Abstract
Enterohemorrhagic Escherichia coli (EHEC) is a food-borne cause of bloody diarrhea and the hemolytic-uremic syndrome (HUS) in humans. Most strains of EHEC belong to a group of bacterial pathogens that cause distinctive lesions on the host intestine termed attaching-and-effacing (A/E) lesions. A/E strains of EHEC, including the predominant serotype, O157:H7, are responsible for the majority of HUS outbreaks worldwide. However, several serotypes of EHEC are not A/E pathogens because they lack the locus of enterocyte effacement (LEE) pathogenicity island. Nevertheless, such strains have been associated with sporadic cases and small outbreaks of hemorrhagic colitis and HUS. Of these LEE-negative organisms, O113:H21 is one of the most commonly isolated EHEC serotypes in many regions. Clinical isolates of LEE-negative EHEC typically express Shiga toxin 2 and carry an ∼90-kb plasmid that encodes EHEC hemolysin, but in the absence of LEE, little is known about the way in which these pathogens colonize the host intestine. In this study we describe the identification of a novel fimbrial gene cluster related to long polar fimbriae in EHEC O113:H21. This chromosomal region comprises four open reading frames, lpfA to lfpD, and has the same location in the EHEC O113:H21 genome as O island 154 in the prototype EHEC O157:H7 strain, EDL933. In a survey of EHEC of other serotypes, homologues of lpfAO113 were found in 26 of 28 LEE-negative and 8 of 11 non-O157:H7 LEE-positive EHEC strains. Deletion of the putative major fimbrial subunit gene, lpfA, from EHEC O113:H21 resulted in decreased adherence of this strain to epithelial cells, suggesting that lpfO113 may function as an adhesin in LEE-negative isolates of EHEC.
Shiga toxin-producing strains of enterohemorrhagic Escherichia coli (EHEC) are a prominent cause of acute gastroenteritis, hemorrhagic colitis, and the hemolytic-uremic syndrome (HUS) in humans (17, 23). HUS is characterized by acute renal failure, thrombocytopenia, and microangiopathic hemolytic anemia, and those most at risk of developing renal disease are children under 10 years and the elderly. In approximately 3 to 5% of affected children, HUS is fatal, while 12 to 30% suffer serious impairment of renal function or neurologic damage (17, 23).
Although the carriage of bacteriophage-encoded Shiga toxin genes (stx) is a defining feature of virulence, strains of EHEC also harbor a large ∼90-kb plasmid, pEHEC, and/or a large chromosomal pathogenicity island, termed the locus for enterocyte effacement (LEE) (6, 14). Strains of EHEC carrying LEE belong to a group of bacterial pathogens that cause distinctive lesions on the host intestine termed attaching-effacing (A/E) lesions. This group of organisms also includes the human pathogen enteropathogenic E. coli (EPEC) and the animal pathogens rabbit-specific enteropathogenic E. coli (REPEC) and Citrobacter rodentium. A/E lesions are characterized by localized destruction (effacement) of intestinal brush border microvilli, intimate attachment of the bacteria to the host cell membrane, and the formation of underlying actin-rich, pedestal-like structures in the host cell (11, 17). The formation of A/E lesions is essential for bacterial colonization of the intestinal mucosa by LEE-positive pathogens. Knockout mutations in eae, espB, tir, and espA abolish the ability of A/E pathogens to cause A/E lesions and seriously impair the capacity of these organisms to colonize the host and cause disease (1, 8, 15, 16, 28, 31). Nevertheless, some serotypes of EHEC do not carry LEE and are not A/E pathogens (9, 22). These strains have regularly been associated with sporadic cases and small outbreaks of hemorrhagic colitis and HUS, but little is known about the way in which these LEE-negative EHEC adhere to and colonize the human intestine. Clinical isolates of LEE-negative EHEC typically express Shiga toxin type 2 and also harbor an ∼90-kb plasmid which encodes EHEC hemolysin (22). However, restriction fragment length polymorphism analysis of the ehxA gene has shown that the large plasmids from LEE-negative EHEC comprise an evolutionarily distinct group compared with the similarly sized plasmids of LEE-positive EHEC (5). EHEC O113:H21 is one of the most commonly isolated LEE-negative EHEC serotypes worldwide, but until recently no virulence-associated genes apart from stx and ehx in these bacteria had been documented. Recently, however, Paton et al. identified a novel plasmid-encoded autoagglutinating adhesin, termed Saa, in LEE-negative EHEC O113:H21 (21). Saa is a 516-amino-acid protein with a low degree of similarity to the adhesin YadA of Yersinia enterocolitica. Mutation of the saa gene or curing of the pO113 plasmid resulted in reduced adhesion to epithelial cells (21). Dytoc et al. have demonstrated the pili on the surfaces of EHEC O113:H21 cells, but the genes coding for pili in these strains or any LEE-negative EHEC have not been described (9).
