Abstract
Commensal bacteria have emerged as an important disease factor in human Crohn's disease (CD) and murine inflammatory bowel disease (IBD) models. We recently isolated I2, a novel gene segment of microbial origin that is associated with human CD and that encodes a T-cell superantigen. To identify the I2 microorganism, BLAST analysis was used to identify a microbial homologue, PA2885, a novel open reading frame (ORF) in the Pseudomonas aeruginosa genome. PCR and Southern analysis identified Pseudomonas fluorescens as the originating species of I2, with homologues detectable in 3 of 13 other Pseudomonas species. Genomic cloning disclosed a locus containing the full-length I2 gene (pfiT) and three other orthologous genes, including a homologue of the pbrA/pvdS iron response gene. CD4+ T-cell responses to recombinant proteins were potent for I2 and pfiT, but modest for PA2885. pfiT has several features of a virulence factor: association with an iron-response locus, restricted species distribution, and T-cell superantigen bioactivity. These findings suggest roles for pfiT and P. fluorescens in the pathogenesis of Crohn's disease.
Human inflammatory bowel disease (IBD) represents a set of a chronic, relapsing, and remitting intestinal inflammatory disorders involving T-cell-mediated mucosal and mural destruction, with polygenic disease susceptibility (4, 13, 38). Although the etiology of these diseases remains uncertain, several lines of evidence implicate commensal bacteria as an important pathogenic element in clinical disease, particularly in Crohn's disease (CD). Patients with CD are sensitive to alteration of fecal flow, and some investigators have reported evidence of a clinical responsiveness to antibiotic therapy (21, 24, 34). In both pharmacologic and genetic animal models of IBD, development of disease is strictly dependent on the presence of enteric microflora (19, 29, 35-37, 40).
The identity of colitigenic bacterial species remains uncertain. In humans, a variety of bacterial and viral species have been implicated in CD, mainly on the basis of seroreactivity (3, 5, 8, 28, 44). However, CD patients typically express elevated levels of antibodies to many bacterial proteins, perhaps due to mucosal disruption and local immunologic challenge by diverse intestinal microflora. Mycobacterium has received attention as a candidate human pathogen, based on serologic evidence, but the presence of mycobacterial DNA in lesions is controversial (9, 12, 31, 32, 43, 47). Bacteroides vulgatus and Helicobacter hepaticus have been evaluated with various rodent IBD models, but other intestinal commensal organisms yet undefined are likely to account for the major colitigenic species (14, 25, 35, 36).
Our laboratory recently introduced subtractive cloning as a fresh approach to identify candidate organisms in human CD (10, 45). A novel microbial gene, I2, was isolated from lesional versus adjacent uninvolved colon tissues of a patient with CD. In a population-based study (45), the I2 gene was selectively detected in lesional CD colonic tissue, although it was also prevalent in both healthy and CD ileum. In that study, antibodies reactive with the recombinantly expressed protein also were detected in CD patients. The recombinant I2 protein also was found to be a murine CD4 T-cell superantigen (11). These lines of evidence suggest that the I2 protein and its originating bacterium might play roles in the pathogenesis of CD. The present study addresses the identity of the originating bacterium and the genomic locus bearing the I2 gene segment.
MATERIALS AND METHODS
Bacterial strains.
The Escherichia coli XL-1 Blue strain (Stratagene, La Jolla, Calif.) and TOP10 strain (Invitrogen, Carlsbad, Calif.) were used for all cloning and recombinant expression experiments. Thirteen clinical isolates of Pseudomonas spp. were obtained from the University of California—Los Angeles (UCLA) Clinical Laboratories (Table 1). In addition to UCLA 268, Pseudomonas fluorescens strains were obtained from the American Type Culture Collection (ATCC 13525; an environmental isolate from a water tower), and clinical isolates were obtained from the University of Illinois Chicago Medical Center (8-1, 8-3, 8-4, 8-5, 19-5, 19-8, 22-23, 25-39, 25-54, 25-55, 25-60, S40628-2). The latter were a generous gift from P. Schreckenberger. The isolates were from sputum, urine, cerebrospinal fluid, and foot wound sources. All Pseudomonas strains were grown on Trypticase soy agar (TSA) plates at 26°C. The identities of the UCLA and ATCC isolates were assessed and coded by biochemical criteria with an API 20NE analyzer (0147555 and 0147514 for the UCLA and ATCC isolates, corresponding to “excellent” and “good” identifications for P. fluorescens, respectively).
TABLE 1.
List of bacterial strains used in this study
| rRNA group and species | Identification by:
|
|
|---|---|---|
| I2 Southern blotting | I2 PCR | |
| Group I (Fluorescens group) | ||
| Pseudomonas aeruginosa | + | − |
| Pseudomonas putida | − | − |
| Pseudomonas fluorescens | + | + |
| Group I (Alcaligenes group) | ||
| Pseudomonas pseudoalcaligenes | + | − |
| Pseudomonas alcaligenes | − | − |
| Group I (Stutzeri group) | ||
| Pseudomonas mendocina | − | − |
| Pseudomonas stutzeri | − | − |
| Group II (Pseudomallei group) | ||
| Ralstonia picketti | − | − |
| Group IV (Diminuta group) | ||
| Bevundimonas diminuta | − | − |
| Bevundimonas vesicularis | − | − |
| Group V (Unknown nucleic acid homology) | ||
| Shewanella putrefaciens | + | − |
| Sphingomonas paucimobilis | − | − |
| Flavimonas oryzihabitans | − | − |
DNA manipulation.
The pCR2.1 TA cloning vector (Invitrogen, Carlsbad, Calif.) was used to clone PCR products according to the manufacturer's instructions. A His-tagged protein expression vector, pQE30 (Qiagen, Valencia, Calif.), was used to express protein in E. coli. Expression plasmids were constructed by inserting cloned fragments into the BamHI and SacI sites of the vector. All restriction enzymes used in this study were purchased from New England Biolabs (Beverly, Mass.).
Sequence analysis.
Plasmid clones were sequenced by dideoxy chain termination, assembled with the University of Wisconsin Genetics Computer Group (GCG) programs and the BLAST program (version 20.11, 20 January 2000), as well as the National Center for Biotechnology Information nonredundant database (2). Amino acid sequence alignment was performed with the ClustalW program at the EMBL Outstation European Bioinformatics Institute and with the GenDoc program (www.psc.edu/biomed/genedoc).
