Abstract
HbhA of Mycobacterium tuberculosis is a multifunctional binding protein, binding to both sulfated sugars such as heparin and to human complement component C3. HbhA may therefore interact with host molecules and/or host cells during M. tuberculosis infection and play a role in the pathogenesis of this bacterium. The purpose of this study was to use allelic exchange to create an M. tuberculosis strain deficient in expression of HbhA to determine whether this protein's C3-binding activity plays a role in the pathogenesis of M. tuberculosis. An in-frame, 576-bp unmarked deletion in the hbhA gene was created using sacB as a counterselectable marker. Southern blotting and PCR analyses confirmed deletion of hbhA in the ΔhbhA mutant. The ΔhbhA mutant strain grew at a rate similar to that of the parent in broth culture and in J774.A1 murine macrophage-like cells but was deficient in growth compared to the parent strain in the lungs, liver, and spleen of infected mice. In addition, the ΔhbhA mutation did not reduce binding of M. tuberculosis to human C3 or to J774.A1 cells in the presence or absence of serum, suggesting that in the absence of HbhA, other molecules serve as C3-binding molecules on the M. tuberculosis surface. Taken together, these data indicate that HbhA is important in the infectivity of M. tuberculosis, but its ability to bind C3 is not required for mycobacterial adherence to macrophage-like cells. Using the ΔhbhA mutant strain, a second M. tuberculosis C3-binding protein similar in size to HbhA was identified as HupB, but the role of HupB as a C3-binding protein in intact organisms remains to be determined.
Mycobacterium tuberculosis is the causative agent of tuberculosis and is a global health threat to humans, infecting approximately one-third of the world's population (9). M. tuberculosis is a facultative intracellular pathogen, and its ability to persistently infect macrophages and other host cells is an important mechanism of pathogenesis, dissemination, latency, and recurrent disease (31). A better understanding of the process of adherence, internalization, intracellular survival, and growth within macrophages is critical in efforts to control mycobacterial infections.
Heparin-binding hemagglutinin (HbhA) is a multifunctional binding protein expressed by M. tuberculosis and other mycobacteria. Menozzi et al. (18, 19) first identified HbhA as a 28-kDa surface-exposed, cell wall- or cell membrane-associated glycoprotein. HbhA binds to several sulfated glycoconjugates, such as heparin, dextran sulfate, fucoidan, and chondroitin sulfate, as well as to decorin (7, 18, 19, 25), but not to fibronectin, bovine serum albumin, ovalbumin, dextran, mannose, or galactose (7, 19). HbhA mediates attachment of M. tuberculosis to Chinese hamster ovary (CHO) cells, as determined by antibody inhibition studies (19). In addition, purified recombinant HbhA binds to human A549 pneumocytes in cell culture (25). Our previous work demonstrated that HbhA can also bind to human complement component C3, and recombinant HbhA mediates attachment of latex beads to murine macrophage-like cells in both a C3-dependent and a C3-independent manner (20). The carboxy terminus of HbhA contains two types of lysine-alanine repeats which have been shown to be responsible for heparin binding (25) and complement component C3 binding (20).
HbhA may play a multifunctional role in promoting entry of M. tuberculosis into host cells. Pethe et al. (24) recently published data indicating that mutation of M. tuberculosis hbhA, in which a kanamycin resistance gene was inserted into hbhA, resulted in decreased binding of M. tuberculosis to pneumocytes and reduced dissemination of M. tuberculosis from the lungs to the spleens of infected mice. This group did not observe any differences between the parent and mutant in adherence to J774.A1 macrophage-like cells, but their studies were done in the absence of complement-sufficient human serum. In the present study, we constructed an unmarked, in-frame deletion of the hbhA gene to determine whether HbhA plays a role in the C3-mediated adherence and/or uptake of M. tuberculosis into mononuclear cells. The ΔhbhA mutant was examined in the mouse model of tuberculosis infection and in murine macrophage-like cells for any differences in adherence, phagocytosis, or survival compared to the parent. Additional studies using the ΔhbhA mutant strain resulted in identification of a second C3-binding protein.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. tuberculosis H37Rv (ATCC 27294) and its derivatives were grown in liquid Middlebrook 7H9 medium supplemented with 0.2% glycerol, 0.05% Tween 80, and albumin-dextrose complex (ADC) (consisting of 0.5% bovine serum albumin [BSA], fraction V, 0.085% NaCl, and 0.2% glucose). Middlebrook 7H10 or 7H11 plates supplemented with either ADC or OADC (ADC with oleic acid; 0.05 g per liter, final concentration), 50 μg of hygromycin/ml, or 2% sucrose were used as indicated for colony isolation and were incubated at 37°C for 3 to 4 weeks under 5.4% CO2. Escherichia coli DH5α1 (Stratagene, La Jolla, Calif.) was used for recombinant DNA studies and plasmid propagation. E. coli strains were grown on solid or liquid Luria-Bertani medium supplemented with 200 μg of hygromycin/ml as indicated and were incubated overnight at 37°C.
Recombinant DNA techniques.
Chromosomal DNA was isolated from M. tuberculosis cells as described by Armitige et al. (1). Molecular cloning and restriction endonuclease digestion were performed by standard techniques (32). Restriction endonucleases and other enzymes were used according to the manufacturers' recommendations. DyNAzyme II DNA polymerase for PCR was purchased from Finnzymes (Waltham, Mass.) and was used in accordance with the instructions provided. Primers used in the studies described in this paper were purchased from Integrated DNA Technologies (Coralville, Iowa) and are listed in Table 1. Unless otherwise indicated, PCR conditions consisted of 96°C for 2 min, 30 cycles of denaturation at 94°C for 40 s, annealing for 40 s at 2°C below the lowest primer melting temperature, and extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min.
TABLE 1.
PCR primers used in the creation and analysis of the hbhA mutants
| Primer name | Primer sequence |
|---|---|
| 102 | 5′-AGTCATCTGGGTGAGGTCGA-3′ |
| 103C | 5′-GCGCGCCGGGATTCTCGAGC-3′ |
| X-103C | 5′-CTTCTAGAGCGCGCCGGGATTCTC-3′ |
| 104D | 5′-CGGTGTGATCGACAAGACGTGGG-3′ |
| X-104D | 5′-GCTCTAGACGGTGTGATCGACAAGACGTG-3′ |
| 105B | 5′-AAGGAATACCCATGGCTGAAAACACCCAGAAGTAGTCGGGCTCCG-3′ |
| 106A | 5′-CGGAGCCCGACTACTTCTGGGTGTTTTCAGCCATGGGTATTCCTT-3′ |
| 110 | 5′-ATGTCGTCGGAGGAGAAGCTGGC-3′ |
| 111 | 5′-GACCAGCGCGGCCACATC-3′ |
| 112 | 5′-TCGGCGGCCTTACGCAGCTC-3′ |
| 113 | 5′-CGAACATTGATGACATCAAGGCTCCGTT-3′ |
| 114 | 5′-GCGGTGTACGCGTTTGTGCATGC-3′ |
| 116 | 5′-GCGCGACTTGCCCTGAATCTCG-3′ |
| 0475 | 5′-GTTGGGTACGGTCGCATCGCAG-3′ |
| 0476 | 5′-CATGCACAAACGCGTACACC-3′ |
| Hyg-F | 5′-CCGGCCCGTACCCTGTGAAT-3′ |
| Hyg-R2 | 5′-GAGCAGCGCGTTCCGGTC-3′ |
| 16s-F | 5′-GCGGTAATACGTAGGGTGCGAGC-3′ |
| 16s-R | 5′-CCCACACCTAGTACCCACCGTT-3′ |
Generation of transforming plasmids.