The most common serotype of EHEC associated with outbreaks and sporadic disease worldwide is O157:H7 (14). Recently, the genome sequences of two EHEC O157:H7 outbreak strains were completed. Analysis of these sequences showed that EHEC O157:H7 has 1.34 Mb of DNA that is not present in nonpathogenic E. coli K-12 (12, 24). The additional regions of DNA (termed O islands) are inserted into a common “backbone” shared by E. coli K-12 and EHEC O157:H7 and vary in size from a few hundred base pairs up to 88 kb. About 33% of the 177 O islands contain only genes of unknown function. Many genes in the other O islands have been classified according to their homology with known virulence factors, although their role in disease is not proven (24). Several O islands contain putative fimbrial biosynthesis operons, including two regions predicted to encode long polar fimbriae (LPF). LPF are related to type I fimbriae in genetic organization and were first identified in Salmonella enterica serovar Typhimurium (3). They form surface structures 7 to 8 nm in diameter and 2 to 10 μm in length extending from the poles of the bacterial cell. In S. enterica serovar Typhimurium, LPF have been shown to facilitate attachment of the bacteria to murine Peyer's patches. Synthesis of LPF is controlled by a phase-variable on/off switch that regulates expression of LPF in different tissues and organs (4). Phase variation of LPF is also a mechanism by which S. enterica serovar Typhimurium evades cross immunity among different Salmonella serotypes (19). In EHEC O157:H7, two putative lpf operons are present as O islands 141 and 154; however, the expression and production of LPF in EHEC O157:H7 has not yet been demonstrated, and their contribution to virulence is unknown (24). In this study, we report the identification of a novel fimbrial gene cluster related to LPF in LEE-negative EHEC serotype O113:H21. We also describe the contribution of these genes to the adherence of EHEC O113:H21 to epithelial cells and the relationship of lpfO113 to lpf gene clusters in other bacteria.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains (except those used for hybridization studies) and plasmids used in this study are listed in Table 1. EHEC O113:H21 (EH41) was isolated from a child with HUS in New South Wales, Australia, and expresses Stx-2 and EHEC hemolysin. Bacteria were grown in Luria-Bertani (LB) broth at 37°C with shaking or on LB agar at 37°C. Where required, antibiotics were added to media at the following concentrations: kanamycin, 100 μg/ml; streptomycin, 50 μg/ml; ampicillin, 100 μg/ml. For the selection of TnphoA mutants, XP (5-bromo-4-chloro-3-indolylphosphate) was added to media at 100 μg/ml. For the induction of lambda red recombinase from plasmid pKD46, 100 mM arabinose was added to LB broth and competent cells were prepared by growth at 30°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| ORN103 | thr-1 leu-6 thi-1 Δ(argF-lac) U169 xyl-7 ara-13 mtl-2 gal-6 rpsL tonA2 fhuA2 minA monB recA13 Δ(fimABCDEFGH) | 33 |
| XL-1 Blue | supE44 hsdR17 recA1 endA1 gyrA46 thi relA1 lac F′ [proAB+lacIqlacZ ΔM15 Tn10] (Tetr) | Stratagene |
| EH41 | Wild-type EHEC O113:H21 HUS isolate (Smr) | This study |
| EH41lpfA− | lpfA deletion mutant of EH41 (Smr Kmr) | This study |
| 15-26 | EH41 pstS::TnphoA (Smr Kmr) | This study |
| 83/39 | Wild-type REPEC O15:H− | 2 |
| Plasmids | ||
| PCR-Script | High-copy-number cloning vector (Ampr) | Stratagene |
| pWSK29 | Low-copy-number cloning vector (Ampr) | 35 |
| pWSK:lpf | pWSK29 carrying lpfABCDO113 | This study |
| pRT733 | Suicide plasmid carrying TnphoA (Ampr) | 32 |
| pKD46 | Low-copy-number thermosensitive plasmid encoding red recombinase | 7 |
| pKD4 | Template for PCR amplification of kanamycin cassette for red recombinase-mediated recombination | 7 |
Bacterial adhesion to epithelial cells.
Qualitative assessment of bacterial adherence to tissue culture cells was performed as described by Vial et al. (34). Washed semiconfluent monolayers of CHO-K1 cells were infected with bacterial strains grown overnight at 37°C in LB broth supplemented with 0.5% glucose. Approximately 105 CHO-K1 cells on glass coverslips were incubated with around 106 bacteria for 4 h in the presence of 0.5% mannose. Following the incubation period, cells were washed five times with phosphate-buffered saline to remove nonadherent bacteria, fixed in methanol, and stained with Giemsa. Mounted coverslips were viewed by bright-field microscopy using a Leica DMLB compound microscope, and digital images were captured with a Leica DC300 digital camera.
Quantitative assessment of bacterial association with CHO-K1 cells was determined as described previously (26). Briefly, washed semiconfluent monolayers of CHO-K1 cells were infected in the presence of 0.5% mannose with around 106 CFU of different bacterial strains for 4 h. Cell monolayers were washed five times with phosphate-buffered saline and lysed in 0.1% digitonin. Following lysis, bacteria were resuspended in LB broth and quantified by serial dilution. Assays were carried out in duplicate, and the results from at least three experiments were expressed as a percentage of the original inoculum (mean ± standard deviation). Differences in adherence were assessed for significance by using Student's two-tailed t test.
DNA manipulations and DNA sequencing.
Routine DNA manipulations (restriction digestion, ligation, agarose gel electrophoresis, transformation, labeling, and Southern and dot blot hybridizations) were performed according to standard techniques described by Sambrook et al. (27). Genomic DNA was prepared by using a DNeasy extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Marker rescue of a 7.1-kb BamHI fragment containing the TnphoA insertion site into PCR-Script cut with BamHI was performed as described previously (32). Plasmid DNA for sequencing was purified using a QIAprep spin miniprep kit (Qiagen). Both strands of the rescued fragment were sequenced with custom-made oligonucleotide primers using a PRISM ready-reaction DyeDeoxy terminator cycle sequencing kit (Applied Biosystems). The DNA sequence was determined by automated DNA sequencing using an Applied Biosystems model 373A DNA sequencing system. DNA sequences were assembled by using Sequencher 3.1.1 (GeneCodes Corp., Ann Arbor, Mich.). BLAST programs were used to determine DNA homology and amino acid similarities and identities in pairwise comparisons of DNA sequences. The neural network promoter prediction (NNPP) program for prokaryotes was used to predict promoter sequences upstream of lpfA (25). The CLUSTAL W program was used to generate multiple amino acid alignments (13).
Identification and cloning of the lpfO113 gene cluster.