Cloning full-length pfiT and its adjacent genes.
Genome-walking (rapid amplification of cDNA ends [RACE] cloning) was employed to clone full-length pfiT gene from P. fluorescens genome (Universal GenomeWalker kit; Clontech, Palo Alto, Calif.). Reverse and forward GSP oligonucleotide pairs were designed to obtain upstream and downstream flanking sequences of the primary I2 gene fragment, respectively (Table 2). Genomic DNA of P. fluorescens was amplified on a GeneAmp PCR system 9700 (PE Applied Biosystem) in 50 μl with 1 μg of genomic DNA, 0.5 U of Taq polymerase, 2 mM deoxynucleoside triphosphates (dNTP), and 1 μM primers. Primary PCR was done with 7 cycles of 94°C for 2 s and 70°C for 3 min, followed by 32 cycles of 94°C for 2 s, 65°C for 3 min, and then 65°C for 4 min. Secondary PCR was performed with 5 cycles of 94°C for 2 s and 70°C for 3 min, followed by 20 cycles of 94°C for 2 s, 65°C for 3 min, and 65°C for 4 min. The prevalence of the I2 gene segment of pfiT and a downstream segment in 13 representative Pseudomonas strains was detected by PCR (Table 1). The PCR was performed for 5 min at 95°C, with 30 cycles of 95°C for 60 s, 65°C for 60 s, 72°C for 60 s, and 72°C for 5 min.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′→3′) | Purpose in this study |
|---|---|---|
| 5′-PA2885 | ATTGGATCCATGCTGGAGCTGGTGGCTACCGGACAGCT | PA2885 protein expression |
| 3′-PA2885 | TATGAGCTCTCAGGCGTTCTTGATCACCA | PA2885 protein expression |
| pfiT-5′ | ATTGGATCCATGCGCACCATGGTCGACAGT | PFTR protein expression |
| pfiT-3′ | TATGAGCTCCTCAGTCCGACTTCAGCACCATCAAC | PFTR protein expression |
| I2-BamHI | ATTGGATCCGATCTGGCCAGCGCCGTGGGCATCCA | I2 protein expression |
| I2-SacI | TATGAGCTCTCAGATCTGCTCATACACGTCA | I2 protein expression |
| I2-5′ | TCTGCTCATACACGTCACG | I2 PCR detection |
| I2-3′ | CCGTGGGCATCCAGTCCG | I2 PCR detection |
| pfiT-DF | TGACCCTTGAGGAGTTGGCCGAAGA | pfiT downstream region |
| pfiT-DR | AATCCAGCGCCGACATATGTGGGTAA | pfiT downstream region |
| 5′-pfiT-GSP1 | ATCATCGCGGTGTTGTAATGGATGGTTTCCTCCAT | pfiT upstream cloning |
| 5′-pfiT-GSP2 | ATCTCATCCTTGCTCTTGAAGTGATGAAAGATGCT | pfiT upstream cloning |
| 3′-pfiT-GSP1 | TGATCCGCTGCGAGTTGCAGTCGATCATG | pfiT downstream cloning |
| 3′-pfiT-GSP2 | AGTGGCGAGGCCATGGCGGTGCTGGTCTACGAAT | pfiT downstream cloning |
| 3′-pfiT-GSP-A | AGCATCTTTCATCACTTCAAGAGCAAGGATGAGAT | pfiT downstream cloning |
| 3′-pfiT-GSP-B | ATGGAGGAAACCATCCATTACAACACCGCGATGAT | pfiT downstream cloning |
Southern blot analysis.
Genomic DNA samples were restricted with EcoRI, electrophoresed on a 0.8% agarose gel, denatured in a mixture of 0.5 M Tris-Cl and 1.5 M NaOH, neutralized, and transferred onto Hybond N+ membrane (Amersham) by capillary blotting. Membranes were equilibrated in Rapid-Hyb buffer (Amersham) and then hybridized at 65°C for 6 h in the same buffer with 32P-labeled primary I2 segment or pfiT downstream fragment (nucleotides 1108 to 2630) prepared from the P. fluorescens genome by I2-5′/I2-3′ and pfiT-DF/pfiT-DR primer pairs, respectively (Table 2). Membranes were washed with 2× standard saline citrate (SSC) in 0.1% sodium dodecyl sulfate (SDS) at room temperature, then washed with 0.1× SSC containing 0.1% SDS at 60°C. Autoradiography was carried out at room temperature with Hyper Film (Amersham).
Expression and purification of recombinant proteins.
Recombinant proteins were constructed for the P. aeruginosa homologue (PA2885), pfiT, and the original I2 gene product into pQE-30 (Table 2). E. coli XL-1 Blue transformants were induced (1 mM isopropyl-β-d-thiogalactopyranoside [IPTG]), and six-His-tagged proteins were purified with a HisTrap column (Amersham) under denatured conditions according to the manufacturer. According to SDS-polyacrylamide gel electrophoresis (PAGE) and gel densitometry, the protein preparations were of similar purities (>85%).
CD4+ T-cell proliferation.
Erythrocyte-depleted splenocytes, used as antigen-presenting cells (APCs), were cultured with 1 or 5 μg of antigen per ml at 5 × 106 cells per ml for 12 to 16 h at 37°C and then washed and irradiated at 3,000 rads (567.7 cGy/min for 5.5 min) prior to T-cell coculture. Splenic CD4+ T cells (>85% pure by flow cytometry) were isolated by depletion with nylon wool and anti-B220 and anti-CD8 magnetic beads. CD4+ T cells (4 × 105) were incubated in triplicate with 4 × 105 antigen-pulsed APCs in wells of a 96-well flat-bottom tissue culture plate at 37°C in 5% CO2 humidified air. After 48 h of incubation, 0.5 μCi of [3H]thymidine was added to each culture for the last 18 h of the incubation period, and then the cells were collected with a Micro 96 harvester (Skatron Instruments, Tranby, Norway). Proliferation was assessed with a Betaplate liquid scintillation counter (Wallac, Gaithersburg, Md.).
