R., and J. E. Samuel. 2001. Characterization of a stress-induced alternate sigma factor, RpoS, of Coxiella burnetii and its expression during the development cycle. Infect. Immun. 69:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simonet, V., M. Mallea, D. Fourel, J.-M. Bolla, and J.-M. Pages. 1996. Crucial domains are conserved in Enterobacteriaceae porins. FEMS Microbiol. Lett. 136:91-97. [DOI] [PubMed] [Google Scholar]
- 37.Smadel, J. E., M. J. Snyder, and F. C. Robbins. 1948. Vaccine against Q fever. Am. J. Hyg. 47:71-78. [DOI] [PubMed] [Google Scholar]
- 38.Snyder, C. E., R. F. Wachter, and J. D. White. 1984. Identification of Coxiella burnetii antigens and attempts at envelope fractionation. Microbiology 1984:263-265. [Google Scholar]
- 39.Sugawara, E., and H. Nikaido. 1992. Pore-forming activity of OmpA protein of Escherichia coli. J. Biol. Chem. 267:2507-2511. [PubMed] [Google Scholar]
- 40.Todt, J., E. Rocque, and E. McGroarty. 1992. Effects of pH on bacterial porin function. Biochemistry 31:10471-10478. [DOI] [PubMed] [Google Scholar]
- 41.Vivona, S., J. P. Lowenthal, S. Berman, A. S. Benenson, and J. E. Smadel. 1964. Report of a field story with Q fever vaccine. Am. J. Hyg. 79:143-153. [DOI] [PubMed] [Google Scholar]
- 42.Waag, D. M., M. J. England, and M. L. M. Pitt. 1997. Comparative efficacy of a Coxiella burnetii chloroform:methanol residue (CMR) vaccine and a licensed cellular vaccine (Q-Vax) in rodents challenged by aerosol. Vaccine 15:1779-1783. [DOI] [PubMed] [Google Scholar]
- 43.Wiebe, M. E., P. R. Burton, and D. M. Shankel. 1972. Isolation and characterization of two cell types of Coxiella burnetii. J. Bacteriol. 110:368-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilhelmsen, C. L., and D. M. Waag. 2000. Guinea pig abscess/hypersensitivity model for study of adverse vaccination reactions induced by Q fever vaccines. Comp. Med. 50:374-378. [PubMed] [Google Scholar]
- 45.Williams, J. C., M. R. Johnston, M. G. Peacock, L. A. Thomas, S. Stewart, and J. L. Portis. 1984. Monoclonal antibodies distinguish phase variants of Coxiella burnetii. Infect. Immun. 43:421-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams, J. C., T. A. Hoover, D. M. Waag, N. Banerjee-Bhatnagar, C. R. Bolt, and G. H. Scott. 1990. Vaccines against coxiellosis and Q fever. Ann. N. Y. Acad. Sci. 590:370-380. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Q., J. C. Meitzler, S. Huang, and T. Morishita. 2000. Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein. Infect. Immun. 68:5679-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]