The transposon TnphoA was introduced into EHEC O113:H21 (EH41) on the suicide plasmid pRT733 by conjugation as described previously (32). Blue (in-frame) TnphoA mutants derived from a streptomycin-resistant derivative of EH41 were selected from LB agar containing streptomycin, kanamycin, and XP and tested for their ability to adhere to a semiconfluent monolayer of CHO-K1 cells. Of the 412 TnphoA mutants screened, only one mutant was identified that showed an apparent reduction in adherence to CHO-K1 cells. The transposon insertion site of the mutant, 15-26, was cloned by marker rescue of a BamHI fragment into PCR-Script. The entire lpf gene cluster from EHEC O113:H21 was amplified from genomic DNA with the Expand long-template PCR system (Roche Diagnostics, Mannheim, Germany) and the oligonucleotide primers O-154F (5′-CTGGCAAAATCGGTAACGGT-3′) and O-154R (5′-CCACCGGAAGAACCGAT-3′). PCR conditions were as follows: 94°C for 2 min followed by 30 cycles of 94°C for 30 s, 50°C for 30 s, and 68°C for 6 min and then an extension period of 7 min at 68°C. The final 5.5-kb product was purified with the Concert nucleic acid purification system (Gibco BRL) and cloned into the T-tailed pDK101 for DNA sequencing. The complete DNA sequence was determined as described above. For adherence studies, the lpf locus was cloned into pWSK29 digested with EcoRV. O-island amplifications on other bacterial strains were performed with O-154F and O-154R or the primers O-141F (5′-AAAAGTGTGGGGAAAGAGTG-3′) and O-141R (5′-AGCAGAAAGTATTGCGTGAG-3′). PCR conditions were identical except that an annealing temperature of 45°C was used for O-141F and O141-R.
Dot blot hybridizations and generation of an lpfAO113 probe.
Hybridization studies were performed with strains from our culture collection as reported previously (18). These strains were isolated from clinical specimens and from animal, food, and environmental sources in a variety of geographic locations. Genomic DNA extracted from these strains was spotted onto Hybond-N membranes (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) and hybridized under stringent conditions with a digoxigenin (DIG)-labeled lpfAO113 probe. Oligonucleotide primers used to generate the lpfAO113 probe were lpfA-F (5′-ATGAAGCGTAATATTATAG-3′) and lpfA-R (5′-TTATTTCTTATATTCGAC-3′), with genomic DNA from EHEC O113:H21 (EH41) as the template. Primer sequences were chosen on the basis that they did not match any DNA sequences in GenBank, and specificity was confirmed by successful amplification of lpfAO113 from EH41 and no amplification from EHEC O157:H7 (EDL933), which is known not to carry lpfO113. By using these primers, DIG-labeled nucleotides (Roche) were incorporated by PCR using the following conditions: 94°C for 2 min followed by 30 cycles of 94°C for 1 min, 52°C for 50 s, and 72°C for 1 min, with an extension period of 5 min at 72°C. Strains that tested positive by hybridization for lpfAO113 were confirmed by PCR analysis using the above-described primers and conditions.
Construction of an lpfA deletion mutant of EHEC O113:H21 (EH41).
To generate an lpfA deletion mutant of strain EH41, the lpfA gene was replaced by a gene encoding kanamycin resistance by using the lambda red recombinase system (7). Long oligonucleotide primers designed for introducing the mutation were lpfAKOF (5′ATAAGTCGATGATTCATGGTAAAGGATATATTATATCAATGTGTAGGCTGGAGCTGCTTC-3′) and lpfAKOR (5′-CTGGCCAAAGCCAACTGAATAAAAAGGCCCTTGATAAATTCATATGAATATCCTCCTTAG-3′). Each primer included 20 bp of sequence homologous to the template plasmid, pKD4, and 40 bp of sequence homologous to regions flanking the lpfA gene. The kanamycin resistance gene was amplified from pKD4 by PCR. The product was treated with DpnI to remove the pKD4 template and purified by using the Concert nucleic acid purification system. The purified PCR product was electroporated into EH41 which had previously been transformed with the lambda red recombinase expression vector, pKD46. Electrocompetent EH41 cells were prepared by culturing the bacteria at 30°C in 100 mM arabinose to induce expression of lambda red recombinase. Following electroporation, transformants of EH41 were recovered at 30°C for 2 h in LB broth supplemented with 100 mM arabinose and plated onto LB agar containing kanamycin for overnight growth at 37°C to induce the loss of pKD46. Kanamycin-resistant colonies were then tested by PCR for replacement of lpfA by the kanamycin resistance marker. The lpfA deletion mutant of EH41 was complemented in trans by introduction of the entire lpfO113 gene cluster on pWSK:lpf.
Nucleotide sequence accession number.
The DNA sequence determined in this study has been assigned GenBank accession no. AY057066.
RESULTS
Identification and cloning of a novel fimbrial gene cluster.
To identify novel factors required for the adherence of LEE-negative EHEC to epithelial cells, we performed TnphoA mutagenesis on a streptomycin-resistant derivative of EHEC O113:H21 (EH41). EH41 was originally isolated from a child with HUS in southeastern Australia. We identified one blue TnphoA mutant (designated 15-26) that showed reduced adherence to CHO-K1 cells compared to the parent strain. We subsequently cloned the TnphoA insertion site from this mutant into PCR-Script by marker rescue of a BamHI fragment. DNA sequencing of the 7.1-kb rescued fragment revealed that 15-26 contained an in-frame insertion of TnphoA into pstS, an E. coli gene predicted to encode a periplasmic phosphate-binding protein. Translated BLAST analysis of the other end of the rescued fragment revealed an open reading frame (ORF) with predicted amino acid similarity to LpfD from S. enterica serovar Typhimurium. In S. enterica serovar Typhimurium, lpfD is part of an operon involved in the expression of LPF and is believed to encode a minor structural component of the fimbriae (3). The genome of EHEC O157:H7 also contains two novel, putative lpf operons. These are present as O islands 141 and 154, where O island 154 is positioned next to pstS and O island 141 (as well as the lpf operon from S. enterica serovar Typhimurium) is located next to yhjW (Fig. 1) (24). The presence of lpfD in the chromosome of EH41 adjacent to pstS suggested that we had identified an LPF operon in EH41. To clone the putative EHEC O113:H21 O-island insertion containing LPF, we designed primers to flanking regions of the E. coli chromosome in the genes pstS and glmS (encoding l-glutamine:d-fructose-6-phosphate aminotransferase) and amplified the O-island insertion by PCR. The 5.5-kb product was cloned into PCR-Script, and DNA sequencing revealed four complete ORFs, lpfA, lpfB, lpfC, and lpfD (Fig. 1A). The overall G+C content was 43.7% (compared with 50.8% for the E. coli K-12 genome), suggesting that the lpf gene cluster (lpfO113) in the chromosome of EHEC O113:H21 (EH41) was acquired by lateral gene transfer. However, we could find no evidence of insertion elements or integrases or evidence of other mobile genetic elements adjacent to the lpfO113 locus.