Detection of P. fluorescens in fecal samples.
Fresh fecal samples from patients with CD were sequentially accrued from the population used in our recent study (10). DNA was isolated with a commercial kit (Stratagene, La Jolla, Calif.), and 0.1 μg was assayed for I2, pfiT, and Omp-W sequences by our previously reported semiquantitative PCR method (10). Doping experiments indicated that the limit of detection for both assays was ∼2,000 molecules per g of fecal sample. Samples from 30 patients with satisfactory quality were subjected to analysis. In parallel, serial dilutions of fecal samples were cultured on TSA plates at 26°C for conventional microbiologic detection of P. fluorescens. Doping with UCLA 268 indicated a limit of detection of 104 CFU/g of sample.
Nucleotide sequence accession number.
The nucleotide sequence data for pfiT and the flanking genomic region in P. fluorescens are available under GenBank accession no. AF173683.
RESULTS
I2 homologues are present in a limited number of species in the family Pseudomonadaceae.
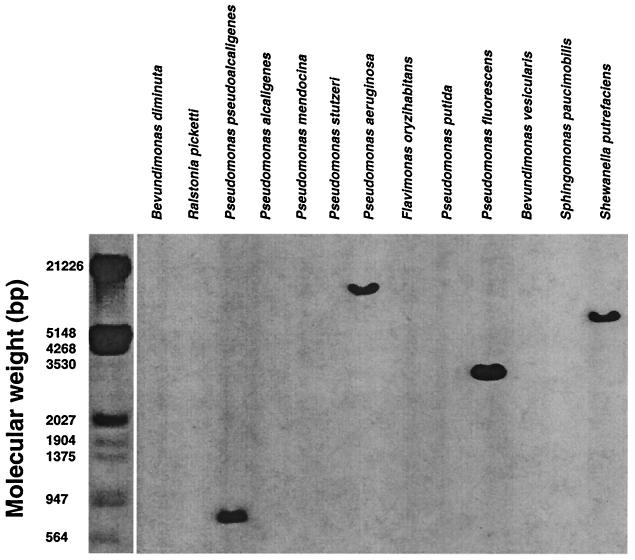
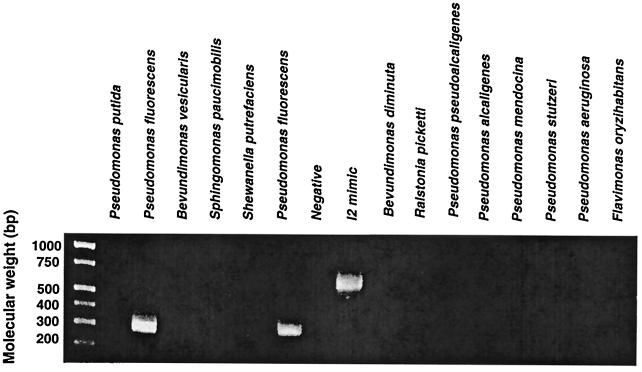
A BLAST search for close homologues of the I2 sequence revealed PA2885, an open reading frame (ORF) of the recently reported P. aeruginosa genome (42). I2 and PA2885 shared 84% nucleotide identity and amino acid sequence identity and similarity of 86 and 93%, respectively. This finding indicated that the I2 gene might have originated from a member of the family Pseudomonadaceae. A panel of 13 Pseudomonas species were examined for the occurrence of the I2 gene segment (Table 1). In Fig. 1, Southern analysis revealed four species positive for the gene (Pseudomonas aeruginosa, Pseudomonas pseudoalcaligenes, P. fluorescens, and Shewanella putrefasciens), among which P. fluorescens gave the strongest hybridization signal. Chromosomal DNA samples were further analyzed for the I2 gene segment with the I2-5′ and I2-3′ primers (Table 2). Among the 13 Pseudomonas species tested, an I2 PCR product was only detected in P. fluorescens (Fig. 2). Sequencing revealed that the sequence of the I2 product from this P. fluorescens strain was nearly identical (1 nucleotide difference) to the index I2 sequence. These findings indicated that the I2 gene segment originated from P. fluorescens.
FIG. 1.
Southern analysis of I2 gene homologues in members of the family Pseudomonadaceae. Genomic DNA (3 μg) from 13 clinical isolates of Pseudomonadaceae were digested with EcoRI, separated on 0.8% agarose gel, and transferred to nitrocellulose membranes. The membranes were hybridized with 32P-labeled I2 gene segment at moderate stringency, washed at high stringency, and visualized by exposure to Hyper Film.
FIG. 2.
P. fluorescens contains the I2 sequence genomic DNA samples from 13 clinical strains of Pseudomonas were analyzed by PCR for the I2 gene segment by using I2-specific primers. The expected product (298 bp) was detected only in P. fluorescens. I2 “mimic” DNA (a 600-bp “stuffer” segment flanked with 5′ and 3′ I2 primer sequences) was used as the positive control.
Cloning and characterization of the full-length pfiT ORF from P. fluorescens.
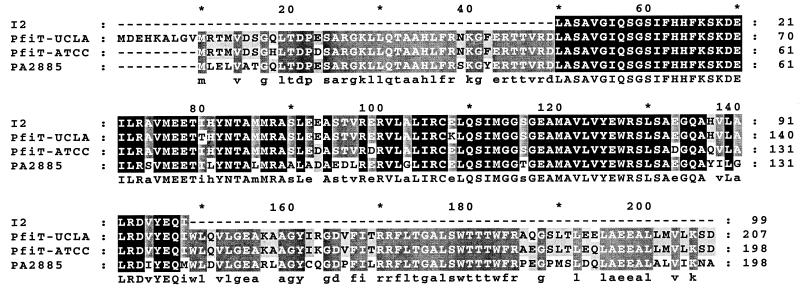
Genome-walking PCR with the I2 sequence was employed to clone the full-length I2 gene and flanking genomic region from P. fluorescens (Fig. 3). A novel 207-amino-acid ORF, pfiT, was identified, which contained the I2 sequence between residues 49 and 148. Compared to the I2 sequence isolated from human CD tissue, this segment had 99% (296 of 300 positions) nucleotide identity and 98 and 99% amino acid identity and similarity, respectively. pfiT was homologous to PA2885 of P. aeruginosa, with 77% nucleotide identity and 78 and 92% amino acid identity and similarity, respectively. These ORFs in P. fluorescens and P. aeruginosa were similar in length, differing by a 9-amino-acid segment at the N terminus.
