Abstract
Salmonella enterica serovar Typhimurium encounters antimicrobial peptides (AP) within the phagosomes of professional phagocytes and at intestinal mucosal surfaces. Salmonella serovar Typhimurium utilizes the two-component regulatory system PmrA-PmrB, which is activated in response to the environmental conditions encountered in vivo, to regulate resistance to several AP, including polymyxin B (PM). Random MudJ transposon mutagenesis was used to identify PmrA-PmrB-regulated genes, as well as genetic loci necessary for PM resistance. Three different phenotypic classes of genes were identified: those necessary for PM resistance and regulated by PmrA, those necessary for PM resistance and not regulated by PmrA, and PmrA-regulated genes not required for PM resistance. Loci identified as necessary for PM resistance showed between 6- and 192-fold increased sensitivities to PM, and transposon insertion sites include surA, tolB, and gnd. PmrA-regulated loci identified included dgoA and yibD and demonstrated 500- and 2,500-fold activation by PmrA, respectively. The role of the identified loci in aminoarabinose modification of lipid A was determined by paper chromatography. The gnd mutant demonstrated a loss of aminoarabinose from lipid A, which was suggested to be due to a polar effect on the downstream gene pmrE. The remaining PMs mutants (surA and tolB), as well as the two PmrA-regulated gene (yibD and dgoA) mutants, retained aminoarabinose on lipid A. yibD, dgoA, and gnd (likely affecting pmrE) played no role in PmrA-regulated resistance to high iron concentrations, while surA and tolB mutations grew poorly on high iron media. All PMs mutants identified in this study demonstrated a defect in virulence compared to wild-type Salmonella serovar Typhimurium when administered orally to mice, while the PmrA-regulated gene (yibD and dgoA) mutants showed normal virulence in mice. These data broaden our understanding of in vivo gene regulation, lipopolysaccharide modification, and mechanisms of resistance to AP in enteric bacteria.
Amphipathic, cationic antimicrobial peptides (AP) are an important component of innate immunity (3, 60) and are present in macrophage phagosomes, granules of neutrophils, and secretions of mucosal epithelia (14, 29, 40). Functionally, AP have antiviral, antifungal, and antibacterial properties. In gram-negative bacteria, many cationic AP associate electrostatically with the bacterial outer membrane, namely the lipopolysaccharide (LPS), via ionic bonds with unsubstituted, anionic phosphate residues present on LPS lipid A and core oligosaccharide (50). Subsequent to binding, the AP permeabilize the outer membrane, translocate to the cytoplasmic membrane, which is the main cytotoxic target of most AP, and then form pores, resulting in leakage of cell constituents and cell death.
Salmonellae cause a diverse array of diseases in humans and animals and are capable of circumventing killing by the host immune system. In Salmonella enterica serovar Typhimurium, evasion of AP killing is directed in part by the PmrA-PmrB two-component regulatory system (17, 19, 40, 44, 47, 51, 52). Activation of PmrA-PmrB confers resistance to polymyxin B (PM), as well as the neutrophil cationic AP CAP-37 (azurocidin) and CAP-57 (bactericidal permeability-increasing protein). PmrA-PmrB accomplishes resistance to these AP by up-regulating genes involved in covalent modifications of the LPS (17, 19). The LPS modifications reduce the negative charge of LPS and consequently decrease attraction and binding of AP to the outer membrane. PmrA-induced modifications to LPS include the addition of 4-amino-4-deoxy-l-arabinose (Ara4N) to the 4′-phosphate of lipid A (and sometimes the 1-phosphate) and addition of phosphoethanolamine (pEtN) to the 1-phosphate of lipid A (and sometimes the 4′-phosphate), to 2-keto-3-deoxyoctulosonic acid, and to the first heptose residue of the core (22, 25, 62). A mutant of Salmonella serovar Typhimurium containing a point mutation in the pmrA gene (pmrA505, PmrAc) that results in a constitutively active protein product expresses an LPS highly modified with Ara4N on lipid A and pEtN on lipid A and core (17, 22, 62). The PmrAc mutant consequently has an increased MIC for PM which is some 80-fold higher than that of the wild type and a PmrA-null mutant.
PmrA-PmrB-regulated genes have been shown to be induced during infection of the vertebrate host (21). The PmrA-PmrB regulon can be activated by PhoP-PhoQ two-component system via the PmrD protein in response to low magnesium concentrations (15, 26). PmrD is thought to act upon the PmrA-PmrB system posttranscriptionally, possibly affecting the phosphorylation state of PmrA. PmrA-PmrB can also be activated independently of PhoP-PhoQ by mild acidic conditions or high iron concentrations, which are thought to be directly sensed by PmrB (42, 43, 57). Moreover, PmrA-PmrB has been shown to be necessary for resistance to high levels of iron (57). In this manner, salmonellae can react to in vivo conditions of low magnesium and mild acid pH, likely encountered within eukaryotic cell vacuoles or phagosomes, by up-regulating LPS modifications in preparation for encountering AP. While high iron environments are uncommon within vertebrate hosts, PmrA-PmrB may play a role in resistance to high iron levels in the environment (57).
Several PmrA-regulated loci involved in modification of LPS have been identified, including pmrE (pagA, ugd), which encodes a putative UDP-glucose dehydrogenase, and the pmrHFIJKLM operon (17). Products of these loci are involved in biosynthesis of the Ara4N modification to lipid A. In the putative pathway for Ara4N biosynthesis, supported by findings for the homologous gene products in Escherichia coli, PmrE converts UDP-glucose to UDP-glucuronate, and products of pmrHFIJKLM participate in the conversion of UDP-glucuronate to UDP-Ara4N and transfer of Ara4N to lipid A (7, 61). Trent et al. have recently identified the pmrK (designated arnT by the authors) gene product as an enzyme that transfers one or two Ara4N moieties from an undecaprenyl phosphate-α-l-Ara4N donor substrate to lipid A and certain lipid A precursors in Salmonella serovar Typhimurium and E. coli (48, 49). Like a pmrA mutant, mutants in pmrE or pmrHFIJKLM exhibit a virulence defect when administered orally to mice, presumably as a result of a lack of Ara4N modification to lipid A and loss of resistance to host AP (19).