FIG. 1.
(A) Genetic organization of the lpf gene clusters from strains of E. coli and S. enterica serovar Typhimurium showing the approximate sizes and number of ORFs of the gene clusters and the E. coli K-12 gene adjacent to the chromosomal insertion site. (B) Analysis of the region upstream from the lpfO113 gene cluster. Putative −10 and −35 promoter sites are underlined, and a putative ribosome binding site is shaded. The amino acid translation is shown above the nucleotide sequence. A potential transcription start point is in bold type and uppercase, and the coding region of lpfA is in bold type. The promoter region was analyzed with the NNPP program for prokaryotes.
Using the NNPP promoter search program for prokaryotes, we identified a putative ribosome binding site and potential −10 and −35 promoter sequences in the nucleotide region upstream from the first ORF, lpfAO113 (Fig. 1B). Potential ribosome binding sites were also identified between coding regions for lpfBO113, lpfCO113, and lpfDO113 (data not shown). The distances of 10 to 50 nucleotides between the four ORFs suggested that these fimbrial biosynthesis genes were part of an operon.
Homology of LpfABCDO113 to other fimbrial proteins.
The lpf operons of S. enterica serovar Typhimurium, EHEC O157:H7, and EHEC O113:H21 carry genes predicted to encode chaperone-usher fimbrial biosynthesis systems and resemble members of the type I pilus family in their organization. By homology to other fimbrial proteins, LpfAO113 is predicted to be a major fimbrial subunit protein, LpfBO113 is predicted to be a putative chaperone, LpfCO113 is predicted to be an outer membrane usher, and LpfDO113 is predicted to be a putative minor fimbrial component. In addition to lpfABCD, the lpf operons from S. enterica serovar Typhimurium and O island 141 from EHEC O157:H7 contain lpfE, which is predicted to encode a minor tip adhesin (Fig. 1A). The properties of the four putative lpfO113 gene products and their amino acid identities and similarities with other related fimbrial proteins are given in Table 2. The putative fimbrial components LpfAO113 and LpfDO113 from EHEC O113:H21 (EH41) exhibited the highest amino acid identity to homologues encoded by O island 154 from EHEC O157:H7, where the genetic loci have the same location in the bacterial chromosome. Across the entire lpfO113 O island, however, no DNA homology to any sequences in GenBank was detected. An amino acid alignment between LpfAO113 and homologues from EHEC O157:H7 and S. enterica serovar Typhimurium is shown in Fig. 2. Like other major fimbrial subunits of the type I family, the putative LpfAO113 product has conserved cysteine residues at positions 19 and 59 of the mature pilin which form a disulfide bridge in other pilins (Cys39 and Cys79) (Fig. 2). LpfAO113 also carries a predicted chaperone-binding domain characterized by an invariant glycine residue (Gly176) and aromatic residue (Tyr188) separated by a series of alternating hydrophobic residues (Fig. 2).
TABLE 2.
Predicted molecular masses and pI of the putative lpfO113 gene products and amino acid identities (and similarities) with related LPF proteins from EHEC O157:H7 EDL933 and S. enterica serovar Typhimurium LT-2
| Gene | Predicted characteristic of product
|
% Amino acid identity (similarity) of lpfO113 with:
|
||||
|---|---|---|---|---|---|---|
| No. of amino acids | Mass (kDa) | pI | EDL933 O island 154 (AE005604a) | EDL933 O island 141 (AE005581a) | LT-2 (AE008868a) | |
| lpfA | 190 | 19.7 | 4.9 | 54 (68) | 30 (44) | 28 (42) |
| lpfB | 243 | 27.2 | 9.6 | 30 (38) | 32 (53) | 34 (50) |
| lpC | 840 | 92.8 | 4.8 | 64 (81) | 23 (35) | 39 (61) |
| lpfD | 265 | 29.1 | 6.2 | 34 (49)[Z5220], 30 (44)[Z5221]b | 24 (37) | 28 (39) |
Accession number.
O island 154 contains two copies of LpfD.
FIG. 2.
Amino acid alignment of the predicted lpfAO113 product and related fimbrial proteins. The alignment was performed with CLUSTAL software, and amino acid numbers are on the right. Gaps are indicated by a dashed line, and conserved amino acids are shaded in black. Cysteine residues (Cys39 and Cys79) predicted to form a disulfide bridge are indicated by an asterisk. The invariant glycine (Gly176) and aromatic (Tyr188) residues predicted to comprise a chaperone-binding domain are also indicated (#).
Distribution of lpfAO113 among other pathogenic E. coli and enteric pathogens.