Plasmid pJQ200mp18 (28) was kindly provided by A. Ballard at Colorado State University and contains the sacB gene, which allows for counterselection in mycobacteria (23). This plasmid encodes for gentamicin resistance, but a hygromycin resistance gene was also added to this plasmid to create plasmid pJQhyg. The hygromycin resistance gene (hyg) was isolated from plasmid p16R1 (ATCC 87120) by treating p16R1 with restriction enzymes BspHI and FspI, and the resulting 1.7-kb DNA fragment containing the hyg gene was purified from a low-melting-point agarose gel and treated with the Klenow fragment of DNase I (New England BioLabs, Beverly, Mass.) to create blunt ends. This fragment was then ligated into plasmid pJQ200mp18 that had been treated with SmaI and shrimp alkaline phosphatase. The resulting plasmid (pJQhyg) was transformed into E. coli DH5α1 as described elsewhere (32).
A PCR technique known as gene splicing by overlap extension (SOEing) (11) was used to create a 1,098-bp fragment containing an in-frame 576-bp deletion in the M. tuberculosis hbhA gene. The 45-bp oligonucleotides 106A and 105B represent the junction region of the deletion. The 5′ portion of primer 105B contained 11 bases of sequence directly upstream of the start codon of hbhA, followed by the first 12 bases of the coding region of hbhA. The 3′ portion of primer 105B contained the final 12 bases of the coding region of hbhA, followed by 10 bases of sequence directly downstream of the stop codon of hbhA. Primer 106A is complementary to primer 105B. Using M. tuberculosis H37Rv chromosomal DNA as template, primers 106A and 103C were used to amplify a 566-bp upstream DNA fragment, and primers 105B and 104D were used to amplify a 562-bp downstream DNA fragment. These PCR products were then used together as template in the following two-step PCR program using the forward primer X-103C and the reverse primer X-104D: 96°C for 2 min, 6 cycles of denaturation at 94°C for 40 s, annealing at 54°C for 40 s, and extension at 72°C for 1 min, followed by 26 cycles at a higher annealing temperature of 60°C, and then by a final extension at 72°C for 10 min. For cloning purposes, 8-bp sequences (underlined in Table 1) were added to the 5′ ends of primers X-103C and X-104D to create XbaI sites. The 1,098-bp PCR product from primers X-103C and X-104D containing a 576-bp deletion in hbhA was purified from a low-melting-point agarose gel, treated with XbaI, and ligated into plasmid pJQhyg that had been treated with XbaI and shrimp alkaline phosphatase. The resulting plasmid (pJQΔhbhA) was transformed into E. coli DH5α1 by standard methods (32). Supercoiled pJQΔhbhA DNA was purified from E. coli DH5α1, and the insert sequence of pJQΔhbhA was verified prior to electroporation of M. tuberculosis.
Deletion of M. tuberculosis hbhA.
M. tuberculosis H37Rv was prepared for electroporation as previously described (12). For deletion of hbhA, supercoiled plasmid pJQΔhbhA (2 μg of DNA) was electroporated at 37°C into 1010 electrocompetent H37Rv cells by using an Electroporator 2510 apparatus (Eppendorf North America, Madison, Wis.) at a setting of 1,250 V and a pulse time of 4 to 5 ms. One milliliter of warm 7H9-ADC broth without antibiotics was added immediately, and the bacteria were incubated at 37°C for 6 h with gentle agitation. Transformants were then plated on 7H10-ADC-hygromycin plates, and subsequent individual hygromycin-resistant colonies were then subcultured onto fresh 7H10-ADC-hygromycin plates. One hygromycin-resistant colony containing a copy of pJQΔhbhA integrated at the hbhA target site was subcultured in 3 ml of 7H9-ADC broth without antibiotics and was grown an additional 3 weeks at 37°C with gentle agitation. Dilutions of the broth culture were then subcultured onto 7H10-ADC plates containing 2% sucrose. Subsequent individual sucrose-resistant colonies were then subcultured onto fresh 7H10-ADC plates containing 2% sucrose prior to further evaluation.
PCR was used to screen for deletion of hbhA. Cells from M. tuberculosis transformants were boiled in Tris-EDTA buffer for 10 min to liberate chromosomal DNA to be used as template for PCR. The primers Hyg-F and Hyg-R2 were internal to the hygromycin resistance gene and were used to screen for the presence of the pJQΔhbhA plasmid on the M. tuberculosis chromosome. Primer pairs 110 and 0476 and 111 and 102 were M. tuberculosis genomic primers that were used to screen for an unmarked deletion in hbhA. Two clones having the desired deletion were designated MtbΔhbhA1 and MtbΔhbhA23.
Southern blot analysis.
To generate an internal hbhA probe, primers 112 and 113 were used together with H37Rv chromosomal DNA as a PCR template to amplify a 240-bp fragment. Fluorescein conjugation of the DNA fragment used as a probe was carried out according to the Gene Images procedure (Amersham-Pharmacia, Arlington Heights, Ill.). M. tuberculosis chromosomal DNA was digested with SmaI, electrophoresed in 0.7% agarose, and transferred to a nylon membrane by using the alkaline transfer procedure (32). The membrane was then hybridized with the fluorescein-labeled probe and processed for chemiluminescent detection as described by the manufacturer (Amersham-Pharmacia).
Preparation of clear cell lysates for Western blot analysis.
The parent strain and the ΔhbhA mutant strain were grown for 24 days in liquid Middlebrook 7H9 broth with supplements at 37°C with shaking. Mycobacterial cells were washed two times with phosphate-buffered saline (PBS) containing 0.05% Tween 20, resuspended in PBS containing a protease inhibitor cocktail (Sigma, St. Louis, Mo.), and lysed by ultrasonic disruption on ice for 5 min using a microtip probe under a laminar flow hood. The lysates were then centrifuged for 8 min at 12,000 × g. The supernatants were removed to new tubes and centrifuged again as above. The supernatants representing clear cell lysates were removed to new tubes. The bicinchoninic acid protein assay reagent (Pierce, Rockford, Ill.), with BSA as a standard, was used to determine the protein concentration of each clear cell lysate. The cell lysates were solubilized at 25°C in an equal volume of solubilization buffer. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis using rabbit antiserum to recombinant HbhA were performed as described previously (20).
RT-PCR analysis.
Total RNA was isolated from the parent and the ΔhbhA mutant strains using RNAzol B (Tel-Test, Inc., Friendswood, Tex.) according to the manufacturer's instructions. The concentration of RNA from each strain was quantitated using a spectrophotometer at an absorbance of 260 nm. RNA samples were pretreated with RQ1 DNase (Promega, Madison, Wis.) for 40 min at 37°C prior to use in reverse transcriptase PCRs (RT-PCRs) to eliminate any contaminating genomic DNA. The Promega Access RT-PCR system was used for the RT-PCRs. Several controls were used for each RT-PCR, including a control without RT to test for genomic DNA contamination of the RNA sample, a control with genomic DNA to amplify the gene in question, and a control containing no template to test for contamination of the reaction mixture. Primer pairs 0475 and 0476, 16S-F and 16S-R, and 114 and 116 were used in RT-PCRs with the following program: 48°C for 1 h for the RT reaction; 94°C for 2 min to inactivate the RT; 40 cycles of denaturation at 94°C, annealing at 60°C for 40 s, extension at 72°C for 30 s; followed by a final extension at 72°C for 10 min.
Sequence analysis.
PCR primer pairs 102 and 0476 were used to amplify the 5′ and 3′ regions, respectively, of the hbhA gene locus in the ΔhbhA mutant, using chromosomal DNA as template. The PCR product was purified and used directly as template for sequencing using primers 102 and 0476. DNA sequencing was performed using an ABI 377 automatic DNA sequencer at Lone Star Labs, Inc., Houston, Tex.