To date, all known PmrA-regulated genes that are involved in modifying LPS affect Ara4N addition to lipid A, leaving a subset of PmrA-regulated genes involved in pEtN addition to the LPS core to be identified. Additionally, PmrA may affect loci with functions altogether distinct from those involved in resistance to AP, such as resistance to high levels of iron. We have undertaken a mutagenesis experiment to identify genes that are regulated by PmrA and/or are necessary for PM resistance. We report in this paper the identification and preliminary characterization of three non-PmrA-regulated loci that are necessary for PM resistance and two PmrA-regulated genes not required for resistance to PM.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are described in Table 1. Salmonellae and E. coli cultures were grown in Luria-Bertani (LB) broth at 37°C with aeration. Antibiotics were used, when appropriate, at the following concentrations: PM, 8 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 45 μg/ml; tetracycline, 25 μg/ml; and ampicillin, 50 μg/ml. The chromogenic substrate for β-galactosidase, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranosidase (X-Gal), was used in LB agar plates at a concentration of 40 μg/ml when needed. For high-iron growth experiments, strains were grown on solid N-minimal medium, pH 5.8, containing 10% Casamino Acids, 38 mM glycerol, 10 μM MgCl2, and 0.1 mM FeSO4 (57).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or relevant phenotype | Source |
|---|---|---|
| Salmonellae | ||
| JSG210 | ATCC 14028s; wild type | ATCC |
| JSG224 | phoN2 zxx::6251 Tn10d-cam (CS019) | 32 |
| TT10288 | MudJ donor strain | 23 |
| JSG425 | Salmonella λ Pir phoP::Tn10d-TET | |
| JSG844 | pmrA505 with streptomycin resistance allele | 19 |
| JSG435 | ATCC 14028s pmrA505 zjd::Tn10d-cam | 18 |
| JSG421 | pmrA::Tn10d | 18 |
| JSG851 | JSG435 with MudJ insertion in imp (affect on surA); PMs | This work |
| JSG895 | JSG435 with MudJ insertion in tolB; PMs | This work |
| JSG1272 | JSG435 with MudJ insertion in gnd; PMs | This work |
| JSG1290 | JSG435 with MudJ insertion in gnd; PMs | This work |
| JSG897 | JSG435 with MudJ insertion in yibD; PmrA regulated | This work |
| JSG898 | JSG421 with MudJ insertion in yibD; PmrA regulated | This work |
| JSG899 | JSG435 with MudJ insertion in dgoA; PmrA regulated | This work |
| JSG900 | JSG421 with MudJ insertion in dgoA; PmrA regulated | This work |
| JSG901 | JSG435 with MudJ insertion in pmrC; PmrA regulated, PMs | This work |
| JSG918 | JSG435 with MudJ insertion in pmrC; PmrA regulated, PMs | This work |
| JSG973 | JSG435 with MudJ insertion in pmrC; PmrA regulated, PMs | This work |
| JSG1036 | JSG435 with MudJ insertion in pmrC; PmrA regulated, PMs | This work |
| JSG923 | JSG435 with MudJ insertion in pmrI; PmrA regulated, PMs | This work |
| JSG1036 | JSG435 with MudJ insertion in pmrI; PmrA regulated, PMs | This work |
| JSG1521 | ATCC 14028s with MudJ insertion in imp (affect on surA) | This work |
| JSG1522 | ATCC 14028s with MudJ insertion in tolB | This work |
| JSG1523 | ATCC 14028s with MudJ insertion in gnd | This work |
| JSG1524 | ATCC 14028s with MudJ insertion in gnd | This work |
| JSG1525 | ATCC 14028s with MudJ insertion in yibD | This work |
| JSG1526 | ATCC 14028s with MudJ insertion in dgoA | This work |
| JSG1325 | ATCC 14028s pmrA505 zjd::Tn10d-cam surA::luc | This work |
| JSG1329 | ATCC 14028s pmrA505 zjd::Tn10d-cam surA::luc imp::MudJ | This work |
| JSG1337 | JSG851 complemented with surA carried on pWSK350 | This work |
| JSG547 | ATCC 14028s pmrA505 pmrE::MudJ | This work |
| JSG1319 | LT2 gnd::Tn10d (Tc) | Gift of D. M. Downs; 11 |
| JSG1330 | pmrE::MudJ gnd::Tn10d (Tc) | This work |
| JSG1686 | JSG844 with pKAS397, single recombinant | This work |
| JSG1724 | JSG844 with in-frame deletion of gnd | This work |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF) U169endA 1 recA 1 hsdR17deoR thi-1 supE441−gyrA96relA1 | BRL |
| SM10λPir | thi-1 thr-1 leuB6 supE44 tonA21lacY1 recA::RP4-2-Tc::Mu | |
| JSG1320 | SM10λPir containing pGPL350 | This work |
| JSG1336 | DH5α containing pWKS350 | This work |
| JSG1543 | DH5α containing pUC370 | This work |
| JSG1548 | DH5α containing pUC372 | This work |
| JSG1567 | SM10λPir containing pCVD370 | This work |
| Plasmids | ||
| pWSK29 | Low-copy expression vector, Ampr | 55 |
| pGPL01 | Firefly luciferase transcriptional fusion suicide vector, Ampr | 18 |
| pUC19 | High-copy cloning vector | 58 |
| pKAS32 | rpsL suicide vector | 41 |
| pWSK350 | pWSK29 containing ∼1.3-kb fragment corresponding to surA gene | This work |
| pGPL350 | pGPL01 containing ∼1.3-kb fragment corresponding to surA gene | This work |
| pUC370 | pUC19 containing ∼2.9-kb fragment consisting of gnd, 800 bp upstream and 80-bp downstream | This work |
| PKAS397 | pKAS32 containing 1.6-kb insert consisting of 800 bp 5′ and 3′ of gnd | This work |
MudJ mutagenesis.
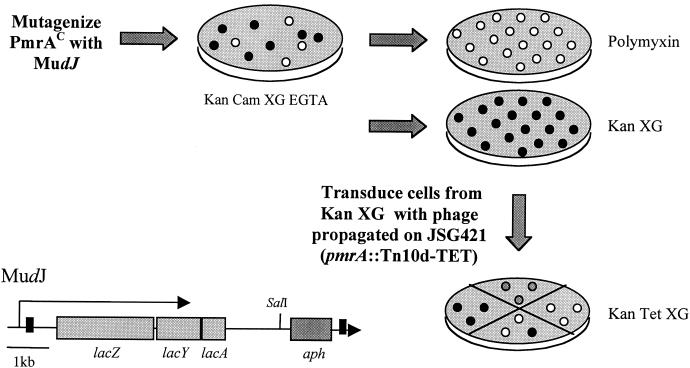
S. enterica serovar Typhimurium strain JSG435 (pmrA505, PmrAc PMr) was mutagenized with the MudJ transposon by infection with P22 bacteriophage that had been propagated with strain TT10288 (Fig. 1). Some MudJ transposon insertions generated active transcriptional fusions to the β-galactosidase-encoding gene contained within the transposon. After several independent transductions, ∼10,000 chloramphenicol- and kanamycin-resistant mutants that showed utilization of the β-galactosidase substrate (resulting in blue colonies) were picked and replica plated on LB agar containing PM or kanamycin-X-Gal (LB-Kan-X-Gal). Colonies that grew on LB-Kan-X-Gal but not on LB-PM were retained as PMs mutants, and the MIC of PM was ascertained. Each colony grown on LB-Kan-X-Gal was transduced with P22 phage propagated with strain JSG421 (pmrA::Tn10d) and selected on LB-Kan-Tet-X-Gal agar, in effect switching the regulatory background from PmrAc to PmrA-null. Colonies that appeared blue in the PmrAc background, but white or light blue in the PmrA-null background, were collected as mutants that contained a MudJ insertion in a PmrA-regulated locus. All mutants were transduced into a fresh PmrAc background to ensure that the phenotype was linked to the MudJ insertion. In addition, mutants were examined by Southern blot analysis to ensure the uniqueness of the MudJ insertion and to eliminate strains containing more than one insertion.
FIG. 1.