The identification of a novel fimbrial gene cluster in an EHEC strain prompted us to examine the distribution of lpfAO113 among other strains of EHEC, EPEC, and REPEC and other pathogenic E. coli strains and enteric pathogens. lpfAO113-specific primers were used to amplify lpfAO113 from genomic preparations of DNA derived from test strains or used to generate a DIG-labeled lpfAO113 probe by PCR from EH41 which was then used for dot blot hybridization. Overall, lpfAO113 was detected by PCR and dot blot hybridization in 26 of 28 LEE-negative EHEC strains tested, including all strains of O113:H21 (Table 3). Of the EHEC O113:H21 strains included in this study, seven strains were clinical isolates from independent cases of HUS, thrombotic thrombocytopenic purpura, or hemorrhagic colitis, three were isolated from food sources, and one was of bovine origin. lpfAO113 was also detected in 8 of 11 non-O157:H7 LEE-positive EHEC strains. In contrast, only 1 of 11 EPEC strains contained lpfAO113. Interestingly, the lpfAO113-positive EPEC strain was O26:H−, which is a serotype common to the EHEC pathotype. lpfAO113 was also detected in four of nine REPEC strains tested (Table 3). In further testing by dot blot hybridization, we found lpfAO113 in one of five strains of enterotoxigenic E. coli but could not detect lpfAO113 in any strains of enteroaggregative E. coli (zero of five), enteroinvasive E. coli (zero of five), S. enterica serovar Typhimurium (zero of five), Y. enterocolitica (zero of three), Pseudomonas aeruginosa (zero of two), Shigella spp. (zero of four), or Klebsiella spp. (zero of three).
TABLE 3.
Prevalence of lpfAO113 among serogroups of EHEC, EPEC, and REPEC
| Pathotype | Serogroup | Presence of eaea | No. of strains positive for lpfAO113/no. tested |
|---|---|---|---|
| EHEC | O157:H7 | + | 0/2 |
| O111:H− | + | 3/4 | |
| O15:H− | + | 1/1 | |
| O26:H11 | + | 1/2 | |
| O147:H− | + | 1/1 | |
| O145:H25 | + | 1/1 | |
| O5:H− | + | 1/1 | |
| O113:H21 | − | 11/11 | |
| O116:H21 | − | 1/1 | |
| O130:H11 | − | 1/1 | |
| O5:H− | − | 2/2 | |
| O1:H7 | − | 0/1 | |
| NT:H7 | − | 1/1 | |
| NT:H− | − | 1/1 | |
| O48:H21 | − | 1/1 | |
| O76:H7 | − | 0/1 | |
| O128:H2 | − | 3/3 | |
| O91:H− | − | 2/2 | |
| O87:H16 | − | 1/1 | |
| OR:H− | − | 1/1 | |
| O123:H− | − | 1/1 | |
| EPEC | O127:H6 | + | 0/1 |
| O111:H2 | + | 0/2 | |
| O114:H2 | + | 0/1 | |
| O125:H2 | + | 0/1 | |
| O128:H2 | + | 0/1 | |
| O142:H6 | + | 0/2 | |
| O55:H7 | + | 0/1 | |
| O26:H− | + | 1/1 | |
| O86:H− | + | 0/1 | |
| REPEC | O15:H− | + | 2/2 |
| O103:H2 | + | 0/3 | |
| O153:H7 | + | 1/1 | |
| O20:H7 | + | 1/1 | |
| O8 | + | 0/1 | |
| O109:H2 | + | 0/1 |
Marker for LEE pathogenicity island.
Contribution of LPFO113 to adherence of EHEC O113:H21 (EH41) to CHO-K1 epithelial cells.
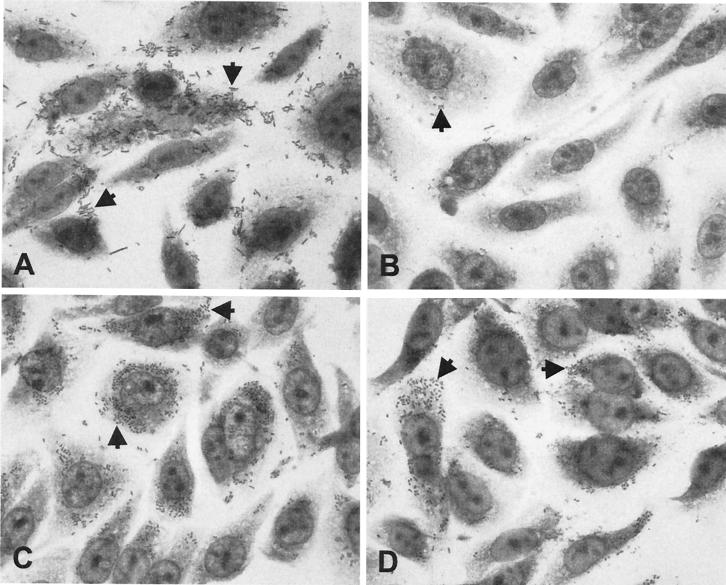
To examine the capacity of LPFO113 to mediate adherence to epithelial cells, the lpfO113 gene cluster was introduced into a nonpiliated fim deletion mutant of E. coli K-12, strain ORN103, on the low-copy-number vector pWSK29. The resulting strain, E. coli ORN103(pWSK:lpf), and a control strain, E. coli ORN103(pWSK29), were used to infect CHO-K1 cell monolayers for 4 h. In a qualitative assay designed to reveal patterns of bacterial adherence, E. coli ORN103(pWSK:lpf) adhered to CHO-K1 cells in clusters and the bacteria aggregated on the cell surface (Fig. 3A). Although the adherence pattern of E. coli ORN103(pWSK:lpf) was not uniform across the microscopic field, this type of adherence was not seen at all for E. coli ORN103(pWSK29) (Fig. 3B). These observations implied that the presence of LPFO113 could alter the surface properties of E. coli ORN103 and promote adherence to epithelial cells and interbacterial interactions. The suggested increase in adherence was confirmed in quantitative adherence assays, where E. coli ORN103(pWSK:lpf) was twice as adherent to CHO-K1 cells as E. coli ORN103(pWSK29) (Table 4).