In vitro growth studies.
The parent strain and the ΔhbhA mutant strain were added to Middlebrook 7H9-ADC liquid broth and incubated with gentle shaking at 37°C for 13 days. On days 0, 1, 4, 8, 10, and 13, aliquots were removed from each broth culture and plated in 10-fold dilutions in duplicate on 7H11-OADC plates for colony counts.
Adherence to J774.A1 cells.
The murine macrophage-like cell line J774.A1 was cultured as described previously (20). Adherent monolayers at a concentration of 106 cells/ml were established in 24-well plates 1 day prior to infection with M. tuberculosis. Suspensions of M. tuberculosis H37Rv and the ΔhbhA mutant were vortexed vigorously prior to addition to Dulbecco's modified Eagle's medium (DMEM) containing either no serum or 5% normal human serum (NHS) and were then incubated at 37°C with gentle shaking for 40 min. The suspensions of the M. tuberculosis strains were then added to the J774.A1 cells, which had been washed two times with warm PBS, in duplicate wells at a multiplicity of infection (MOI) of 1:1 for 1 hour at 37°C with gentle shaking. The monolayers were then washed three times for 5 min at 37°C with gentle shaking with warm PBS to remove nonadherent bacteria, and then the J774.A1 cells were lysed with 0.05% SDS. Serial 10-fold dilutions of the lysates were made and plated out on 7H11-OADC agar plates for colony counts.
Binding of human C3 to M. tuberculosis organisms.
Whole M. tuberculosis H37Rv or ΔhbhA mutant cells were washed with sodium carbonate buffer, pH 9.6, and were then resuspended in this same buffer at a concentration of 108 bacteria per ml. Immulon I 96-well plates (Dynex Technologies, Chantilly, Va.) were coated overnight at 4°C with bacteria at a concentration of 107 cells per well. The next day, nonspecific binding sites on the M. tuberculosis cells were blocked by incubating the wells with 5% BSA for 1 h at 37°C with gentle shaking. The wells were washed twice with PBS containing 0.05% Tween 20 (PBST). The wells were then incubated with either 2.5% NHS or with 2.5% heat-inactivated human serum (HIS) (serum heated at 56°C for 30 min to inactivate C3) for 30 min at 37°C with gentle shaking. Human serum was obtained from a purified protein derivative-negative laboratory volunteer. The wells were washed six times with PBST. The wells were incubated with a horseradish peroxidase-conjugated goat anti-human C3 antibody (ICN Pharmaceuticals, Inc., Costa Mesa, Calif.) at a dilution of 1:20,000 for 1 h at room temperature with gentle shaking. The wells were then washed six times with PBST. Detection of bound C3 was achieved by incubating the wells with 3,3′,5,5′-tetramethylbenzidine (TMB) liquid substrate system (Sigma) for 15 min at room temperature. A 1 M HCl stop solution was then added to each well. The optical density was read at 405 nm with a SpectraMAX 250 plate reader (Molecular Dynamics, Sunnyvale, Calif.).
Intracellular survival in J774.A1 cells.
Adherent monolayers of J774.A1 cells at a concentration of 106 cells/ml were established in 24-well plates 1 day prior to infection with M. tuberculosis. Suspensions of M. tuberculosis H37Rv and the ΔhbhA mutant were dispersed by gentle sonication and were added to the J774.A1 cells in triplicate in DMEM containing 2% heat-inactivated fetal bovine serum at an MOI of 1:1 for 4 h at 37°C. The monolayers were then washed three times with warm PBS. Twenty-four-well plates with infected J774.A1 cells were incubated at 37°C in DMEM containing 2% heat-inactivated fetal bovine serum for various lengths of time. Infected J774.A1 cells were lysed with 0.05% SDS on days 0 (baseline CFU), 1, 3, and 7. Serial 10-fold dilutions of the lysates were made and plated out on 7H11-OADC agar plates for colony counts.
Mouse infection.
Four- to 8-week-old female C57BL/6 mice were purchased from the University of Texas M.D. Anderson Cancer Center, Houston, Tex., and were housed within a BSL-3 vivarium and given food and water ad libitum. All manipulations were conducted under Institutional Animal Care and Use Committee-approved procedures, which require that moribund mice be sacrificed to prevent unnecessary suffering. Mice were infected with M. tuberculosis via the aerosol route using a Middlebrook aerosol apparatus (Glas-Col., Terre Haute, Ind.) at a concentration of 50 to 1,000 CFU/mouse lung. The number of bacteria delivered to the lungs after aerosol infection was determined by plating whole-lung homogenate 4 h after infection on 7H11-OADC agar plates. The number of viable bacteria present in the lungs, liver, and spleen was assessed 7, 14, and 21 days postinfection by plating serial dilutions of whole-organ homogenates on 7H11-OADC agar plates. The data were expressed as the log10 value of the mean number of bacteria recovered ± the standard deviation (SD) in each group of four mice.
C3 ligand affinity blotting.
The C3 ligand affinity blotting protocol was used to detect C3 bound to M. tuberculosis proteins as described previously (20).
Ion-exchange affinity chromatography.
MtbΔhbhA23 cells grown in liquid culture were washed in 200 mM Tris-HCl (pH 8.0), 1 mM EDTA. The cells were resuspended in 200 mM Tris-HCl (pH 8.0), 1 mM EDTA, 10% sucrose and lysed by ultrasonic disruption using a microtip probe in a laminar flow hood for 7 min on ice. A 10% Triton X-114 solution was added to the lysate to make a final concentration of 1.8% Triton X-114, and the lysate was incubated overnight at 4°C on a rotator. The next day, the lysate was centrifuged at 13,000 × g for 10 min at 4°C. The supernatant was transferred to a new tube and was centrifuged again in the same manner. The supernatant was again transferred to a new tube, incubated at 37°C for 10 min, and then centrifuged at 5,000 × g for 10 min at 37°C. The aqueous phase was applied to a 1-ml HiTrap SP Sepharose fast-performance liquid chromatography (FPLC) column (Amersham-Pharmacia) equilibrated with 25 mM Tris-HCl, pH 8.1. The flowthrough from the column was collected, and the column was then washed with the equilibration buffer to remove proteins that bound nonspecifically. Bound proteins were then eluted with a continuous gradient of NaCl in 25 mM Tris-HCl, pH 8.1. The distribution of the C3-binding protein was determined by testing the elution fractions using the C3 ligand affinity blotting protocol.
Protease digestion and mass spectrometry analysis.
The purified C3-binding protein was excised from a Coomassie blue-stained SDS-12% PAGE gel and submitted to Richard G. Cook at the Baylor College of Medicine Protein Chemistry Core Facility for protease digestion and amino acid sequencing. The protein gel slice was digested with Lys-C purified from Achromobacter lyticus (Wako Chemicals, Richmond, Va.), and two peptides were analyzed by mass spectrometry.
Statistical analyses.
Student's t test was used to analyze data for statistical significance.
RESULTS
Deletion of M. tuberculosis hbhA.