FIG. 3.
Sequence comparison of PfiT and PA2885. The predicted amino acid sequences of I2, PfiT, and PA2885 were aligned by using the ClustalW multiple-sequence alignment program and displayed by using the GenDoc program. The two PfiT sequences were derived from P. fluorescens ATCC 13525 and UCLA 268, respectively. The darker-shaded areas in each column indicate identical residues among four sequences that are listed underneath as capitalized letters. Lighter-shaded areas indicate the identical residues in three sequences shown as lowercase characters. I2 and pfiT (UCLA) share 99% nucleotide identity and 98 and 99% amino acid identity and similarity, respectively. The ATCC and UCLA pfiT genes share 89% nucleotide identity and 94 and 99% amino acid identity and similarity, respectively. The pfiT (UCLA) and PA2885 genes share 77% nucleotide identity and 78 and 92% amino acid identity and similarity, respectively.
In order to further confirm the presence of pfiT in P. fluorescens, PCR (with I2-5′ and I2-3′ amplimers) was performed on genomic DNA from 14 P. fluorescens isolates (1 environmental isolate and 13 clinical isolates; see Materials and Methods). Among these isolates, all were positive (data not shown). For comparison to our clinical isolate, a second pfiT gene was cloned by PCR from a standard P. fluorescens strain (ATCC 13525). The pfiT genes in ATCC and UCLA strains shared 89% nucleotide identity and 94 and 99% amino acid identity and similarity, respectively (Fig. 3). These findings reflect modest molecular divergence of P. fluorescens strains at this locus.
The pfiT locus includes an iron-response gene and a genetically unstable element.
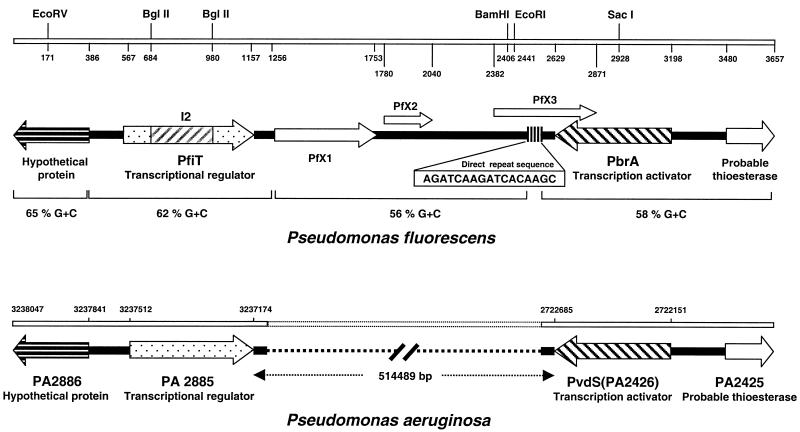
RACE cloning of pfiT flanking sequence disclosed a previously undefined locus in P. fluorescens. As shown in Fig. 4, P. fluorescens and P. aeruginosa were homologous for several colinear ORFs. Cloning of the region upstream of pfiT reached a hypothetical ORF that was homologous to PA2886, a novel ORF in the same genomic position relative to PA2885 in P. aeruginosa.
FIG. 4.
Comparison of the homologue genomic regions in P. aeruginosa and P. fluorescens The P. fluorescens was cloned by genome-walking PCR. The top of the figure lists predicted restriction enzyme sites and nucleotide numbering beginning at the 5′ end of the cloned region. The homologous region in P. aeruginosa was identified by BLAST analysis of the recently reported genome of this microorganism (42); the numbering corresponds to the genomic nucleotide position. Sequence analysis revealed significant sequence homology between the two genomes for each of the colinear ORFs. A direct repeat (hexamer element, 11 repeats) within PFX3 of P. fluorescens is also indicated (nucleotide positions 2473 to 2565).
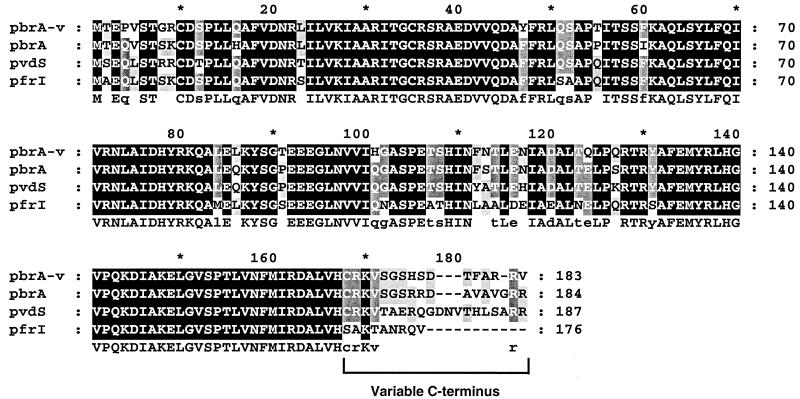
Cloning of the region downstream of pfiT disclosed two notable elements. First, we identified pbrA, a previously reported iron-regulated transcription factor in P. fluorescens (41). In P. aeruginosa, this genomic position is occupied by pvdS, a homologous iron starvation sigma factor and is also homologous to the pfrI gene in P. putida (26, 49). A comparison of pbrA from UCLA 268 to other genes in this family demonstrates strong conservation at the peptide level, with the exception of a divergent C-terminal region (Fig. 5).
FIG. 5.
Alignment analysis of pbrA, pvdS, and pfrI proteins The predicted pbrA sequence isolated from UCLA 268 (pbrA-v) was aligned and compared to those of the genes pbrA of P. fluorescens, pvdS of P. aerugonosas, and pfrI of P. putida. Alignment was performed with ClustalW, and the results are displayed by the GenDoc program, as described in the legend to Fig. 4. The variable region at the extreme C terminus of these iron-responsive genes is indicated.