FIG. 3.
Adherence of derivatives of E. coli ORN103 and EHEC O113:H21 (EH41) to Chinese hamster ovary cells (CHO-K1). (A) E. coli ORN103(pWSK:lpf); (B) E. coli ORN103(pWSK29); (C) parent strain EHEC O113:H21 (EH41); (D) EH41lpf−. Arrows indicate adherent bacteria.
TABLE 4.
Adherence of derivatives of E. coli strains EH41 (parent) and ORN103 (nonfimbriated) to CHO-K1 cells
| Strain | % Adherence to CHO-K1 cellsa |
|---|---|
| ORN103(pWSK29) | 1.1 ± 0.54 |
| ORN103(pWSK:lpf) | 2.2 ± 1.3b |
| EH41 (wild type) | 49.2 ± 34.2 |
| EH41lpfA− | 10.6 ± 6.7c |
| EH41lpfA−(pWSK:lpf) | 47.1 ± 22.6 |
Percentage of original inoculum; mean ± standard deviation of at least three experiments in duplicate wells.
Significantly more than ORN103(pWSK29) (P = 0.01).
Significantly less than EH41 (wild type) (P = 0.005).
To investigate the putative contribution of LPFO113 to the adherence of EHEC O113:H21 (EH41) to CHO-K1 cells, we introduced an lpfAO113 deletion using the recently described lambda red recombinase system. Successful replacement of lpfAO113 with a gene encoding kanamycin resistance was confirmed by PCR analysis (data not shown). We could find no clear difference in the adherence patterns of wild-type EHEC O113:H21 (EH41) and EH41lpfA− (Fig. 3C and D), although fewer bacteria per cell were observed for EH41lpfA. Quantitative adherence assays confirmed that EHEC O113:H21 (EH41) was significantly more adherent to CHO-K1 cells than the lpfA mutant (Table 4). trans complementation of EH41lpfA− with the lpfO113 gene cluster carried on pWSK:lpf restored adherence of the mutant to CHO-K1 cells to a level comparable to that of wild-type EH41 (Table 4).
Analysis of the genome insertion site of lpfO113 and the presence of O-island insertions near pstS and yhjW in other E. coli pathogens.
DNA sequence comparison between the chromosomal positions of the lpf loci from EHEC O113:H21 and EHEC O157:H7 revealed that the positions of lpfO113 and O island 154 in the E. coli backbone were virtually identical, with only a 2-bp difference at the pstS end of the O island (Fig. 4). In view of this coincidence, we looked for the presence of an O-island insertion in the chromosome of EHEC O113:H21 next to yhjW which corresponds to O-island position 141 in EHEC O157:H7. Primers were designed based on flanking regions of the E. coli chromosome in the genes yhjW and yhjX. A 200-bp product corresponding to the E. coli K-12 sequence was amplified, indicating that this site in EHEC O113:H21 does not contain an O island. We then tested a number of other serotypes of LEE-negative and LEE-positive EHEC strains and the related pathogens EPEC and REPEC for O-island insertions next to pstS and yhjW and were able to amplify O islands from one or both of these regions in 10 of 11 strains (Table 5). Cloning and sequencing of the O island next to pstS from REPEC 83/39 revealed that this region in the chromosome also contained an lpf gene cluster identical to lpfO113, which we have designated lpfR154. Nucleotide sequencing of the remaining O islands is currently in progress.
FIG. 4.
Nucleotide sequence of the regions flanking different lpf sites adjacent to pstS. Conserved nucleotides are shaded in black, and two nucleotides unique to EHEC O157: H7 (EDL933) are shaded in grey.
TABLE 5.
Presence of insertions in the chromosome of different strains of E. coli
| Strain | Presence of eae | Product size (kb)a
|
|
|---|---|---|---|
| O island 141 position (yhjW) | O island 154 position (pstS) | ||
| E. coli K-12 (HB101) | − | —b | — |
| EHEC O113:H21 (EH41) | − | — | 5.5 |
| EHEC O113:H21 (EH53) | − | — | 5.5 |
| EHEC O113:H21 (EH71) | − | — | 5.5 |
| EHEC O116:H21 (EH42) | − | — | 5.5 |
| EHEC O130:H11 (EH43) | − | 6.0 | 5.5 |
| EHEC O157:H7 (EDL933) | + | 6.0 | 6.9 |
| EHEC O111:H− (EH39) | + | 6.0 | 5.5 |
| EPEC O55:H7 | + | 6.0 | 6.9 |
| EPEC O127:H6 (E2348/69) | + | — | — |
| REPEC O103:H2 (84/110) | + | 6.0 | — |
| REPEC O15:H− (83/39) | + | 6.0 | 5.5 |
Approximate sizes from amplification of the regions corresponding to O islands 141 and 154 in the genome of EHEC O157:H7. In both PCRs, a 200-bp product is generated from E. coli K-12.
—, no O island was detected.
DISCUSSION
Although a wide variety of Shiga toxin-producing strains of E. coli are associated with disease in humans, the predominant causative agent of hemorrhagic colitis and HUS worldwide is EHEC O157:H7 (14, 23). As a result, much of the research effort into EHEC has focused on the characterization of virulence determinants in this pathogen, in particular the role of the O157:H7 LEE pathogenicity island in colonization and disease. Nevertheless, a significant proportion of EHEC strains isolated from patients with hemorrhagic colitis and HUS do not carry the LEE pathogenicity island yet are capable of causing severe, even life-threatening disease (9, 10, 22). Aside from the production of EHEC hemolysin and Stx-2, little is known about the virulence determinants of these pathogens. In particular, in the absence of LEE, the mechanisms by which these strains adhere to the host mucosal epithelium are unknown. Recently, Srimanote et al. described a pO113-encoded type IV pilus that promotes plasmid conjugation but does not appear to contribute to bacterial adherence (30). In contrast, mutation of the saa gene encoded by pO113 or curing of the large pO113 plasmid from wild-type EHEC O113:H21 resulted in a reduction in bacterial adherence to HEp-2 cells. Adherence was not completely abolished, however, indicating that pO113-independent factors also contribute to the adherence phenotype in vitro (21).