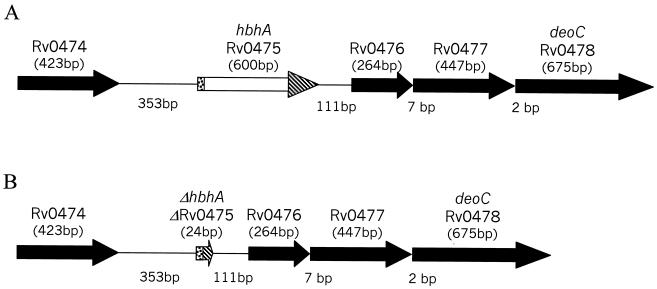
Previously, our group demonstrated that HbhA of M. tuberculosis H37Rv binds the human complement component C3 and enhances the adherence of HbhA-coated beads to J774.A1 murine macrophage-like cells (20). In the present study, an in-frame, markerless deletion of hbhA in H37Rv was constructed to determine whether this mutation affected C3 binding, adherence, and virulence of M. tuberculosis. As depicted in Fig. 1A, hbhA appears to be part of a four-gene operon. The open reading frame directly downstream of hbhA, designated Rv0476 by the Sanger Centre (5), is predicted to encode an 87-amino-acid protein. This hydrophobic protein has similarity to a region of a 113-amino-acid Streptomyces coelicolor predicted protein and also contains a domain similar to the active site of β-ketoacyl synthases (GenBank accession number 3261606). RT-PCR using the primers 0475 and 0476 (Table 1) on either side of the hbhA-Rv0476 intergenic region yielded a product of the expected size (358 bp), whereas a control in which the RT step was omitted did not result in a detectable product (data not shown). These results indicate that hbhA and Rv0476 are cotranscribed. Furthermore, Rv0476, Rv0477 (encoding a protein of unknown function), and Rv0478 (predicted to encode the deoxyribose phosphate aldolase DeoC) are separated by only 7 and 2 bp, respectively, consistent with cotranscription. Rv0479 is oriented in the opposite direction (data not shown). Thus, hbhA and the three open reading frames downstream are likely to represent an operon.
FIG. 1.
Representation of the M. tuberculosis genomic region surrounding hbhA. (A) Gene arrangement in the wild-type H37Rv strain. (B) Alteration in the ΔhbhA mutant strain. The ΔhbhA mutant contains a 576-bp in-frame deletion in the hbhA gene. The hbhA gene in the ΔhbhA mutant consists of the first 12 bases of the coding region of hbhA followed by the final 12 bases of the coding region of hbhA. The designations for the gene names were assigned by the Sanger Centre. hbhA is also known as Rv0475. The sizes of the genes are given in base pairs below the gene names, and the number of nucleotides separating the genes is also given in base pairs.
An unmarked, in-frame deletion within hbhA was created in H37Rv to avoid potential pleiotropic effects due to altered transcription of downstream genes. The deletion was achieved in a two-step process (21, 22), utilizing the suicide vector pJQΔhbhA containing a hygromycin cassette as a positive selection marker and the sacB gene encoding the enzyme levansucrase as a counterselectable marker (see Materials and Methods). Following electroporation, the transformation mixtures were plated on 7H10-ADC-hygromycin plates to select for hygromycin-resistant (Hygr) colonies. One Hygr clone (MtbΔ16) showed integration of pJQΔhbhA at the hbhA target site by PCR analysis (data not shown). MtbΔ16 was grown to saturation in nonselective 7H9-ADC broth. Dilutions of the MtbΔ16 nonselective culture were plated on 7H10-ADC-sucrose plates to select for loss of the pJQΔhbhA plasmid, leaving either an intact hbhA gene or a deleted hbhA gene on the chromosome. Based on PCR analysis, two sucrose-resistant colonies contained a 576-bp deletion in the hbhA gene (data not shown). These two clones were named MtbΔhbhA1 and MtbΔhbhA23, and they both appeared to contain an unmarked in-frame deletion in the hbhA gene (Fig. 1B). MtbΔhbhA23 was selected for further analysis.
Southern blot hybridization, immunoblotting, and sequence determination were used to confirm the functional deletion of hbhA in MtbΔhbhA23. According to the genomic sequence, treatment of wild-type H37Rv chromosomal DNA with the restriction enzyme SmaI generated a 2.5-kb hbhA gene fragment. Hybridization of wild-type H37Rv chromosomal DNA treated with SmaI with an internal 240-bp hbhA gene fragment yielded the expected 2.5-kb band, whereas MtbΔhbhA23 chromosomal DNA treated with SmaI produced no hybridizing band (Fig. 2A.) Western blot analysis of cell lysates from wild-type H37Rv and the ΔhbhA mutant revealed that the ΔhbhA mutant did not produce detectable HbhA (Fig. 2B and C). Finally, a 490-bp PCR product encompassing the deletion site was amplified from the ΔhbhA mutant and sequenced. As expected, the hbhA region in the ΔhbhA mutant contained the first 12 nucleotides followed by the last 12 nucleotides of the coding region (data not shown). Taken together, these results confirm that hbhA was effectively deleted in-frame in MtbΔhbhA23.
FIG. 2.
Confirmation of hbhA deletion in M. tuberculosis H37Rv clone MtbΔhbhA23. (A) Southern blot analysis. Genomic DNA digested with SmaI was hybridized with an internal 240-bp fragment of hbhA. Lane 1, H37Rv DNA; lane 2, MtbΔhbhA23 DNA. Size standards are indicated on the left. (B) Coomassie-stained SDS-PAGE gel of H37Rv cell lysate (lane 1) and an MtbΔhbhA23 cell lysate (lane 2). (C) Western blot of gel pattern identical to that shown in panel B reacted with rabbit antiserum against recombinant HbhA. Molecular mass markers are indicated on the left.
Transcriptional analysis of the M. tuberculosis strains.
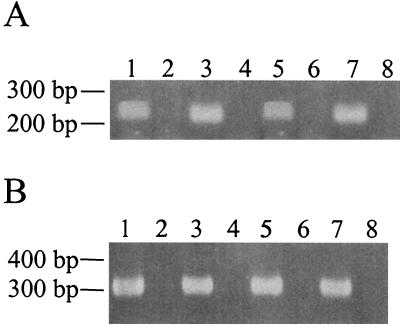
To ensure that no polar effects were caused by the in-frame deletion of hbhA, RT-PCR analysis was used to examine message levels for the gene directly downstream of hbhA, Rv0476, in the parental and ΔhbhA mutant strains. The M. tuberculosis 16S rRNA gene was amplified first as a control to normalize the RNA levels among the two M. tuberculosis strains (Fig. 3B.) An equal concentration of total RNA was used for each RT-PCR. Once the total RNA concentrations between the two strains were found to be equal (Fig. 3B, compare lanes 1 and 5), message for Rv0476 was amplified using primers 114 and 116. The transcript for Rv0476 was present in both the parent and ΔhbhA mutant (Fig. 3A), indicating, as expected, that the deletion of hbhA did not have a polar effect on Rv0476 transcription.
FIG. 3.
In-frame deletion of hbhA does not disrupt transcription of the downstream gene, Rv0476. (A) RT-PCR results obtained using primers within Rv0476 and either H37Rv or MtbΔhbhA23 as the source of RNA. (B) Controls utilizing primers for the M. tuberculosis 16S rRNA. Lanes 1 to 4, H37Rv; lanes 5 to 8, MtbΔhbhA23. Lanes 1 and 5, reactions containing RT and RNA; lanes 2 and 6, reactions containing RNA but no RT; lanes 3 and 7, reactions containing chromosomal DNA; lanes 4 and 8, reactions containing no template.
In vitro growth studies.
The parent and ΔhbhA mutant were cultured in Middlebrook 7H9-ADC broth containing glycerol and Tween 80 to test for any difference in the in vitro growth rate. Both strains grew at similar rates (data not shown), indicating that lack of HbhA expression is not deleterious to the in vitro growth of M. tuberculosis under these conditions.
Adherence of the M. tuberculosis strains to macrophage-like cells.