Second, we identified a novel 1.5-kb downstream region (nucleotide positions 1108 to 2630) between pfiT and pbrA. This region encoded three putative proteins (PFX1, PFX2, and PFX3) without database homology (Fig. 4). The G+C content of this downstream region (56%) was different from those of the adjacent regions (hypothetical protein, 68%; pfiT, 62%; pbrA, 58%). Notably, five 17-bp direct repeats (AGATCAAGATCACAAGC) were found within pfx3. This sequence includes a core hexamer (AGATCA), repeated 11 times in the direct repeat region spanning nucleotide positions 2473 to 2565. These features suggested that the novel region was created by deletion mutagenesis.
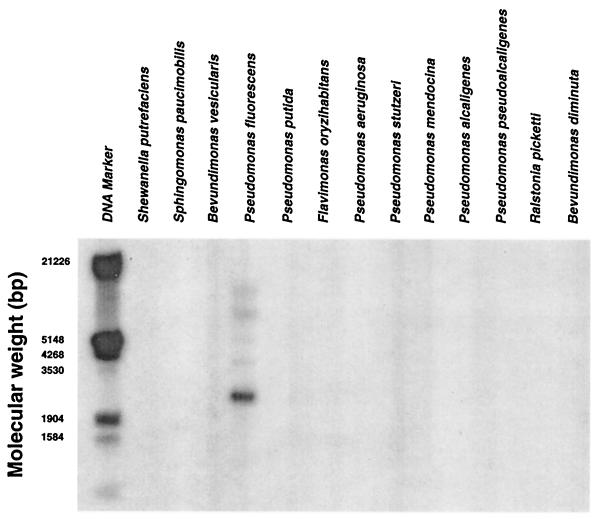
The presence of the novel pfiT downstream region was evaluated with a DNA probe (nucleotide positions 1108 to 2630) in other Pseudomonas species. This region was detected only in P. fluorescens by Southern analysis (Fig. 6). With the pfiT-DF and pfiT-DR primers, a PCR product was detected in UCLA 268, but not in ATCC 13525 or other P. fluorescens clinical isolates (data not shown). These findings indicate that this region is polymorphic among different P. fluorescens isolates.
FIG. 6.
The UCLA 268 downstream segment is a strain-specific locus in P. fluorescens. Genomic DNA from different species in the family Pseudomonadaceae were hybridized with a probe within the pfiT downstream region of UCLA 268 (nucleotide positions 1108 to 2630). By Southern analysis, a positive band was detected only in P. fluorescens.
In the P. aeruginosa genome, the orthologous region between PA2885 and pvdS/PA2426) was much larger (∼515 kb). This region encoded 459 putative ORFs, with diverse functional categories of these genes, including transcriptional regulators, two-component regulatory systems, small molecule transport, and energy metabolism. Fifty-two percent of the hypothetical proteins in this region lacked definable biological function. In comparison to this region in P. aeruginosa, the features of the pfiT downstream region in the UCLA 268 strain of P. fluorescens (short length, replacement with a novel gene segment, and featured repeat sequence) suggest that it represents a recombination-mediated deletional process (39).
CD4+ T-cell activation by PfiT protein homologues.
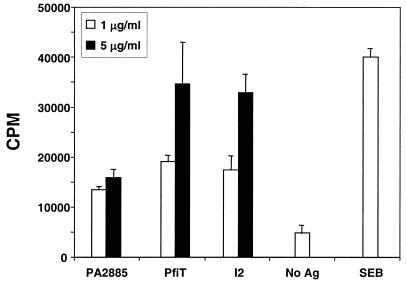
The I2 protein activates murine T cells, displaying features most consistent with a T-cell superantigen (11). We were therefore interested in evaluating whether the full-length parent protein (PfiT) or its P. aeruginosa homologue (PA2885) was also immunostimulatory. In the previous report, recombinant proteins were expressed as glutathione S-transferase (GST) fusion proteins. In this study, a His-tagged protein expression vector was used to express I2, PfiT, and PA2885, and the recombinant proteins were purified by nickel chromatography. The proteins were preincubated with APCs and tested for their ability to stimulate CD4+ T-cell proliferation (Fig. 7). Both I2 and PfiT strongly stimulated proliferation, at levels similar to each other and to that of the superantigen control (SEB). In contrast, PA2885 stimulated a weaker response. This difference was not due detectable differences in protein purity. First, the proteins were of similar purities by SDS-PAGE criteria. Second, experiments combining I2 (1 or 5 μg/ml) with PA2885 (0.5, 1, and 2.5 μg/ml) revealed the same or additive proliferation compared to experiments with I2 alone. Thus, there did not appear to be an inhibitor in the PA2885 preparation for I2-induced CD4+ T-cell stimulation (data not shown). These results indicated that PfiT induces CD4+ T-cell proliferation and that the central third of the PfiT protein (I2) is the functional domain for this bioactivity. Sequence polymorphisms between PfiT and PA2885 may be structurally important for CD4+ T-cell stimulation.
FIG. 7.
PfiT and PA2885 stimulation of CD4 T-cell proliferation. CD4+ T cells from C57BL/6J mice were cultured with syngeneic APCs previously pulsed with recombinant I2, PfiT, or PA2885 protein at 1 and 5 μg/ml. SEB (1 μg/ml) was the positive control. According to a two-tailed Student's t test, proliferation was significantly lower with PA2885 than with PfiT and I2 (P < 0.03 and 0.01 at 5 μg/ml; P < 0.05 at 1 μg/ml). Proliferation in response to all three antigens (PA2885, PfiT, and I2) at both concentrations was significantly greater than that with the negative control (no antigen [No Ag]) (P < 0.03 or less).
Absence of detectable P. fluorescens in CD fecal samples.
Two assays were used to assess the presence of P. fluorescens in fresh fecal samples sequentially accrued from 30 CD patients. First, samples were assessed for Pseudomonas colony formation by conventional microbiologic culture. However, no growth was detected (limit of detection, 10,000 colonies per g of fecal material). Second, samples were assessed for P. fluorescens DNA by PCR for the species-specific I2 and pfiT sequences. However, no sequence was detected in any of the 30 specimens (limit of detection, copy number of 2,000/g of fecal material).