Schematic of the strategy used to mutagenize JSG435 (PmrAc) with MudJ in order to obtain mutants in PmrA-regulated genes and/or genes involved in PM resistance. The MudJ transposon encodes genes involved in β-galactosidase production, as well as a kanamycin resistance cassette. The SalI site used in cloning the MudJ-chromosomal DNA junction for sequencing is shown. Black circles represent blue colonies resulting from active fusion of lacZ in the MudJ transposon to an ORF. White circles symbolize Kanr colonies that contain inactive fusions. Light grey colonies represent a light blue phenotype corresponding to low-level transcription of lacZ. Abbreviations: Kan, kanamycin; Tet, tetracycline; Cam, chloramphenicol; XG, X-Gal.
DNA cloning procedures and generation of mutants.
In order to clone the genomic DNA adjacent to the MudJ insertion site for each mutant, a SalI library of the mutant was constructed in plasmid pWSK29. A SalI restriction site is present in MudJ, which allowed the kanamycin resistance marker to be used in selection of constructs containing a MudJ-chromosomal DNA junction (Fig. 1). The libraries were transformed into DH5α, and kanamycin- and ampicillin-resistant clones were sequenced using primer JG49 specific to MudJ DNA corresponding to the sequence at the 3′ end of the transposon (5′ CTA ATC CCA TCA GAT CCC G 3′). The sequence of the insertion site of one PMs mutant, JSG1290, was obtained using direct genomic sequencing with primer JG49. Routine methods of determining the site of transposon insertion failed for identification of the MudJ insertion site in JSG1272. As an alternative, a “vectorette” method of obtaining the sequence for the MudJ-chromosomal DNA junction of JSG1272 was utilized (38). Chromosomal DNA was obtained using (i) the genomic DNA isolation protocol previously described, with proteinase K used in lieu of pronase (18); or (ii) the UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Inc.). Restriction enzymes were purchased from New England Biolabs (Beverly, Mass.)
For generation of an independent fusion to surA, the surA gene was amplified from wild-type Salmonella serovar Typhimurium (JSG210) chromosomal DNA using primers JG350 (5′ GGAATTCGAACACCGAAGCGAAG 3′) and JG352 (5′ GAGGGTACCCCTTTATGCAGCTTCG 3′), which introduced EcoRI and KpnI restriction sites (underlined), respectively. The PCR product was cloned into pGPL01 (18) to give plasmid pGPL350. pGPL350 was propagated in SM10λPir during cloning (JSG1320) and then introduced into wild-type Salmonella serovar Typhimurium JSG224 by mating, creating a luciferase fusion to the genomic copy of surA (JSG1321). The surA::luc fusion was then moved into PmrAc JSG435 by P22 phage-mediated transduction to yield JSG1325. For complementation studies, the surA gene was amplified from wild-type genomic DNA with primers JG355 (5′ GGAATTCCAACGTAATCCGCATTGCG 3′) and JG356 (5′ GGGGTACCGTTGCGCACTGCTCATTAG 3′), which introduced EcoRI and KpnI restriction sites, respectively, for cloning into the expression vector pWKS29. The resulting construct was transformed into JSG851 by electroporation, creating JSG1327(pWKS355).
For generation of an in-frame deletion in gnd, the open reading frame (ORF) of gnd, plus 800 bp of DNA flanking both the 5′ and 3′ ends of the ORF, were amplified from wild-type (JSG210) chromosomal DNA by PCR using JG370 (5′ GTCGAGCTCCGTTCTGTTACTGTAGTCCCCTCG 3′) and JG371 (5′ ACATGCATGCCGACTCGCATAGCGAGATAAGTATTGG 3′). The primers introduced SacI and SphI restriction sites, respectively, which allowed for cloning of the gnd-containing PCR fragment into pUC19 to yield pUC370 in JSG1543. Primers JG372 (5′ GGACTAGTCGTACTGATAAAGAAGGC 3′) and JG373 (5′ GGACTAGTCATCACTGCCATACCGACGAC 3′) were used for PCR amplification with pUC370 as a template, resulting in a product 4.3 kb in size consisting of all pUC370 DNA except the majority of gnd. Primers JG372 and JG373 each introduced SpeI sites, permitting circularization of the amplified product (pUC372, JSG1548). The gnd deletion fragment was then subcloned into rpsL suicide vector pKAS32 (41), creating pKAS397. pKAS397 was propagated in SM10λPir (JSG1607) and moved into a streptomycin-resistant, PmrAc Salmonella serovar Typhimurium background (JSG844) by conjugation to generate JSG1686. Colonies obtained from the transduction were screened for double homologous recombination on LB agar containing 1 mg of streptomycin/ml. Strains incorporating the deletion were identified by PCR (JSG1724).
For experiments concerning cotranscription of gnd and pmrE, a gnd::Tn10d mutation was moved via P22 phage-mediated transduction into JSG547, which displays the PmrAc background and contains a pmrE::MudJ fusion.
MIC and transcriptional assays.
The MIC of PM for each mutant was obtained as previously described (46), testing serial dilutions of PM (U.S. Biochemicals; 8,040 U/mg) in 0.2% bovine serum albumin-0.01% acetic acid. β-Galactosidase assays were performed as described previously (18). Firefly luciferase assays were performed using 100 μl of cells grown to mid-logarithmic phase (optical density at 600 nm, ∼0.6). Cells were suspended in luciferase buffer (25 mM Tris phosphate, pH 7.8; 2 mM dithiothreitol; 2 mM EDTA, pH 8.0; 10% glycerol; 1% Triton X-100) and then sonicated briefly. Cell debris was removed by centrifugation. Assays were performed using 10 μl of lysate, using an EG&G Berthold Lumat LB 9507 luminometer and luciferase assay reagents from Promega (Luciferase Assay System). All MIC and transcriptional assays were performed in triplicate.
Virulence assays.
The MudJ fusions identified in the mutagenesis experiment were moved into a wild-type Salmonella serovar Typhimurium background via P22 phage-mediated transduction (JSG1521 to -1526). Survival assays were performed as described previously (19). Briefly, each mutant, as well as a PmrA-null control, was grown to stationary phase (16 h) at 37°C. Approximately 5 × 106 stationary-phase bacteria (1 log unit above the 50% lethal dose) were washed and resuspended in 20 μl of phosphate-buffered saline, pH 7.4. Female BALB/c mice (weighing 16 to 18 g) were inoculated orally using the swallowing reflex of the mouse. Dilutions of the stationary-phase cultures were plated to determine the number of bacteria present in the inocula. Infected mice were observed for 21 days postinoculation.
LPS and lipid A isolation and paper chromatography.
LPS isolation was accomplished by a hot phenol-water method as previously described (2). Lipid A was obtained from whole LPS by hydrolysis in 1% sodium dodecyl sulfate-10 mM sodium acetate, pH 4.5, for 18 h at 100°C (8). One-milligram samples were loaded onto Whatmann paper and allowed to run in descending fashion overnight in a solvent consisting of isopropanol-ethyl acetate-water (7:1:2). Dry chromatograms were treated with ninhydrin spray reagent (Sigma) to detect aminoarabinose as yellow spots.
RESULTS
MudJ mutagenesis.