Murine macrophage-like J774.A1 cells were used to determine if the ΔhbhA mutant differed in its ability to adhere to macrophage-like cells compared to the parental strain. The two M. tuberculosis strains were used to infect J774.A1 cells in the presence of either no serum or 5% NHS. No differences were observed among the two strains in terms of their ability to adhere to the J774.A1 cells (Table 2.) NHS significantly enhanced the binding of both M. tuberculosis strains to the J774.A1 cells compared to the adherence observed in the absence of serum (P ≤ 0.04) (Table 2). These data indicate that HbhA is not necessary for the adherence of M. tuberculosis to macrophage-like J774.A1 cells.
TABLE 2.
Deletion of hbhA does not affect the adherence of M. tuberculosis to J774.A1 cellsa
| M. tuberculosis strain | Serum treatment | Mean log10 CFU ± SD per J774.A1 cell | % of inoculum bound to J774.A1 cells |
|---|---|---|---|
| H37Rv parent | None, inoculum | 6.11 ± 0.31 | NA |
| H37Rv parent | None | 4.25 ± 0.24 | 1 |
| H37Rv parent | NHS | 5.76 ± 0.24 | 45 |
| ΔhbhA mutant | None, inoculum | 5.90 ± 0.10 | NA |
| ΔhbhA mutant | None | 4.34 ± 0.25 | 3 |
| ΔhbhA mutant | NHS | 5.46 ± 0.25 | 36 |
H37Rv and the ΔhbhA mutant were incubated in either no serum or 5% NHS for 40 min at 37°C. The M. tuberculosis cells, still in their respective serum treatments, were then used to infect J774.A1 cells at an MOI of 1:1 for 1 h at 37°C. After 1 h, the J774.A1 cells were washed, lysed, and plated for mycobacteria CFU counts on 7H11 agar. The experiment was performed in duplicate wells in each of two separate experiments. Mean log10 CFU ± SD for one representative experiment is shown. The values for the no serum treatment and the NHS treatment are statistically significant within each strain (P ≤ 0.04) but not between strains (P > 0.10). Student's t test was used for statistical analysis. NA, not applicable.
Binding of human C3 to the intact M. tuberculosis strains.
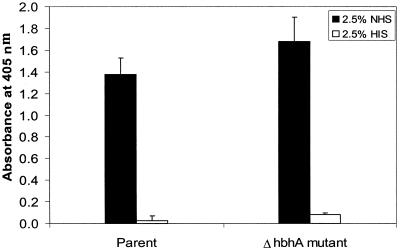
Our group has previously shown that HbhA binds human complement component C3 (20). One explanation as to why the ΔhbhA mutant was not impaired in its ability to bind macrophage-like cells in the presence of NHS is that the mutant is still capable of binding human C3. To assess this possibility, a 96-well plate was coated with the ΔhbhA mutant and wild-type H37Rv whole cells and subsequently incubated with either 2.5% NHS or 2.5% HIS to compare the ability of both M. tuberculosis strains to bind human C3. HIS, in which C3 has been heat inactivated, was used as a negative control for the binding assay. As shown in Fig. 4, the ΔhbhA mutant and the parental strain bound similar amounts of C3 (P = 0.48). C3 was not deposited nonspecifically in the enzyme-linked immunosorbent assay wells, since control wells that lacked M. tuberculosis cells but that were incubated with both NHS and antibody to C3 had absorbance readings similar to those of control wells that received no bacteria and no serum (data not shown). These data indicate that other molecules besides HbhA on the surface of M. tuberculosis are capable of binding human C3.
FIG. 4.
Binding of human C3 to intact M. tuberculosis H37Rv and MtbΔhbhA23. A 96-well plate was coated with whole wild-type H37Rv cells and ΔhbhA mutant cells overnight at 4°C and then incubated with either 2.5% NHS (black bars) or 2.5% HIS (white bars) for 30 min at 37°C. Bound C3 was detected by first incubating the wells for 1 h with a horseradish peroxidase-conjugated goat anti-human C3 antibody, followed by a 15-min incubation with TMB liquid substrate. The optical density at 405 nm was measured in a plate reader. The experiment was performed two times in triplicate. Means ± SD for one representative experiment are shown.
Intracellular survival of the M. tuberculosis strains in macrophage-like cells.
The macrophage-like J774.A1 cells were used to determine if the ΔhbhA mutant differed in its ability to survive in an intracellular environment compared to the parental strain. The parent and the ΔhbhA mutant demonstrated very similar survival rates within the J774.A1 cells (data not shown). Thus, HbhA expression does not appear to be critical for the survival of M. tuberculosis in this intracellular environment.
Survival of the M. tuberculosis strains in mice.
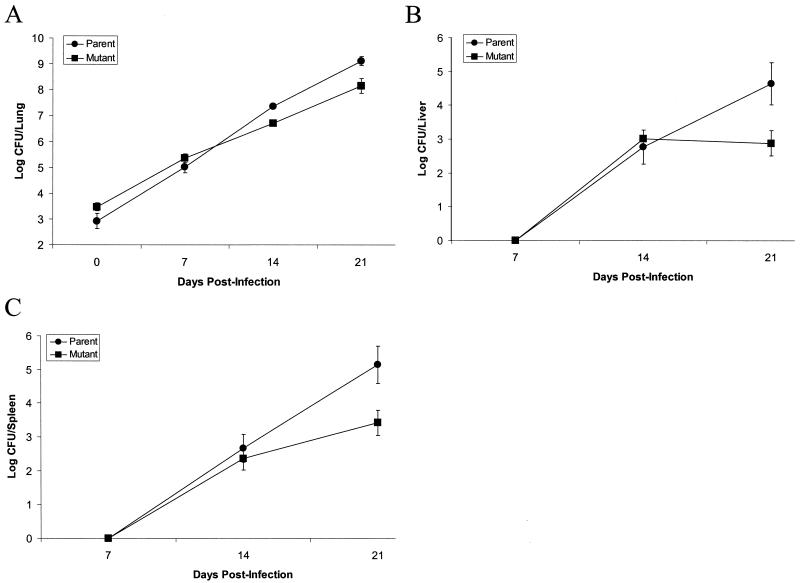
Aerosol inoculation of C57BL/6 mice was employed to determine if the ΔhbhA mutant differed in its ability to survive in vivo compared to the parental strain. The ΔhbhA mutant strain was deficient in growth in the lungs, liver, and spleen compared to the parental strain (Fig. 5.) The initial lung inoculum was slightly greater for the ΔhbhA mutant strain compared to the parental strain; however, by 21 days postinfection lung tissue from the ΔhbhA mutant-inoculated animals contained a full log less organisms than those inoculated with the parental strain (P < 0.01) (Fig. 5A). By 21 days postinfection, mouse livers contained 1.8 logs more of the parental strain than the ΔhbhA mutant strain (P < 0.01) (Fig. 5B). Likewise, after 21 days of infection, mouse spleens infected with the parental strain had 1.7 logs more bacteria compared to spleens that were infected with the ΔhbhA mutant strain (P < 0.01) (Fig. 5C). The reduced numbers of ΔhbhA mutant bacteria in infected mice suggest that HbhA plays a role in the infectivity of M. tuberculosis.
FIG. 5.
Survival and dissemination of M. tuberculosis H37Rv and MtbΔhbhA23 in the organs of C57BL/6 mice infected via aerosol inhalation. The mice were infected with either wild-type H37Rv (circles) or the ΔhbhA mutant (squares). On the indicated days after infection, the mice were sacrificed and their organs were collected and homogenized. The number of bacteria present in the lungs (A), liver (B), and spleen (C) were determined by serial dilution and plating of organ homogenates onto 7H11 agar plates. The mean log CFU ± SD per organ for one experiment with four mice per group at each time point is shown.
Characterization of a second C3-binding protein.