DISCUSSION
I2 is a novel microbial gene recently associated with colon lesions in CD and encodes a T-cell superantigen (11, 45). The present study demonstrates that I2 is derived from pfiT, a newly identified gene of P. fluorescens. The pfiT homologues are restricted mainly to the RNA group 1 family of Pseudomonadaceae, and the genomic region harboring this gene in P. fluorescens has features of a virulence locus. Immunologic analysis indicates that the I2 segment of the PfiT protein fully accounts for its T-cell superantigen activity. The discussion addresses the phylogeny of pfiT, the nature of its genomic locus, and its potential immunologic role in CD pathogenesis.
I2, PfiT, and P. fluorescens.
Several lines of evidence indicate the I2 is derived from the pfiT gene of P. fluorescens. Southern and PCR analyses of 13 bacterial species in the family Pseudomonadaceae demonstrated that the I2 sequence was present in P. fluorescens alone. Molecular cloning of this genomic region from a clinical isolate of P. fluorescens disclosed a new gene, pfiT, which was detected in all of the 14 P. fluorescens isolates available for examination. The physiologic role of PfiT and its P. aeruginosa homologue (PA2885) is unknown. On structural grounds, they are predicted to be a tetR-like component of transcription complexes (42, 45).
Analysis of the genomic region surrounding pfiT in P. fluorescens revealed overall homology with the corresponding region of P. aeruginosa with respect to both the organization and complement of predicted ORFs. One of these adjacent genes was pbrA, an iron-responsive gene of P. fluorescens, with homologues pvdS and pfrI in P. aeruginosa and Pseudomonas putida, respectively (26, 41, 49). PbrA protein upregulates certain siderophores and exotoxins facilitating the iron deprivation response, and in some systems, it is an important virulence factor (50).
I2 was recently shown in murine assays to be a new structural class of T-cell superantigen, activating CD4+ T cells particularly by using the T-cell receptor (TCR) Vβ5 gene (11). In this study, PfiT was also found to activate CD4+ T cells, with a potency comparable to that of the I2 protein. This indicates that the central region of PfiT (the I2 sequence) fully accounts for the immunologic activity of the protein. Compared to PfiT, PA2885 was only modestly stimulatory, suggesting that PA2885 may differ in sequences affecting the avidity of major histocompatibility complex (MHC) or TCR binding.
T-cell superantigens are an important virulence trait of certain pathogenic bacteria, due to direct and indirect tissue damage mediated by the cognately activated T-cell population (27, 30). Among species known to express T-cell superantigens, gram-negative bacteria are notably absent, with the exception of an apparent T-cell superantigen in Yersinia pseudotuberculosis (1). PfiT is thus distinguished both as a distinct structural class of T-cell superantigens and as the first example of such bioactivity in gram-negative intestinal commensals. The modest immunoactivity of PA2885 makes it uncertain whether P. aeruginosa may express significant T-cell superantigen activity. In view of the important pathogenicity of P. aeruginosa in several clinical settings, the issue of PA2885 immunoactivity may deserve further study.
Members of the family Pseudomonadaceae are rare components of fecal bacteria of the colon, usually below the limits of culture detection (16, 48). There are reports associating fecal Pseudomonadaceae with CD (17, 18), but the findings are inconsistent (3, 20, 33). Similarly, the present study did not detect fecal P. fluorescens in CD patients, but the limits of detection were 10,000 CFU or 2,000 gene copies per g of stool, respectively, for culture or PCR assay. However, a PCR assay for the P. fluorescens-specific I2 sequence is usually positive for mucosa of the ileum (healthy as well as with IBD) and colon (CD) (45). This suggests that P. fluorescens may be a normal, low-level commensal of ileal mucosa and may broaden its colonization to susceptible colonic mucosa in CD.
Swidsinski et al. recently reported a striking elevation of colonic mucosa-associated bacteria of diverse genera in IBD, suggesting that IBD may involve a disorder in the regulation of bacterial-mucosal adherence (46). That study did not detect Pseudomonas in their culture assay (perhaps due to the limit of detection) and observed the disordered adherence found in both ulcerative colitis and CD. However, it points to the idea that disordered mucosal adherence in CD may be a predisposing factor for the association of mucosal P. fluorescens in CD. Colonization by P. aeruginosa and its pathogenicity of P. aeruginosa are opportunistic, with dependence on local host factors and microbial traits modifying host-bacterial interaction (15, 23, 42). In mouse experimental and genetic models, diverse modes of immunologic or structural disturbance of the intestinal mucosa promote a CD-like disease (6, 7). While P. fluorescens rarely involves opportunistic infection (22), the disordered mucosal environment in genetically susceptible individuals may suit P. fluorescens opportunism and through expression of the PfiT superantigen may contribute a role in CD mucosal damage.
Acknowledgments
We are grateful for the advice of Paul Colonna and Jeffrey H. Miller and the able and cordial technical assistance of Kathleen Lechowitzc and Kevin Ward. We appreciate the gift of P. fluorescens isolates from Paul C. Schrenckenberger (Director of Clinical Microbiology, University of Illinois Chicago Medical Center). We thank Dominica Salvatore for assistance with preparation of the manuscript.
This work was supported by NIH DK46763 (J.B.), the Jonsson Comprehensive Cancer Center (J.B.), and the Crohn's and Colitis Foundation of America (W.B.).