Salmonella serovar Typhimurium strain JSG435 was used for mutagenesis because it expresses a constitutively active PmrA, rendering the use of inducing conditions for growth unnecessary. The PmrAc strain also expresses LPS modified with Ara4N and pEtN, which results in resistance to PM (17, 22, 62). PM-sensitive mutants and mutants in PmrA-regulated genes were obtained using MudJ mutagenesis of JSG435, as detailed in Materials and Methods. The strategy for identification of mutants is depicted in Fig. 1. Kanamycin-resistant colonies containing an active transcriptional fusion to lacZYA, i.e., blue colonies, were chosen for further analysis as described in Materials and Methods. A total of ∼10,000 β-galactosidase-positive mutants were screened for both PM sensitivity and regulation by PmrA. Mutants were divided into three categories: those with an insertion in a PmrA-regulated gene not affecting PM resistance, those with an insertion in a gene regulated by PmrA but not involved in PM resistance, and those with a mutation in a PmrA-regulated gene necessary for PM resistance (see Table 2 for a summary of the results).
TABLE 2.
Mutants identified in the MudJ transposon mutagenesis screen
| Description | Strain | Tn insertion site |
|---|---|---|
| PM sensitive | JSG851 | imp/surA |
| JSG895 | tolB | |
| JSG1272 | gnd | |
| JSG1290 | gnd | |
| PmrA regulated | JSG897 | yibD |
| JSG899 | dgoA | |
| PmrA regulated, PM sensitive | JSG901 | pmrC |
| JSG918 | pmrC | |
| JSG973 | pmrC | |
| JSG1036 | pmrC | |
| JSG923 | pmrI | |
| JSG925 | pmrI |
Two PmrA-regulated gene mutants that retained PM resistance were identified: JSG897 and JSG899. In the former, MudJ was inserted in yibD, which encodes a putative glycosyl transferase (31). In JSG899, the insertion occurred in dgoA, which is predicted to encode 2-oxo-3-deoxygalactonate-6-phosphate aldolase-galactonate dehydratase (31).
Four PM-sensitive mutants with insertions in non-PmrA-regulated genes were obtained. In JSG851, MudJ was inserted near the 3′ end of imp, an ortholog of an E. coli gene involved in resistance to organic solvents (31). JSG895 contained a MudJ insertion in tolB, which plays a role in tolerance to colicins and affects overall membrane stability in a variety of bacteria (5, 6, 9, 10, 24, 30, 36, 37, 39). The third PM-sensitive mutant obtained, JSG1272, contained an insertion in gnd, which encodes a 6-phosphogluconate dehydrogenase with a primary role in the pentose-phosphate pathway (45). The PM-sensitive mutant JSG1290 also contained a MudJ insertion within the gnd gene.
Mutations identified in loci that are both PmrA regulated and necessary for PM resistance were in previously identified genes, namely pmrC and pmrI (also called arnA and pbgP3) (17, 49, 57). Four independent mutations in pmrC, which is upstream of and cotranscribed with pmrAB, were identified: JSG901, JSG918, JSG973, and JSG1036. Two additional mutants, JSG923 and JSG925, contained insertions in pmrI, a gene included in the pathway for Ara4N biosynthesis and addition to lipid A.
Analysis of PmrA-regulated genes.
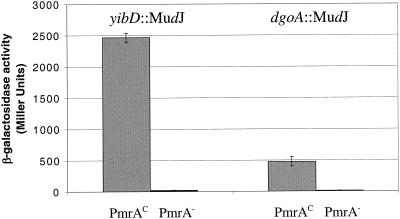
The regulatory effect of PmrA on newly identified PmrA-activated genes was examined. Transcription of the lacZ gene fusions created by insertion of the MudJ transposon was measured for yibD::MudJ and dgoA::MudJ in PmrAc and PmrA-null regulatory backgrounds. Figure 2 shows the results of these assays. Transcription of yibD was activated approximately 2,500-fold in the presence of PmrA. Similarly, PmrA activated transcription of the dgoA locus by nearly 500-fold. In PmrA-null backgrounds, transcription of these genes was negligible.
FIG. 2.
Regulation of yibD and dgoA by PmrA. Transcription of the loci was measured by the production of β-galactosidase from the lacZ gene in the MudJ transposon. Strains compared were yibD::MudJ in a PmrAc and a PmrA-null background and dgoA::MudJ in a PmrAc and a PmrA-null background. Loci were transcribed effectively in a PmrAc background (grey bars); transcription of the loci was essentially eliminated in a PmrA-null background (black bars, barely visible).
The PmrA-regulated gene mutants JSG897 (yibD) and JSG899 (dgoA) were also analyzed for sensitivity to PM by MIC assay. The MIC of PM for each mutant was 12 μg/ml (data not shown), equivalent to the MIC for the parental PmrAc Salmonella serovar Typhimurium strain, indicating that the PmrA-regulated genes identified in the screen may have functions other than those ascribed to genes regulated by PmrA, i.e., they may not be required for PM resistance.
Wosten et al. have demonstrated that PmrA is necessary for resistance of Salmonella serovar Typhimurium to high iron concentrations (57). To address the potential role of the PmrA-regulated genes identified in the screen in iron resistance, JSG897 (yibD) and JSG899 (dgoA) were grown on solid N-minimal medium supplemented with 100 μM FeSO4. While a PmrA-null mutant was sensitive to this level of iron, JSG897 and JSG899 were able to grow as well as a PmrAc strain (data not shown), demonstrating that neither of these PmrA-regulated genes is essential for resistance to the toxic effects of iron.
For further characterization of the PmrA-regulated gene mutants, the promoter regions of yibD and dgoA were analyzed for the presence of a putative PmrA-binding site, which was defined by the occurrence of the consensus binding sequence for PmrA, YTTAAK-N5-YTTAAK (1). For yibD, a strong match to the consensus binding sequence was identified 50 bp upstream of the translational start codon (Fig. 3). Moreover, the YTTAAK repeats described by Aguirre et al. (1) are conserved. No consensus PmrA-binding sequence was identified in this manner for dgoA, neither within the putative promoter region upstream of dgoA nor within the putative promoter upstream of the predicted dgoKAT operon. Lack of a consensus PmrA-binding site for JSG899 suggests that regulation of this gene by PmrA may be indirect.
FIG. 3.
Promoter analysis of PmrA-regulated loci identified in screen. The consensus PmrA-binding sites shown for pmrE (ugd) and pmrCAB were identified previously by Aguirre and colleagues (1). The YTTAAK direct repeats are marked in bold and underlined. The predicted −10 regions are italicized and marked in bold. Transcriptional start sites are labeled with arrows for pmrCAB and pmrE (56). For yibD, the putative PmrA consensus binding site begins 67 bp upstream of the translational start site. Ambiguous codes used are as follows: Y, C/T; K, G/T.
Analysis of PM-sensitive mutants.
The degree of PM sensitivity of PMs, non-PmrA-regulated gene mutants JSG851 (imp::MudJ), JSG895 (tolB::MudJ), JSG1272 (gnd::MudJ), and JSG1290 (gnd::MudJ) was obtained by MIC assay. Results are summarized in Table 3. The MIC of PM for these mutants ranged from 0.0625 to 2.0 μg/ml, which equates to a 6- to 192-fold increased sensitivity to PM when compared to the PmrAc parental strain.
TABLE 3.