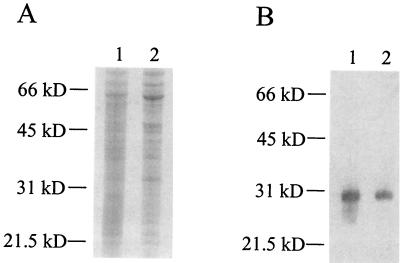
A C3 ligand affinity blotting protocol was previously used by our group to demonstrate binding of C3 to HbhA present in cell lysates of M. tuberculosis (20). Since the ΔhbhA mutant was still capable of binding human C3 (Fig. 4), we wanted to use the C3 ligand affinity blotting protocol to detect other possible C3-binding proteins in a cell lysate of the ΔhbhA mutant. As shown in Fig. 6, C3 bound to a 30-kDa molecule in the ΔhbhA mutant cell lysate just as in the parental strain cell lysate, indicating that a second, comigrating C3-binding protein similar in size to HbhA was present in the M. tuberculosis cell lysates.
FIG. 6.
Detection of a second C3-binding protein in an MtbΔhbhA23 cell lysate by C3 ligand affinity blotting analysis. Cell lysates of M. tuberculosis H37Rv (lane 1) and MtbΔhbhA23 (lane 2) were either stained with Coomassie blue R-250 (A) or transferred to polyvinylidene difluoride membrane and subjected to the C3 ligand affinity blotting technique, using NHS as the source of C3 (B). Molecular mass markers are indicated on the left.
Ion-exchange chromatography was utilized to partially purify the second C3-binding protein from the ΔhbhA mutant. Cells from the ΔhbhA mutant were used for the purification to eliminate any possible comigrating effects from HbhA. Cells were sonicated for 7 min on ice in the absence of protease inhibitors to create a cell lysate, and then Triton X-114 phase partitioning was used to separate the cell lysate into a detergent phase and an aqueous phase. Under these conditions, like HbhA, the second C3-binding protein localized to the aqueous phase. An SP Sepharose column and FPLC were used to purify the second C3-binding protein from the aqueous phase using a continuous gradient of NaCl. The elution fractions were tested for the presence of the second C3-binding protein, using the C3 ligand affinity blotting protocol (data not shown). The fraction containing the C3-binding protein in highest quantity was electrophoresed on a large SDS-12% PAGE gel to better separate the proteins present in the sample, and the protein band corresponding to the 30-kDa C3-binding protein was cut out of the Coomassie-stained gel (data not shown). The protein in the gel slice was treated with the proteinase Lys-C, and two of the resulting peptides were analyzed by mass spectrometry.
The amino acid sequences of the two peptides were determined to be AELIDVLTQK and PTSVPAFRPGAQF. These two sequences matched perfectly to a putative M. tuberculosis HupB ortholog (accession number G70673), which has also been identified as Hlp (accession number P95109) (27). M. tuberculosis HupB is 214 amino acids in length with a predicted molecular mass of 22.2 kDa and a pI of 11.95. This protein has regions that show sequence homology to both bacterial histone-like proteins (HU) and eukaryotic H1 histones. PSORT, which predicts protein localization sites, did not find an N-terminal signal sequence or transmembrane domain and predicted that HupB would be localized in the cytoplasm. TMpred, which predicts protein hydrophobicity, did not identify any transmembrane domains or hydrophobic stretches. PLOTSTRUCTURE (GCG, Wisconsin Package, version 10.0), which examines secondary structure, predicted HupB to have a high α-helical content. Taken together, these computer analyses predict HupB to be located in the cytoplasm of M. tuberculosis, to have a high α-helical content, and to have a highly positive charge with a pI of 11.95.
DISCUSSION
In this study, we created a 576-bp in-frame, unmarked deletion in the 600-bp hbhA gene of M. tuberculosis H37Rv. PCR, Southern blotting, and Western blotting analyses confirmed deletion of the hbhA gene and subsequent loss of HbhA expression. The ΔhbhA mutant and the parental strain did not differ in their abilities to bind human C3 or to adhere to murine J774.A1 macrophage-like cells in either the presence or absence of human serum, indicating that HbhA is not necessary for the binding of C3 to M. tuberculosis or for the adherence of M. tuberculosis to J774.A1 macrophage-like cells. The ΔhbhA mutant grew at the same rate as the parental strain in broth culture; thus, HbhA is not required for the in vitro growth of M. tuberculosis under typical culture conditions. In addition, survival of the ΔhbhA mutant in J774.A1 cells was similar to that of wild-type H37Rv. However, the ΔhbhA mutant exhibited reduced infectivity in C57BL/6 mice. The lungs from infected mice contained 10-fold-fewer ΔhbhA mutant organisms than wild-type organisms 3 weeks after aerosol infection, and the livers and spleens of infected mice contained 84-fold- and 66-fold-fewer ΔhbhA mutant organisms than wild-type organisms, respectively. These data indicate that HbhA is necessary for full virulence of M. tuberculosis in vivo. The decrease in the number of ΔhbhA mutant organisms in the lungs of infected mice could be due to a slower in vivo growth rate of the mutant or to a decrease in the ability of the ΔhbhA mutant to colonize or persist in the lungs. The reduced number of ΔhbhA mutant organisms in the livers and spleens of infected mice could also indicate a decreased ability of the mutant to disseminate to these organs via hematogenous spread.
Recently, Pethe et al. (24) published data describing their mutation of hbhA in M. tuberculosis strain 103 in which a kanamycin resistance gene (aph) was inserted into the hbhA gene. The disruption of hbhA had no effect on the growth of M. tuberculosis in broth culture or in J774.A1 cells or on the adherence of M. tuberculosis to J774.A1 cells, similar to our results. However, binding of the mutant to A549 pneumocytes was reduced 60% compared to the parental strain. The mutant showed decreased numbers of organisms in the spleens, but not the lungs, of BALB/c mice that had been infected via the intranasal route. BALB/c mice were then infected intravenously with the parental and mutant strains, and equal numbers of the parent and mutant were recovered from the spleens, indicating that HbhA plays a role in the dissemination of M. tuberculosis from the lungs to other organs. The hbhA::aph mutant was successfully complemented with an intact hbhA gene inserted at the attB site on the chromosome, which restored all phenotypes to the level of the parental strain. Our data regarding the ΔhbhA mutant agree with that of Pethe et al. (24) in most aspects. Unlike Pethe et al., we showed that mice infected with the ΔhbhA mutant had significantly decreased numbers of bacteria in both lung and spleen tissue (livers were not examined by Pethe et al.) compared to mice infected with the parental strain. We used C57BL/6 mice, whereas the other group used BALB/c mice, which may account for the differences observed in the lungs.
M. tuberculosis can bind to complement receptors (CR1, CR3, and CR4) on mononuclear cells (6, 10, 33, 34, 38, 40), and the binding of M. tuberculosis to mononuclear cells is enhanced in the presence of human complement component C3 (10, 33, 34). Our group previously demonstrated that HbhA has the ability to bind human complement component C3 and mediate attachment of latex beads to macrophage-like cells (20). Our working hypothesis was that binding of C3 to HbhA on M. tuberculosis was an essential step to the binding and internalization of the bacteria in mononuclear cells. However, the absence of HbhA did not affect the ability of the ΔhbhA mutant to bind human C3 or to adhere to murine J774.A1 macrophage-like cells (Table 2; Fig. 4). It is possible that HbhA is only one of several human complement component C3-binding molecules present on the surface of M. tuberculosis. HupB may represent one of these molecules, although it is not predicted to be surface-localized (see below). Other molecules that may be important in C3 binding include trehalose dimycolate (S. L. Mueller, S. J. Norris, and A. R. Wanger, Abstr. 98th Gen. Meet. Am. Soc. Microbiol., abstr. U-98, p. 511-512, 1998) or proteins inactivated by SDS-PAGE (and hence not detected in our C3 ligand binding assay).