In another recent study of LEE-negative Shiga toxin-producing E. coli (STEC), Schmidt et al. uncovered a novel genomic island in STEC O91:H− (29). This region is located next to the selC locus and encodes a serine protease autotransporter of Enterobacteriaceae protein EspI, and a homologue of the putative adhesin, Iha. However, the contribution of these genomic factors to bacterial adherence was not examined, and despite having tested a large number of LEE-negative STEC strains, the authors did not report the presence of this putative pathogenicity island in EHEC O113:H21 (29).
In this study, we describe the identification of a novel chromosomal fimbrial gene cluster in LEE-negative EHEC O113:H21 that is related to LPF. Nucleotide sequencing of this novel region, lpfO113, revealed four putative ORFs predicted to encode homologues of long polar fimbrial proteins LpfA, LpfB, LpfC, and LpfD. In S. enterica serovar Typhimurium, LPF evidently contribute to the tropism of the bacteria for Peyer's patches of mice (4). The contribution of the homologues of these genes to the virulence of EHEC O157:H7 and O113:H21, however, is not known. Like other lpf gene clusters, lpfO113 exhibited similarity to the type I chaperone-usher family of fimbriae but lacked genes encoding regulatory elements (3). A comparison of the genetic organization among different lpf gene clusters showed that those located next to yhjW carry an additional gene, lpfE, encoding a putative fimbrial adhesin, which is not present in the fimbrial gene clusters adjacent to pstS (24).
Evidence that the lpfO113 genes are functional was obtained by performing site-specific deletion of the putative major fimbrial subunit gene, lpfAO113, which resulted in significantly reduced adherence of EHEC O113:H21 (EH41) to CHO-K1 cells. Complementation of the EH41 lpfA deletion mutant with lpfO113 in trans restored adherence to wild-type levels. We attempted to visualize LPFO113 on the surface of E. coli ORN103(pWSK:lpf) by using negative staining and electron microscopy, but without success. This may be explained by poor expression of LPFO113 by ORN103, as evidenced by the relatively weak adherence of E. coli ORN103(pWSK:lpf) to CHO-K1 cells, despite being significantly more adherent than E. coli ORN103(pWSK29). In addition, E. coli ORN103 may not be capable of supporting the correct assembly of LPFO113 on its surface, because genes other than the four ORFs described here are required for the full expression and normal assembly of the fimbriae. This is in contrast to the lpf operon from S. enterica serovar Typhimurium, which, when introduced into a nonfimbriated strain of E. coli, ORN172, encoded structures 7 to 8 nm in width and 2 to 10 μm in length in approximately one-third of the bacterial population (3). However, these structures were not confirmed as LPF by immunogold electron microscopy and may represent the activation of a cryptic fimbrial pathway in E. coli ORN172. The production of specific antibodies recognizing native LpfAO113 may aid the visualization of LPFO113 in EHEC O113:H21 as well as in E. coli ORN103(pWSK:lpf).
The lpf gene clusters from EHEC, REPEC, and S. enterica serovar Typhimurium are present in one or two locations in the E. coli chromosome, namely, next to yhjW (S. enterica serovar Typhimurium and EHEC O157:H7 O island 141) or abutting pstS (EHEC O113:H21, REPEC O15:H−, EHEC O157:H7 O island 154, and S. enterica serovar Typhi). The presence in several pathogens of additional DNA at these two positions suggested that they might be common sites for chromosomal insertions. Indeed, PCR amplification of these regions in several EHEC, EPEC, and REPEC isolates revealed insertions in one or other of these positions in 10 of 11 strains tested. While several of the insertions next to pstS are likely to be identical to lpfO113, insertions adjacent to yhjW were also present in several EHEC and REPEC strains, but the DNA sequence of these regions has yet to be determined. Sequence analysis of the lpf operons positioned next to pstS in three different pathogens revealed that the sites of insertion in the E. coli chromosome were identical. This suggests that the putative lateral acquisition of lpf in different bacterial chromosomes was precise and specific for this site, or that lpf was an integral part of ancestral E. coli strains but was subsequently lost from some of its progeny.
In phylogenetic analyses, the predicted LpfAO113 subunit clusters with a putative fimbrial subunit from S. enterica serovar Typhi, StgA, and the putative major fimbrial subunit from EHEC O157:H7 O island 154 (data not shown) (20, 24). The three fimbrial gene clusters are all located between pstS and glmS in the chromosomal backbone and thus are related in amino acid sequence and genome position. However, the putative usher gene, stgC, is a pseudogene, suggesting that this fimbrial operon is not functional in S. enterica serovar Typhi (20). Likewise, LpfA from S. enterica serovar Typhimurium is less related to LpfAO113 than it is to the putative major fimbrial subunit from EHEC O157:H7 O island 141, where these two gene clusters have the same position in the chromosome.
An examination of the distribution of lpfO113 among other pathogenic E. coli strains and enteric pathogens revealed that lpfO113 is closely associated with EHEC strains and in particular with different serogroups of LEE-negative EHEC. This is in contrast to saa, which was associated only with LEE-negative strains of EHEC (21). lpfO113 was also found in four strains of REPEC. The presence of a lpf gene cluster identical to lpfO113 in REPEC O15:H− (83/39) provides us with an opportunity to test the contribution of these fimbriae to virulence. The construction of a REPEC (83/39) lpfA mutant is currently in progress for testing in a rabbit model of infection (2). We propose that the putative LPF of EHEC may facilitate colonization of humans and/or the persistence of E. coli pathogens in animal reservoirs of infection. Further studies are required to elucidate fully the role of lpf gene clusters in the pathogenesis of E. coli-mediated diarrheal disease.