MICs of PM for the PMs mutants
| Strain | Description | MIC (μg/ml) |
|---|---|---|
| JSG435 | PmrAc | 12 |
| JSG421 | PmrA-null | 0.0625 |
| JSG851 | imp::MudJ (surA) | 2.0 |
| JSG895 | tolB::MudJ | 0.0625 |
| JSG1272 | gnd::MudJ | 0.125 |
| JSG1290 | gnd::MudJ | 0.125 |
β-Galactosidase assays were used to determine if the PM-sensitive mutants contained insertions in non-PmrA-regulated genes, as suggested by the preliminary screen (Fig. 1). The β-galactosidase activity of imp::MudJ, tolB::MudJ, and gnd::MudJ fusions were measured in PmrAc and PmrA-null backgrounds. Each of these loci expressed essentially the same levels of β-galactosidase with or without a constitutively active PmrA, with less-than-twofold increases in expression in a PmrAc background (data not shown). These data confirmed that PmrA did not affect transcription of these loci.
The transposon insertion that occurred in JSG851 interrupted the imp gene 23 bp from the termination codon. Because imp is immediately followed by surA, with only a 43-bp intervening sequence, and surA has been shown to affect membrane integrity, the possibility was investigated that the PMs phenotype of the mutant resulting from the MudJ insertion in imp was a consequence of a polar effect on surA. An intact surA gene was reintroduced into the PMs mutant JSG851 on a low-copy expression vector to create JSG1337. Full complementation for PM resistance was obtained in JSG1337, which displayed a PM MIC of 12 μg/ml, which was equivalent to that displayed by the PmrAc strain. To further support that the insertion in imp is actually affecting production of SurA, cotranscription of imp and surA was demonstrated by measuring transcription of a surA::luc fusion in a strain with an intact imp gene (JSG1325) or a strain with an interrupted imp (JSG1329). Disruption of imp caused a 46-fold reduction of surA transcription (data not shown.)
JSG1272 and JSG1290 contained MudJ insertions in Salmonella serovar Typhimurium gnd. The ORF for pmrE is 237 bp downstream of gnd, with no other ORFs present in the intervening DNA. The possibility arose that the two genes are cotranscribed and that the PMs phenotype resulting from the MudJ insertion may be due to a polar effect on pmrE rather than a disruption of gnd itself, particularly since a pmrE mutant has been demonstrated to be PMs (17). To address the issue of cotranscription of gnd and pmrE, transcription of a pmrE::MudJ fusion was measured in a strain containing an intact gnd gene and in a strain containing a gnd::Tn10d mutation (11). Transcription of pmrE::MudJ was consistently about 1.5-fold higher in a background with gnd intact (data not shown), but this result may not indicate operonic arrangement of these genes; this is supported by the fact that pmrE is strongly activated by PmrA, but gnd is not PmrA regulated. To further examine the role of gnd in PM resistance, an independent, in-frame deletion in gnd was created and introduced into JSG844 (PmrAc Strr). The new Δgnd mutant, JSG1724, was examined for PM resistance. JSG1724 demonstrated a PM MIC of 12 μg/ml, equivalent to the PM resistance exhibited by the PmrAc parental strain. Deletion of gnd therefore does not result in PM sensitivity, indicating that the MudJ insertion in gnd in JSG1290 is likely affecting pmrE.
The four PMs mutants were tested for sensitivity to high iron concentrations, as mutations that result in loss of LPS modifications that affect sensitivity to cationic AP may also result in sensitivity to ferric ions (57). JSG851 (surA) and JSG895 (tolB) grew poorly on high iron, whereas JSG1272 (gnd) and JSG1290 (gnd) were as resistant to 100 μM FeSO4 as was PmrAc Salmonella serovar Typhimurium. It may be that surA and tolB mutations cause a destabilization of the bacterial envelope and in this manner lead to increased sensitivity to high iron concentrations. A transposon insertion in gnd, however, which may affect pmrE transcription and in this way alter LPS structure, does not affect resistance to high iron concentrations.
Analysis of lipid A for presence of Ara4N.
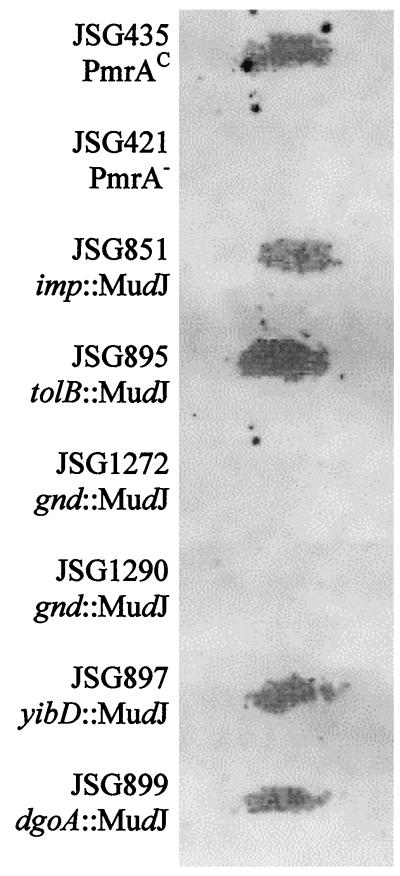
Ara4N modification of lipid A, which is a consequence of the function of a number of PmrA-regulated genes, is associated with a PM-resistant phenotype of gram-negative bacteria. Accordingly, the lipid A of each mutant identified in the screen, including four PMs, non-PmrA-regulated gene mutants and two PMr, PmrA-regulated gene mutants, was analyzed for the presence of Ara4N. Purified lipid A was examined using a paper chromatography method in which ninhydrin is the detection agent. Staining of Ara4N with ninhydrin results in a unique yellow color, whereas other lipid A components containing amino groups yield a purple color. Both of the PMs, non-PmrA-regulated gene mutants with insertions in gnd (JSG1272 and JSG1290) lacked Ara4N modification to lipid A (Fig. 4). However, based on the data derived from the gnd deletion strain, it is likely that the Ara4N loss can be attributed to an effect of the MudJ insertion on pmrE transcription. PMs mutants JSG851 (surA) and JSG895 (tolB) retained Ara4N on lipid A in similar amounts to the parental PmrAc strain, suggesting that the MudJ insertions in these mutants affect PM resistance without influencing Ara4N modification to LPS. JSG897 and JSG899, with mutations in PmrA-regulated genes yibD and dgoA, respectively, also still possessed Ara4N modification of lipid A in similar amounts to the parental PmrAc strain. Therefore, these genes fall into a minor class of PmrA-regulated genes that alone do not affect LPS modification or AP resistance.
FIG. 4.
Analysis of lipid A for the presence of aminoarabinose. Paper chromatography was performed on lipid A from PmrAc and PmrA-null Salmonella serovar Typhimurium, as well as lipid A from the four PMs mutants and two PmrA-regulated gene mutants identified in this study. The chromatograms were run with a solvent system of isopropanol-ethyl acetate-water (7:1:2). Yellow color upon development with ninhydrin is characteristic of 4-aminoarabinose (yellow color was converted to grey).
Virulence analysis of mutants.