M. tuberculosis can also bind to epithelial cells, such as HEp-2 laryngeal cells, alveolar epithelial cells or pneumocytes, CHO cells, and specialized epithelial cells called M cells (2-4, 16, 17, 19, 24, 29, 39). McDonough and Kress (16) also showed that M. tuberculosis is cytotoxic for A549 human lung epithelial cells and that entry of M. tuberculosis into these cells is increased by intracellular passage through macrophages. Likewise, Bermudez and Goodman (2) and Mehta et al. (17) demonstrated that M. tuberculosis has the ability to not only invade A549 human lung epithelial cells but also replicate inside these cells. Teitelbaum et al. (39) showed that M. tuberculosis binds to and invades M cells of mice that have been infected either by the intratracheal route or the intranasal route. Therefore, it is possible that the decreased virulence of the ΔhbhA mutant is due to changes in the interactions with epithelial cells or other host cell types. This hypothesis is supported by the data of Pethe et al. (24), who showed that the M. tuberculosis hbhA::aph mutant was significantly impaired in its ability to bind to pneumocytes.
HbhA has been shown to bind to several extracellular matrix proteins (7, 18, 19, 25). Purified HbhA binds to human pneumocytes in a manner dependent on the presence of heparin (25), and antibody to HbhA inhibits the binding of M. tuberculosis to CHO cells by 50% (19). Reddy and Hayworth (29) and Reddy and Kumar (30) recently demonstrated that M. tuberculosis HbhA and a Mycobacterium avium 31-kDa protein with 89% similarity to M. tuberculosis HbhA bind to HEp-2 cell extracts. The binding of HbhA to host epithelial cells and/or host extracellular matrix proteins may be critical for the persistence and/or dissemination of M. tuberculosis in vivo.
In our previous studies (20), we identified a 30-kDa C3-binding protein from an M. tuberculosis cell lysate using a C3 ligand affinity blotting protocol, and by performing ion-exchange chromatography we were able to purify a single C3-binding protein that was sequenced and identified as HbhA. In our present studies, we were surprised to find that the ΔhbhA mutant continued to bind human C3 despite the absence of HbhA. When we repeated the C3 ligand affinity blotting protocol using the ΔhbhA mutant, we discovered that the mutant still contained a 30-kDa C3-binding protein. We used the ΔhbhA mutant to purify and identify this second C3-binding protein as HupB. HupB and HbhA are similar in size, with HupB predicted to be approximately 0.7 kDa larger than HbhA. HupB has a higher predicted pI than HbhA, 11.95 compared to 8.98. HupB may not have been identified in previous studies due to its high isoelectric point, which may have resulted in its migration outside the pH range of our two-dimensional gels (20).
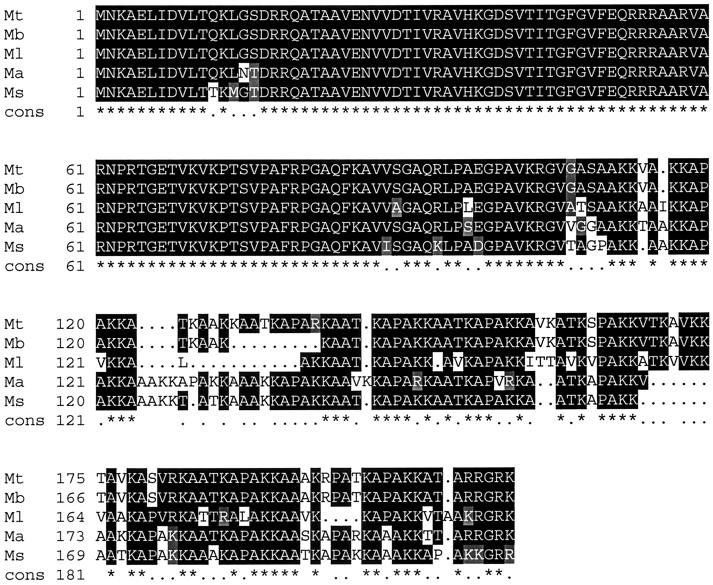
HupB homologs are found in other mycobacteria such as M. bovis (accession number BAA78330.1), M. avium (www.tigr.org), M. smegmatis (accession number AAD13809.1), and M. leprae (accession number O33125). The N-terminal half of the proteins is highly conserved, with less conservation seen in the C-terminal half (Fig. 7.) Based on a computer prediction program called PSORT, HupB is predicted to be located in the cytoplasm of M. tuberculosis. HupB does not contain an N-terminal signal sequence for export out of the cytoplasm, nor does it contain a transmembrane domain for insertion into the cell membrane or cell wall. HupB from M. tuberculosis has been shown to have DNA-binding activity (27), which would also suggest that the protein is predominantly located in the cytoplasm of M. tuberculosis. This localization would obviously prevent binding to host molecules such as human C3. The HupB homologs in M. smegmatis (14, 26, 36) and M. leprae (8, 35) have been characterized as laminin-binding proteins and were reported to be accessible on the cell surface of M. smegmatis and M. leprae, as determined by immunoelectron microscopy using whole bacteria mounted on grids (26, 35). However, this approach does not address the relative proportion of the protein on the cell surface compared to a possible cytoplasmic location; it is possible, for example, that the HupB detected on the surface of mycobacterial cells is due to lysis of bacteria and “decoration” of the surface of neighboring cells with the protein. In addition, the binding of C3 to HupB may be biologically irrelevant, due to the fact that C3 has a broad range of binding activities and is able to form covalent bonds with OH or NH2 side chains on target molecules. As a result, C3 can bind to serine, threonine, tyrosine, and lysine residues, and to carbohydrates (13, 15, 37). HupB, like HbhA, has an abundance of lysine residues and has been shown to be a glycoprotein in M. smegmatis (24). Expression and purification of recombinant HupB in E. coli, as well as gene inactivation in M. tuberculosis, should shed light on the C3-binding characteristics of this protein and its potential role in mycobacterial pathogenesis.
FIG. 7.
Comparison of the amino acid sequences of M. tuberculosis HupB and the M. bovis, M. leprae, M. avium, and M. smegmatis HupB homologs. Periods (.) within the alignment itself represent gaps introduced for optimal alignment. A consensus sequence is shown below each line of the alignment, with identical amino acids indicated by an asterisk (*) and similar amino acids indicated by a period (.). Mt = M. tuberculosis; Mb = M. bovis; Ml = M. leprae; Ma = M. avium; and Ms = M. smegmatis.
Acknowledgments
We thank L. Y. Armitige for helpful discussions and suggestions, and we also thank A. Ballard for providing plasmid pJQ200mp18.