Editor: J. D. Clements
REFERENCES
- 1.Abe, J., M. Onimaru, S. Matsumoto, S. Noma, K. Baba, Y. Ito, T. Kohsaka, and T. Takeda. 1997. Clinical role for a superantigen in Yersinia pseudotuberculosis infection. J. Clin. Investig. 99:1823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lippman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Auer, I. O., A. Roder, F. Wensinck, J. P. van de Merwe, and H. Schmidt. 1983. Selected bacterial antibodies in Crohn's disease and ulcerative colitis. Scand. J. Gastroenterol. 18:217-223. [DOI] [PubMed] [Google Scholar]
- 4.Bhan, A. K., E. Mizoguchi, R. N. Smith, and A. Mizoguchi. 1999. Colitis in transgenic and knockout animals as models of human inflammatory bowel disease. Immunol. Rev. 169:195-207. [DOI] [PubMed] [Google Scholar]
- 5.Blaser, M. J., R. A. Miller, J. Lacher, and J. W. Singleton. 1984. Patients with active Crohn's disease have elevated serum antibodies to antigens of seven enteric bacterial pathogens. Gastroenterology 87:888-894. [PubMed] [Google Scholar]
- 6.Blumberg, R. S., L. J. Saubermann, and W. Strober. 1999. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr. Opin. Immunol. 11:648-656. [DOI] [PubMed] [Google Scholar]
- 7.Bregenholt, S., D. Delbro, and M. H. Claesson. 1997. T-cell transfer and cytokine TCR gene deletion models in the study of inflammatory bowel disease. APMIS 105:655-662. [DOI] [PubMed] [Google Scholar]
- 8.Cohavy, O., D. Bruckner, L. K. Gordon, R. Misra, B. Wei, M. E. Eggena, S. R. Targan, and J. Braun. 2000. Colonic bacteria express an ulcerative colitis pANCA-related protein epitope. Infect. Immun. 68:1542-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohavy, O., G. Harth, M. Horwitz, M. Eggena, C. Landers, C. Sutton, S. R. Targan, and J. Braun. 1999. Identification of a novel mycobacterial histone H1 homologue (HupB) as an antigenic target of pANCA monoclonal antibody and serum immunoglobulin A from patients with Crohn's disease. Infect. Immun. 67:6510-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalwadi, H., B. Wei, and J. Braun. 2000. Defining new pathogens and non-culturable infectious agents: the case of inflammatory bowel disease. Curr. Opin. Gastroenterol. 16:56-59. [DOI] [PubMed] [Google Scholar]
- 11.Dalwadi, H., B. Wei, M. Kronenberg, C. L. Sutton, and J. Braun. 2001. The Crohn's disease-associated bacterial protein I2 is a novel enteric T cell superantigen. Immunity 15:149-158. [DOI] [PubMed] [Google Scholar]
- 12.Del Prete, R., M. Quaranta, A. Lippolis, V. Giannuzzi, A. Mosca, E. Jirillo, and G. Miragliotta. 1998. Detection of Mycobacterium paratuberculosis in stool samples of patients with inflammatory bowel disease by IS900-based PCR and colorimetric detection of amplified DNA. J. Microbiol. Methods 33:105-114. [Google Scholar]
- 13.De Winter, H., H. Cheroutre, and M. Kronenberg. 1999. Mucosal immunity and inflammation. II. The yin and yang of T cells in intestinal inflammation: pathogenic and protective roles in a mouse colitis model. Am. J. Physiol. 276:G1317-G1321. [DOI] [PubMed] [Google Scholar]
- 14.Dieleman, L. A., A. Arends, S. L. Tonkonogy, M. S. Goerres, D. W. Craft, W. Grenther, R. K. Sellon, E. Balish, and R. B. Sartor. 2000. Helicobacter hepaticus does not induce or potentiate colitis in interleukin-10-deficient mice. Infect. Immun. 68:5107-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epelman, S., T. F. Bruno, G. G. Neely, D. E. Woods, and C. H. Mody. 2000. Pseudomonas aeruginosa exoenzyme S induces transcriptional expression of proinflammatory cytokines and chemokines. Infect. Immun. 68:4811-4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finegold, S. M., V. L. Sutter, P. T. Sugihara, H. A. Elder, S. M. Lehmann, and R. L. Phillips. 1977. Fecal microbial flora in Seventh Day Adventist populations and control subjects. Am. J. Clin. Nutr. 1:1781-1792. [DOI] [PubMed] [Google Scholar]
- 17.Foreman, N. K., W. C. Wang, E. J. Cullen, G. L. Stidham, T. A. Pearson, and J. L. Shenep. 1991. Endotoxic shock after transfusion of contaminated red blood cells in a child with sickle cell disease. Pediatr. Infect. Dis. J. 10:624-626. [PubMed] [Google Scholar]
- 18.Franzetti, F., M. Cernuschi, R. Esposito, and M. Moroni. 1992. Pseudomonas infections in patients with AIDS and AIDS-related complex. J. Intern. Med. 231:437-443. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Lafuente, A., M. Antolin, F. Guarner, E. Crespo, A. Salas, P. Forcada, and J. Malagelada. 1998. Derangement of mucosal barrier function by bacteria colonizing the rat colonic mucosa. Eur. J. Clin. Investig. 28:1019-1026. [DOI] [PubMed] [Google Scholar]
- 20.Graham, D. Y., H. H. Yoshimura, and M. K. Estes. 1983. DNA hybridization studies of the association of Pseudomonas maltophilia with inflammatory bowel diseases. J. Lab. Clin. Med. 101:940-954. [PubMed] [Google Scholar]
- 21.Gui, G. P. H., P. R. S. Thomas, M. L. V. Tizard, J. Lake, J. D. Sanderson, and J. Hermon-Taylor. 1997. Two-year-outcomes analysis of Crohn's disease treated with rifabutin and macrolide antibiotics. J. Antimicrob. Chemother. 39:393-400. [DOI] [PubMed] [Google Scholar]
- 22.Hsueh, P.-R., L.-J. Teng, H.-J. Pan, Y.-C. Chen, C.-C. Sun, S.-W. Ho, and K.-T. Luh. 1998. Outbreak of Pseudomonas fluorescens bacteremia among oncology patients. J. Clin. Microbiol. 36:2914-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ichikawa, J. K., A. Norris, M. G. Bangera, G. K. Geiss, A. B. Wout, R. E. Bumgarner, and S. Lory. 2000. Interaction of Pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc. Natl. Acad. Sci. USA 97:9659-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janowitz, H. D., E. C. Croen, and D. B. Sachar. 1998. The role of the fecal stream in Crohn's disease: an historical and analytic review. Inflamm. Bowel Dis. 4:29-39. [DOI] [PubMed] [Google Scholar]