The PmrA-null mutant has been demonstrated to have a virulence defect when administered orally to mice (19). Additionally, mutants that are sensitive to AP, including those lacking Ara4N on lipid A, possess similar virulence defects (17). Consequently, the mutants identified by MudJ mutagenesis were examined for attenuation of virulence in mice. Experiments were performed using strains with MudJ insertion mutations in a wild-type background rather than a PmrAc background. PmrA-regulated gene mutants JSG1525 (yibD) and JSG1526 (dgoA) were as virulent as wild-type Salmonella serovar Typhimurium, so the genes interrupted have no essential role in the infection process (Table 4). However, mutants JSG1521 (surA), JSG1522 (tolB), JSG1523 (gnd, affecting pmrE), and JSG1524 (gnd, affecting pmrE), all of which showed increased sensitivity to PM, were less virulent than the wild type. Since these mutants (surA, tolB, and gnd, affecting pmrE) are also PM sensitive, these data may support the hypothesis that AP resistance is required for Salmonella serovar Typhimurium virulence. However, the membrane-destabilizing effects of the surA and tolB mutants may result in a more generalized in vivo sensitivity.
TABLE 4.
Virulence of PmrA-regulated gene mutants and mutants with increased sensitivity to PM
| Strain | Description | No. of mice survivinga |
|---|---|---|
| JSG210 | Wild type | 1/4 |
| JSG1521 | imp::MudJ (surA), PMs | 4/4 |
| JSG1522 | tolB::MudJ, PMs | 4/4 |
| JSG1523 | gnd::MudJ, PMs | 3/4 |
| JSG1524 | gnd::MudJ, PMs | 3/4 |
| JSG1525 | yibD::MudJ, PmrA-regb | 0/4 |
| JSG1526 | dgoA::MudJ, PmrA-reg | 0/4 |
Mice were inoculated orally with 5 × 106 bacteria.
PmrA-reg, PmrA-regulated mutant.
DISCUSSION
Modifications to LPS have been suggested to play an important role in pathogenesis of Salmonella serovar Typhimurium by increasing resistance to cationic AP (4, 12, 13, 16, 20, 33, 47, 53, 54, 59). Salmonella serovar Typhimurium extensively modifies its LPS in response to signals that the organism is likely to encounter in the host (15, 42, 43, 57). These modifications are mediated by the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems (4, 16-19, 22, 52, 62). The PmrA-PmrB two-component system plays a key role in responding to the host microenvironment by up-regulating the genes involved in adding Ara4N to lipid A in response to mild acidic pH and alterations in the external concentrations of iron and magnesium (17, 18, 42, 57). Mutants in pmrE or pmrF, two PmrA-activated loci, lack Ara4N modification to lipid A and demonstrate reduced virulence when administered orally to mice (19). The Ara4N modification results in neutralization of the net negative charge of LPS, resulting in a decreased affinity of cationic AP to LPS. While several PmrA-regulated loci identified to date are involved in addition of Ara4N to lipid A, other potential targets for regulation by PmrA are proposed: PmrA-regulated genes responsible for pEtN addition to LPS have not been found, and PmrA-regulated genes involved in resistance to high iron (in addition to pmrL/pbgE2 and pmrM/pbgE3) remain to be identified (57).
MudJ transposon mutagenesis was utilized to find PmrA-regulated genes, genes necessary for PM resistance, and genes that exhibit both properties. Three transposon insertion sites were identified as affecting PM resistance. JSG851 contained a MudJ insertion within the imp ORF. Cotranscriptional and complementation analyses demonstrated that the transposon insertion in this mutant caused the PM-sensitive phenotype by affecting surA, the gene downstream of imp. In E. coli, SurA has been shown to be required for correct folding of certain outer membrane proteins, including OmpA, OmpF, and LamB (27). The inability of a surA mutant to efficiently fold these important outer membrane proteins would likely cause a destabilization of the outer membrane. For instance, an E. coli surA mutant shows sensitivity to compounds such as vancomycin, bacitracin, and bile salts (27); a stable outer membrane contributes to resistance to these compounds. The envelope defects of a Salmonella serovar Typhimurium surA mutant may thus result in increased sensitivity to PM. Similarly, the finding that JSG851 grew poorly on high concentrations of iron suggests that deficiencies in outer membrane integrity can affect sensitivity to ferric ions. The fact that JSG851 retained its ability to modify its lipid A with Ara4N and still showed PM sensitivity supports the conclusion that factors other than LPS modifications are responsible for the PM sensitivity of this mutant.
The PMs mutant JSG895 contained an insertion in tolB. The Tol-peptidoglycan-associated lipoprotein (Pal) system of E. coli is necessary for uptake of group A colicins and is believed to anchor the outer membrane to the cytoplasmic membrane (5). The various components of the Tol/Pal complex are associated with the cytoplasmic membrane (TolQ, TolR, and TolA), the periplasm (TolB), and the outer membrane (PAL) (28). These proteins interact with the various membrane components and with one another to stabilize the envelope of several gram-negative bacteria, and homologues have been studied in E. coli, Pseudomonas putida, Pseudomonas aeruginosa, and Haemophilus influenzae (5, 6, 9, 10, 24, 30, 36, 37, 39). Mutations in the Salmonella serovar Typhimurium tol genes result in increased sensitivity to antibiotics and detergents such as bile salts (35). The transposon insertion in Salmonella serovar Typhimurium tolB could therefore result in hypersensitivity to PM as a consequence of the loss of outer membrane stability. In addition, JSG895 showed sensitivity to high iron. Like the surA mutant, iron sensitivity may be due to outer membrane structural defects. Susceptibility to PM and high iron concentrations is not due to loss of Ara4N substitutions to lipid A, as this modification is still present in JSG895.
In two PM-sensitive MudJ mutants, JSG1272 and JSG1290, the transposon insertion site was determined to interrupt gnd, which encodes a gluconate-6-phosphate dehydrogenase involved in the pentose-phosphate metabolic pathway (45). gnd is the final gene in the rfb locus involved in LPS biosynthesis (34) and is located upstream of pmrE, a PmrA-regulated gene shown to be necessary for PM resistance and wild-type virulence (17). Previous analyses of the rfb locus suggest the presence of a strong transcriptional terminator following gnd and preceding pmrE (34). Nonetheless, the potential for a polar effect of the transposon insertion in gnd on the transcription of pmrE was investigated using cotranscription studies and an independent mutation in gnd. Assays analyzing the effect of a MudJ insertion in gnd on transcription of a pmrE::MudJ fusion showed a slight decrease in pmrE transcription in a gnd::Tn10d background. Because these results are not conclusive, we proceeded to generate an in-frame deletion of gnd to determine whether elimination of gnd specifically causes sensitivity to PM. MIC analysis of the Δgnd mutant (JSG1724) showed resistance to PM equivalent to that of the parental PmrAc strain, suggesting that gnd does not play a role in PM resistance. It may be postulated that the MudJ transposon insertion in gnd was capable of interrupting pmrE transcription, thus causing PM sensitivity, while a deletion of gnd precluded this effect. This interpretation is supported by previous studies showing that a PmrAc strain containing a pmrE mutation does not express Ara4N modification of lipid A (17), as this study demonstrated a lack of Ara4N on lipid A in both gnd::MudJ mutants.