This work was supported in part by the Gilson Longenbaugh Foundation of Houston, Tex.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Armitige, L. Y., C. Jagannath, A. R. Wanger, and S. J. Norris. 2000. Disruption of the genes encoding antigen 85A and antigen 85B of Mycobacterium tuberculosis H37Rv: effect on growth in culture and in macrophages. Infect. Immun. 68:767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bermudez, L. E., and J. Goodman. 1996. Mycobacterium tuberculosis invades and replicates within type II alveolar cells. Infect. Immun. 64:1400-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bermudez, L. E., K. Shelton, and L. S. Young. 1995. Comparison of the ability of Mycobacterium avium, M. smegmatis and M. tuberculosis to invade and replicate within HEp-2 epithelial cells. Tuber. Lung Dis. 76:240-247. [DOI] [PubMed] [Google Scholar]
- 4.Birkness, K. A., M. Deslauriers, J. H. Bartlett, E. H. White, C. H. King, and F. D. Quinn. 1999. An in vitro tissue culture bilayer model to examine early events in Mycobacterium tuberculosis infection. Infect. Immun. 67:653-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 6.Cywes, C., H. C. Hoppe, M. Daffe, and M. R. Ehlers. 1997. Nonopsonic binding of Mycobacterium tuberculosis to complement receptor type 3 is mediated by capsular polysaccharides and is strain dependent. Infect. Immun. 65:4258-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Delogu, G., and M. J. Brennan. 1999. Functional domains present in the mycobacterial hemagglutinin, HBHA. J. Bacteriol. 181:7464-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Melo Marques, M. A., S. Mahapatra, D. Nandan, T. Dick, E. N. Sarno, P. J. Brennan, and M. C. Vidal Pessolani. 2000. Bacterial and host-derived cationic proteins bind α2-laminins and enhance Mycobacterium leprae attachment to human Schwann cells. Microbes Infect. 2:1407-1417. [DOI] [PubMed] [Google Scholar]
- 9.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. W.H.O. Global Surveillance and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch, C. S., J. J. Ellner, D. G. Russell, and E. A. Rich. 1994. Complement receptor-mediated uptake and tumor necrosis factor-alpha-mediated growth inhibition of Mycobacterium tuberculosis by human alveolar macrophages. J. Immunol. 152:743-753. [PubMed] [Google Scholar]
- 11.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 12.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 13.Law, S. K., T. M. Minich, and R. P. Levine. 1984. Covalent binding efficiency of the third and fourth complement proteins in relation to pH, nucleophilicity, and availability of hydroxyl groups. Biochemistry 23:3267-3272. [DOI] [PubMed] [Google Scholar]
- 14.Lee, B. H., B. Murugasu-Oei, and T. Dick. 1998. Upregulation of a histone-like protein in dormant Mycobacterium smegmatis. Mol. Gen. Genet. 260:475-479. [DOI] [PubMed] [Google Scholar]
- 15.Levine, R. P., R. Finn, and R. Gross. 1983. Interactions between C3b and cell-surface macromolecules. Ann. N. Y. Acad. Sci. 421:235-245. [DOI] [PubMed] [Google Scholar]
- 16.McDonough, K. A., and Y. Kress. 1995. Cytotoxicity for lung epithelial cells is a virulence-associated phenotype of Mycobacterium tuberculosis. Infect. Immun. 63:4802-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta, P. K., C. H. King, E. H. White, J. J. Murtagh, Jr., and F. D. Quinn. 1996. Comparison of in vitro models for the study of Mycobacterium tuberculosis invasion and intracellular replication. Infect. Immun. 64:2673-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menozzi, F. D., R. Bischoff, E. Fort, M. J. Brennan, and C. Locht. 1998. Molecular characterization of the mycobacterial heparin-binding hemagglutinin, a mycobacterial adhesin. Proc. Natl. Acad. Sci. USA 95:12625-12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menozzi, F. D., J. H. Rouse, M. Alavi, M. Laude-Sharp, J. Muller, R. Bischoff, M. J. Brennan, and C. Locht. 1996. Identification of a heparin-binding hemagglutinin present in mycobacteria. J. Exp. Med. 184:993-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller-Ortiz, S. L., A. R. Wanger, and S. J. Norris. 2001. Mycobacterial protein HbhA binds human complement component C3. Infect. Immun. 69:7501-7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 22.Pavelka, M. S., Jr., and W. R. Jacobs, Jr. 1999. Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis bacillus Calmette-Guerin, and Mycobacterium tuberculosis H37Rv by allelic exchange. J. Bacteriol. 181:4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol. Microbiol. 20:919-925. [DOI] [PubMed] [Google Scholar]
- 24.Pethe, K., S. Alonso, F. Biet, G. Delogu, M. J. Brennan, C. Locht, and F. D. Menozzi. 2001. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature 412:190-194. [DOI] [PubMed] [Google Scholar]
- 25.Pethe, K., M. Aumercier, E. Fort, C. Gatot, C. Locht, and F. D. Menozzi. 2000. Characterization of the heparin-binding site of the mycobacterial heparin-binding hemagglutinin adhesin. J. Biol. Chem. 275:14273-14280. [DOI] [PubMed] [Google Scholar]
- 26.Pethe, K., V. Puech, M. Daffe, C. Josenhans, H. Drobecq, C. Locht, and F. D. Menozzi. 2001. Mycobacterium smegmatis laminin-binding glycoprotein shares epitopes with Mycobacterium tuberculosis heparin-binding haemagglutinin. Mol. Microbiol. 39:89-99. [DOI] [PubMed] [Google Scholar]
- 27.Prabhakar, S., P. S. Annapurna, N. K. Jain, A. B. Dey, J. S. Tyagi, and H. K. Prasad. 1998. Identification of an immunogenic histone-like protein (HLPMt) of Mycobacterium tuberculosis. Tuber. Lung Dis. 79:43-53. [DOI] [PubMed] [Google Scholar]
- 28.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 29.Reddy, V. M., and D. A. Hayworth. 2002. Interaction of Mycobacterium tuberculosis with human respiratory epithelial cells (HEp-2). Tuberculosis 82:31-36. [DOI] [PubMed] [Google Scholar]
- 30.Reddy, V. M., and B. Kumar. 2000. Interaction of Mycobacterium avium complex with human respiratory epithelial cells. J. Infect. Dis. 181:1189-1193. [DOI] [PubMed] [Google Scholar]
- 31.Riley, L. W. 1996. Phagocytosis of M. tuberculosis, p. 281-289. In W. N. Rom and S. M. Garay (ed.), Tuberculosis. Little, Brown and Company, New York, N.Y.
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 33.Schlesinger, L. S. 1993. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J. Immunol. 150:2920-2930. [PubMed] [Google Scholar]
- 34.Schlesinger, L. S., C. G. Bellinger-Kawahara, N. R. Payne, and M. A. Horwitz. 1990. Phagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3. J. Immunol. 144:2771-2780. [PubMed] [Google Scholar]
- 35.Shimoji, Y., V. Ng, K. Matsumura, V. A. Fischetti, and A. Rambukkana. 1999. A 21-kDa surface protein of Mycobacterium leprae binds peripheral nerve laminin-2 and mediates Schwann cell invasion. Proc. Natl. Acad. Sci. USA 96:9857-9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shires, K., and L. Steyn. 2001. The cold-shock stress response in Mycobacterium smegmatis induces the expression of a histone-like protein. Mol. Microbiol. 39:994-1009. [DOI] [PubMed] [Google Scholar]
- 37.Shohet, J. M., P. Pemberton, and M. C. Carroll. 1993. Identification of a major binding site for complement C3 on the IgG1 heavy chain. J. Biol. Chem. 268:5866-5871. [PubMed] [Google Scholar]
- 38.Stokes, R. W., L. M. Thorson, and D. P. Speert. 1998. Nonopsonic and opsonic association of Mycobacterium tuberculosis with resident alveolar macrophages is inefficient. J. Immunol. 160:5514-5521. [PubMed] [Google Scholar]
- 39.Teitelbaum, R., W. Schubert, L. Gunther, Y. Kress, F. Macaluso, J. W. Pollard, D. N. McMurray, and B. R. Bloom. 1999. The M cell as a portal of entry to the lung for the bacterial pathogen Mycobacterium tuberculosis. Immunity 10:641-650. [DOI] [PubMed] [Google Scholar]
- 40.Zaffran, Y., L. Zhang, and J. J. Ellner. 1998. Role of CR4 in Mycobacterium tuberculosis-human macrophage binding and signal transduction in the absence of serum. Infect. Immun. 66:4541-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]