- 25.Kullberg, M. C., J. M. Ward, P. L. Gorelick, P. Caspar, S. Hieny, A. Cheever, D. Jankovic, and A. Sher. 1998. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect. Immun. 66:5157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leoni, L., N. Orsi, V. de Lorenzo, and P. Visca. 2000. Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa. J. Bacteriol. 182:1481-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, H., A. Llera, E. L. Malchiodi, and R. A. Mariuzza. 1999. The structural basis of T cell activation by superantigens. Annu. Rev. Immunol. 17:435-466. [DOI] [PubMed] [Google Scholar]
- 28.Liu, Y., H. J. Van Kruiningen, A. B. West, R. W. Cartun, A. Cortot, and J.-F. Colombel. 1995. Immunocytochemical evidence of Listeria, Escherichia coli, and Streptococcus antigens in Crohn's disease. Gastroenterology 108:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madsen, K. L., J. S. Doyle, M. M. Tavernini, L. D. Jewell, R. P. Rennie, and R. N. Fedorak. 2000. Antibiotic therapy attenuates colitis in interleukin 10 gene-deficient mice. Gastroenterology 118:1094-1105. [DOI] [PubMed] [Google Scholar]
- 30.Marrack, P., and J. Kappler. 1994. Subversion of the immune system by pathogens. Cell 76:323-332. [DOI] [PubMed] [Google Scholar]
- 31.Moss, M. T., E. P. Green, M. L. Tizard, Z. P. Malik, and J. Hermon-Taylor. 1991. Specific detection of Mycobacterium paratuberculosis by DNA hybridisation with a fragment of the insertion element IS900. Gut 32:395-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naser, S. A., K. Hulten, I. Shafran, D. Y. Graham, and F. A. El Zaatari. 2000. Specific seroreactivity of Crohn's disease patients against p35 and p36 antigens of M. avium subsp. paratuberculosis. Vet. Microbiol. 77:497-504. [DOI] [PubMed] [Google Scholar]
- 33.Parent, K., and P. Mitchell. 1978. Cell wall-defective variants of Pseudomonas-like (group Va) bacteria in Crohn's disease. Gastroenterology 75:368-372. [PubMed] [Google Scholar]
- 34.Prantera, C., F. Zannoni, M. L. Scribano, E. Berto, A. Andreoli, A. Kohn, and C. Luzi. 1996. An antibiotic regimen for the treatment of active Crohn's disease: a randomized, controlled clinical trial of metronidazole plus ciprofloxacin. Am. J. Gastroenterol. 91:328-332. [PubMed] [Google Scholar]
- 35.Rath, H. C., H. H. Herfarth, J. S. Ikeda, W. B. Grenther, T. E. Hamm, Jr., E. Balish, J. D. Taurog, R. E. Hammer, K. H. Wilson, and R. B. Sartor. 1996. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J. Clin. Investig. 98:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rath, H. C., M. Schultz, R. Freitag, L. A. Dieleman, F. Li, H.-J. Linde, J. Schölmerich, and R. B. Sartor. 2001. Different subsets of enteric bacteria induce and perpetuate experimental colitis in rats and mice. Infect. Immun. 69:2277-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saparov, A., L. A. Kraus, Y. Cong, J. Marwill, X. Y. Xu, C. O. Elson, and C. T. Weaver. 1999. Memory/effector T cells in TCR transgenic mice develop via recognition of enteric antigens by a second, endogenous TCR. Int. Immunol. 11:1253-1264. [DOI] [PubMed] [Google Scholar]
- 38.Sartor, R. B. 1997. Pathogenesis and immune mechanisms of chronic inflammatory bowel diseases. Am. J. Gastroenterol. 92:5S-11S. [PubMed] [Google Scholar]
- 39.Scott, J. R., and G. G. Churchward. 1995. Conjugative transposition. Annu. Rev. Microbiol. 49:367-397. [DOI] [PubMed] [Google Scholar]
- 40.Sellon, R. K., S. Tonkonogy, M. Schultz, L. A. Dieleman, W. Grenther, E. Balish, D. M. Rennick, and R. B. Sartor. 1998. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 66:5224-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sexton, R., P. R. Gill, D. N. Dowling, and F. O'Gara. 1996. Transcriptional regulation of the iron-responsive sigma factor gene pbrA. Mol. Gen. Genet. 250:50-58. [DOI] [PubMed] [Google Scholar]
- 42.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 43.Suenaga, K., Y. Yokoyama, I. Nishimori, S. Sano, M. Morita, K. Okazaki, and S. Onishi. 1999. Serum antibodies to Mycobacterium paratuberculosis in patients with Crohn's disease. Dig. Dis. Sci. 44:1202-1207. [DOI] [PubMed] [Google Scholar]
- 44.Sutton, C., H.-Y. Yang, J. I. Rotter, S. R. Targan, and J. Braun. 2000. Familial expression of anti-Saccharomyces cerevisiae mannan antibodies (ASCA) in affected and unaffected relatives of Crohn's disease patients. Gut 46:58-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutton, C. L., J. Kim, A. Yamane, H. Dalwadi, B. Wei, C. Landers, S. R. Targan, and J. Braun. 2000. Identification of a novel bacterial sequence associated with Crohn's disease. Gastroenterology 119:23-28. [DOI] [PubMed] [Google Scholar]
- 46.Swidsinski, A., A. Ladhoff, A. Pernthaler, S. Swidsinski, V. Loening-Baucke, M. Ortner, J. Weber, U. Hoffmann, S. Schreiber, M. Dietel, and H. Lochs. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44-54. [DOI] [PubMed] [Google Scholar]
- 47.Van Kruiningen, H. J. 1999. Lack of support for a common etiology in Johne's disease of animals and Crohn's disease in humans. Inflamm. Bowel. Dis. 5:183-191. [DOI] [PubMed] [Google Scholar]
- 48.Wilson, K. H., J. S. Ikeda, and R. B. Blitchington. 1997. Phylogenetic placement of community members of human colonic biota. Clin. Infect. Dis. 25(Suppl. 2):S114-S116. [DOI] [PubMed] [Google Scholar]
- 49.Wilson, M. J., B. J. McMorran, and I. L. Lamont. 2001. Analysis of promoters recognized by PvdS, an extracytoplasmic-function sigma factor protein from Pseudomonas aeruginosa. J. Bacteriol. 183:2151-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong, Y. Q., M. L. Vasil, Z. Johnson, U. A. Ochsner, and A. S. Bayer. 2000. The oxygen- and iron-dependent sigma factor pvdS of Pseudomonas aeruginosa is an important virulence factor in experimental infective endocarditis. J. Infect. Dis. 181:1020-1026. [DOI] [PubMed] [Google Scholar]