Virulence analysis of the PMs mutants showed that insertions in imp (affecting surA), tolB, and gnd (likely affecting pmrE) resulted in a decrease in virulence (i.e., fewer mice died) in the mouse model, compared to wild-type Salmonella serovar Typhimurium. In light of the findings suggesting that the PM sensitivity shown by the gnd::MudJ mutants (JSG1272 and JSG1290) is likely a result of a polar effect of MudJ on pmrE, it may be interpreted that the virulence defect in JSG1272 and JSG1290 can be attributed to the polarity of the transposon insertion as well. As discussed above, mutations in tolB and surA may cause a destabilization of the bacterial envelope. In addition to the resulting sensitivity to PM and high iron concentrations, the potential effect of the mutations on membrane integrity may also affect the ability of these strains to survive within the host, due to increased sensitivities to other membrane-active agents or osmotic changes.
Several mutants were identified in which the transposon insertion occurred in PM resistance genes regulated by PmrA. However, these insertions occurred in known loci, including the pmrCAB and pmrHFIJKLM operons. The fact that these loci were repeatedly identified in the screen suggests completeness of the mutagenesis. However, there was specificity for MudJ insertion in both the pmrCAB and pmrHFIJKLM operons, as all insertions recovered were in pmrC or pmrI.
The transposon insertion in JSG897 identified a PmrA-regulated gene, yibD, which encodes a putative glycosyl transferase. Sequencing of the MudJ insertion site of JSG899 identified dgoA as a PmrA-regulated gene. dgoA is predicted to encode a 2-oxo-3-deoxygalactonate-6-phosphate aldolase-galactonate dehydratase involved in galactonate metabolism. Minimal research has been performed on these two loci. Neither dgoA nor yibD affect PM resistance, which is supported by the finding that JSG897 and JSG899 are capable of modifying lipid A with Ara4N. In addition, neither yibD nor dgoA affect resistance to high concentrations of iron or virulence in the mouse model. Therefore, it is not clear if these genes affect unknown functions of the PmrA-PmrB regulon or if redundant functions exist for these genes and they do affect AP resistance and/or Ara4N modification. While it is possible that these PmrA-regulated genes (as well as the genes involved in PM resistance) could affect the other known PmrA-induced LPS modification, pEtN, we feel that this is unlikely based on what is known about the identified genes (surA, tolB, and pmrE) and because yibD and dgoA do not affect virulence or PM resistance, which are predicted phenotypes of the loss of pEtN modification from the core (59).
A strong match to the consensus PmrA-binding site was identified upstream of yibD, indicating direct regulation by PmrA. However, no consensus site was apparent in dgoA, suggesting indirect activation by PmrA. PmrA has been shown to bind the promoters of other PmrA-regulated loci, including pmrCAB, pmrHFIJKLM, and pmrE (ugd) (1, 56). Future studies involving Salmonella serovar Typhimurium yibD and dgoA mutants will entail determining whether PmrA is capable of directly binding the promoters of these loci and identifying the putative regulatory binding sites in these regions.
Acknowledgments
This work was supported by grant AI43521 from The National Institutes of Health to J.S.G.
Editor: V. J. DiRita
REFERENCES
- 1.Aguirre, A., S. Lejona, E. G. Vescovi, and F. C. Soncini. 2000. Phosphorylated PmrA interacts with the promoter region of ugd in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:3874-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apicella, M. A., J. M. Griffiss, and H. Schneider. 1994. Isolation and characterization of lipopolysaccharides, lipooligosaccharides, and lipid A. Methods Enzymol. 235:242-252. [DOI] [PubMed] [Google Scholar]
- 3.Bevins, C. L. 1994. Antimicrobial peptides as agents of mucosal immunity. Ciba Found. Symp. 186:250-260. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, R. E., H. S. Gibbons, T. Guina, M. S. Trent, S. I. Miller, and C. R. Raetz. 2000. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 19:5071-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouveret, E., H. Benedetti, A. Rigal, E. Loret, and C. Lazdunski. 1999. In vitro characterization of peptidoglycan-associated lipoprotein (PAL)-peptidoglycan and PAL-TolB interactions. J. Bacteriol. 181:6306-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouveret, E., R. Derouiche, A. Rigal, R. Lloubes, C. Lazdunski, and H. Benedetti. 1995. Peptidoglycan-associated lipoprotein-TolB interaction. A possible key to explaining the formation of contact sites between the inner and outer membranes of Escherichia coli. J. Biol. Chem. 270:11071-11077. [DOI] [PubMed] [Google Scholar]
- 7.Breazeale, S. D., A. A. Ribeiro, and C. R. Raetz. 2002. Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. Origin of lipid A species modified with 4-amino-4-deoxy-l-arabinose. J. Biol. Chem. 277:2886-2896. [DOI] [PubMed] [Google Scholar]
- 8.Caroff, M., A. Tacken, and L. Szabo. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175:273-282. [DOI] [PubMed] [Google Scholar]
- 9.Clavel, T., P. Germon, A. Vianney, R. Portalier, and J. C. Lazzaroni. 1998. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol. Microbiol. 29:359-367. [DOI] [PubMed] [Google Scholar]
- 10.Duan, K., E. R. Lafontaine, S. Majumdar, and P. A. Sokol. 2000. RegA, iron, and growth phase regulate expression of the Pseudomonas aeruginosa tol-oprL gene cluster. J. Bacteriol. 182:2077-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enos-Berlage, J. L., and D. M. Downs. 1996. Involvement of the oxidative pentose phosphate pathway in thiamine biosynthesis in Salmonella typhimurium. J. Bacteriol. 178:1476-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst, R. K., T. Guina, and S. I. Miller. 1999. How intracellular bacteria survive: surface modifications that promote resistance to host innate immune responses. J. Infect. Dis. 179(Suppl. 2):S326-S330. [DOI] [PubMed] [Google Scholar]
- 13.Farley, M. M., W. M. Shafer, and J. K. Spitznagel. 1988. Lipopolysaccharide structure determines ionic and hydrophobic binding of a cationic antimicrobial neutrophil granule protein. Infect. Immun. 56:1589-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganz, T., M. E. Selsted, D. Szklarek, S. S. Harwig, K. Daher, D. F. Bainton, and R. I. Lehrer. 1985. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 76:1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groisman, E. A., J. Kayser, and F. C. Soncini. 1997. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J. Bacteriol. 179:7040-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunn, J. S., R. K. Ernst, A. J. McCoy, and S. I. Miller. 2000. Constitutive mutations of the Salmonella enterica serovar Typhimurium transcriptional virulence regulator phoP. Infect. Immun. 68:3758-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 18.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunn, J. S., S. S. Ryan, J. C. Van Velkinburgh, R. K. Ernst, and S. I. Miller. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 21.Heithoff, D. M., C. P. Conner, P. C. Hanna, S. M. Julio, U. Hentschel, and M. J. Mahan. 1997. Bacterial infection as assessed by in vivo gene expression. Proc. Natl. Acad. Sci. USA 94:934-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helander, I. M., I. Kilpelainen, and M. Vaara. 1994. Increased substitution of phosphate groups in lipopolysaccharides and lipid A of the polymyxin-resistant pmrA mutants of Salmonella typhimurium: a 31P-NMR study. Mol. Microbiol. 11:481-487. [DOI] [PubMed] [Google Scholar]