Acknowledgments
We are grateful to the Australian Paediatric Surveillance Unit for the provision of EHEC strains.
This study was supported in part by the Australian National Health and Medical Research Council, ANZ Charitable Trusts, Ramaciotti Foundations for Biomedical Research, Monash University, and the Murdoch Children's Research Institute.
Editor: B. B. Finlay
REFERENCES
- 1.Abe, A., U. Heczko, R. G. Hegele, and B. B. Finlay. 1998. Two enteropathogenic Escherichia coli type III secreted proteins, EspA and EspB, are virulence factors. J. Exp Med. 188:1907-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, L. M., C. P. Simmons, L. Rezmann, R. A. Strugnell, and R. M. Robins-Browne. 1997. Identification and characterization of a K88- and CS31A-like operon of a rabbit enteropathogenic Escherichia coli strain which encodes fimbriae involved in the colonization of rabbit intestine. Infect. Immun. 65:5222-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumler, A. J., and F. Heffron. 1995. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J. Bacteriol. 177:2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumler, A. J., R. M. Tsolis, and F. Heffron. 1996. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer's patches. Proc. Natl. Acad. Sci. USA 93:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boerlin, P., S. Chen, J. K. Colbourne, R. Johnson, S. De Grandis, and C. Gyles. 1998. Evolution of enterohemorrhagic Escherichia coli hemolysin plasmids and the locus for enterocyte effacement in Shiga toxin-producing E. coli. Infect. Immun. 66:2553-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnenberg, M. S., C. O. Tacket, S. P. James, G. Losonsky, J. P. Nataro, S. S. Wasserman, J. B. Kaper, and M. M. Levine. 1993. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J. Clin. Investig. 92:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dytoc, M. T., A. Ismaili, D. J. Philpott, R. Soni, J. L. Brunton, and P. M. Sherman. 1994. Distinct binding properties of eaeA-negative verocytotoxin-producing Escherichia coli of serotype O113:H21. Infect. Immun. 62:3494-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott, E. J., R. M. Robins-Browne, E. V. O'Loughlin, P. Henning, G. G. Hogg, J. Knight, H. Powell, D. Redmond, and V. Bennett-Wood. 2001. Nationwide surveillance of haemolytic uraemic syndrome in Australia: a comparison of endemic cases with those in a single epidemic. Arch. Dis. Child. 85:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:47-52. [DOI] [PubMed] [Google Scholar]
- 13.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 14.Kaper, J. B. 1998. Enterohemorrhagic Escherichia coli. Curr. Opin. Microbiol. 1:103-108. [DOI] [PubMed] [Google Scholar]
- 15.Krejany, E. O., T. H. Grant, V. Bennett-Wood, L. M. Adams, and R. M. Robins-Browne. 2000. Contribution of plasmid-encoded fimbriae and intimin to capacity of rabbit-specific enteropathogenic Escherichia coli to attach to and colonize rabbit intestine. Infect. Immun. 68:6472-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marches, O., J. P. Nougayrede, S. Boullier, J. Mainil, G. Charlier, I. Raymond, P. Pohl, M. Boury, J. De Rycke, A. Milon, and E. Oswald. 2000. Role of tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect. Immun. 68:2171-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 19.Norris, T. L., and A. J. Baumler. 1999. Phase variation of the lpf operon is a mechanism to evade cross-immunity between Salmonella serotypes. Proc. Natl. Acad. Sci. USA 96:13393-13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 21.Paton, A. W., P. Srimanote, M. C. Woodrow, and J. C. Paton. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paton, A. W., M. C. Woodrow, R. M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3357-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 25.Reese, M. G., N. L. Harris, and F. H. Eeckman. 1996. Large scale sequencing specific neural networks for promoter and splice recognition, p. 737-738. In L. Hunter and T. Klein (ed.), Biocomputing: proceedings of the 1996 Pacific Symposium. World Scientific Publishing Company, Singapore.
- 26.Robins-Browne, R. M., and V. Bennett-Wood. 1992. Quantitative assessment of the ability of Escherichia coli to invade cultured animal cells. Microb. Pathog. 12:159-164. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 28.Schauer, D. B., and S. Falkow. 1993. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect. Immun. 61:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt, H., W. L. Zhang, U. Hemmrich, S. Jelacic, W. Brunder, P. I. Tarr, U. Dobrindt, J. Hacker, and H. Karch. 2001. Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect. Immun. 69:6863-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srimanote, P., A. W. Paton, and J. C. Paton. 2002. Characterization of a novel type IV pilus locus encoded on the large plasmid of locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 70:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tacket, C. O., M. B. Sztein, G. Losonsky, A. Abe, B. B. Finlay, B. P. McNamara, G. T. Fantry, S. P. James, J. P. Nataro, M. M. Levine, and M. S. Donnenberg. 2000. Role of EspB in experimental human enteropathogenic Escherichia coli infection. Infect. Immun. 68:3689-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor, R. K., C. Manoil, and J. J. Mekalanos. 1989. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J. Bacteriol. 171:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tewari, R., T. Ikeda, R. Malaviya, J. I. MacGregor, J. R. Little, S. J. Hultgren, and S. N. Abraham. 1994. The PapG tip adhesin of P fimbriae protects Escherichia coli from neutrophil bactericidal activity. Infect. Immun. 62:5296-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vial, P. A., J. J. Mathewson, H. L. DuPont, L. Guers, and M. M. Levine. 1990. Comparison of two assay methods for patterns of adherence to HEp-2 cells of Escherichia coli from patients with diarrhea. J. Clin. Microbiol. 28:882-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]