- 23.Hughes, K. T., and J. R. Roth. 1988. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics 119:9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Journet, L., E. Bouveret, A. Rigal, R. Lloubes, C. Lazdunski, and H. Benedetti. 2001. Import of colicins across the outer membrane of Escherichia coli involves multiple protein interactions in the periplasm. Mol. Microbiol. 42:331-344. [DOI] [PubMed] [Google Scholar]
- 25.Kanipes, M. I., S. Lin, R. J. Cotter, and C. R. Raetz. 2001. Ca2+-induced phosphoethanolamine transfer to the outer 3-deoxy-d-manno- octulosonic acid moiety of Escherichia coli lipopolysaccharide. A novel membrane enzyme dependent upon phosphatidylethanolamine. J. Biol. Chem. 276:1156-1163. [DOI] [PubMed] [Google Scholar]
- 26.Kox, L. F., M. M. Wosten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazar, S. W., and R. Kolter. 1996. SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 178:1770-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazzaroni, J. C., P. Germon, M. C. Ray, and A. Vianney. 1999. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol. Lett. 177:191-197. [DOI] [PubMed] [Google Scholar]
- 29.Lehrer, R. I., T. Ganz, and M. E. Selsted. 1991. Defensins: endogenous antibiotic peptides of animal cells. Cell 64:229-230. [DOI] [PubMed] [Google Scholar]
- 30.Llamas, M. A., J. L. Ramos, and J. J. Rodriguez-Herva. 2000. Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for maintenance of outer membrane stability. J. Bacteriol. 182:4764-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 32.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nummila, K., I. Kilpelainen, U. Zahringer, M. Vaara, and I. M. Helander. 1995. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 16:271-278. [DOI] [PubMed] [Google Scholar]
- 34.Paton, A. W., and J. C. Paton. 1999. Molecular characterization of the locus encoding biosynthesis of the lipopolysaccharide O antigen of Escherichia coli serotype O113. Infect. Immun. 67:5930-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prouty, A. M., J. C. Van Velkinburgh, and J. S. Gunn. 2002. Salmonella enterica serovar Typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J. Bacteriol. 184:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray, M. C., P. Germon, A. Vianney, R. Portalier, and J. C. Lazzaroni. 2000. Identification by genetic suppression of Escherichia coli TolB residues important for TolB-Pal interaction. J. Bacteriol. 182:821-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rigal, A., E. Bouveret, R. Lloubes, C. Lazdunski, and H. Benedetti. 1997. The TolB protein interacts with the porins of Escherichia coli. J. Bacteriol. 179:7274-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riley, J., R. Butler, D. Ogilvie, R. Finniear, D. Jenner, S. Powell, R. Anand, J. C. Smith, and A. F. Markham. 1990. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 18:2887-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen, K., D. J. Sikkema, and T. F. Murphy. 1996. Isolation and characterization of the Haemophilus influenzae tolQ, tolR, tolA and tolB genes. Gene 178:75-81. [DOI] [PubMed] [Google Scholar]
- 40.Shafer, W. M., L. E. Martin, and J. K. Spitznagel. 1984. Cationic antimicrobial proteins isolated from human neutrophil granulocytes in the presence of diisopropyl fluorophosphate. Infect. Immun. 45:29-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 42.Soncini, F. C., E. Garcia Vescovi, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soncini, F. C., and E. A. Groisman. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178:6796-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spitznagel, J. K. 1990. Antibiotic proteins of human neutrophils. J. Clin. Investig. 86:1381-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sprenger, G. A. 1995. Genetics of pentose-phosphate pathway enzymes of Escherichia coli K-12. Arch. Microbiol. 164:324-330. [DOI] [PubMed] [Google Scholar]
- 46.Steinberg, D. A., M. A. Hurst, C. A. Fujii, A. H. Kung, J. F. Ho, F. C. Cheng, D. J. Loury, and J. C. Fiddes. 1997. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob. Agents Chemother. 41:1738-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stinavage, P., L. E. Martin, and J. K. Spitznagel. 1989. O antigen and lipid A phosphoryl groups in resistance of Salmonella typhimurium LT-2 to nonoxidative killing in human polymorphonuclear neutrophils. Infect. Immun. 57:3894-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trent, M. S., A. A. Ribeiro, W. T. Doerrler, S. Lin, R. J. Cotter, and C. R. Raetz. 2001. Accumulation of a polyisoprene-linked amino sugar in polymyxin-resistant Salmonella typhimurium and Escherichia coli: structural characterization and transfer to lipid A in the periplasm. J. Biol. Chem. 276:43132-43144. [DOI] [PubMed] [Google Scholar]
- 49.Trent, M. S., A. A. Ribeiro, S. Lin, R. J. Cotter, and C. R. Raetz. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276:43122-43131. [DOI] [PubMed] [Google Scholar]
- 50.Vaara, M. 1992. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56:395-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaara, M. 1981. Increased outer membrane resistance to ethylenediaminetetraacetate and cations in novel lipid A mutants. J. Bacteriol. 148:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaara, M., T. Vaara, M. Jensen, I. Helander, M. Nurminen, E. T. Rietschel, and P. H. Makela. 1981. Characterization of the lipopolysaccharide from the polymyxin-resistant pmrA mutants of Salmonella typhimurium. FEBS Lett. 129:145-149. [DOI] [PubMed] [Google Scholar]
- 53.Vaara, M., T. Vaara, and M. Sarvas. 1979. Decreased binding of polymyxin by polymyxin-resistant mutants of Salmonella typhimurium. J. Bacteriol. 139:664-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaara, M., and P. Viljanen. 1985. Binding of polymyxin B nonapeptide to gram-negative bacteria. Antimicrob. Agents Chemother. 27:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 56.Wosten, M. M., and E. A. Groisman. 1999. Molecular characterization of the PmrA regulon. J. Biol. Chem. 274:27185-27190. [DOI] [PubMed] [Google Scholar]
- 57.Wosten, M. M., L. F. Kox, S. Chamnongpol, F. C. Soncini, and E. A. Groisman. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113-125. [DOI] [PubMed] [Google Scholar]
- 58.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 59.Yethon, J. A., J. S. Gunn, R. K. Ernst, S. I. Miller, L. Laroche, D. Malo, and C. Whitfield. 2000. Salmonella enterica serovar Typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence in vivo. Infect. Immun. 68:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zasloff, M. 1992. Antibiotic peptides as mediators of innate immunity. Curr. Opin. Immunol. 4:3-7. [DOI] [PubMed] [Google Scholar]
- 61.Zhou, Z., S. Lin, R. J. Cotter, and C. R. Raetz. 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K-12. Detection of 4-amino-4-deoxy- l-arabinose, phosphoethanolamine and palmitate. J. Biol. Chem. 274:18503-18514. [DOI] [PubMed] [Google Scholar]
- 62.Zhou, Z., A. A. Ribeiro, S. Lin, R. J. Cotter, S. I. Miller, and C. R. Raetz. 2001. Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PmrA-dependent 4-amino-4-deoxy-l-arabinose, and phosphoethanolamine incorporation. J. Biol. Chem. 276:43111-43121. [DOI] [PubMed] [Google Scholar]