Abstract
Infection of human macrophages with Salmonella enterica serovar Typhimurium or Salmonella enterica serovar Dublin produces delayed cytotoxicity characterized by cell detachment and associated apoptosis. Using a site-specific mutant in the SpvB active site, we verify that the ADP-ribosylation activity of SpvB is required for delayed cytotoxicity in human macrophages infected with Salmonella. SipB and the type III protein secretion system (TTSS) encoded by Salmonella pathogenicity island 1 (SPI1) are not involved, whereas the SPI2 TTSS is absolutely required for SpvB-dependent cytotoxicity. Furthermore, we show that infection of macrophage cultures with wild-type or sipB mutant bacteria led to a complete loss of polymerized actin in over half of the cells after 24 h. In contrast, macrophages infected with the spvB or SPI2 (ssaV or ssaJ) mutant strain retained normal F-actin filaments, despite similar numbers of intracellular bacteria. We conclude that SpvB and a functional SPI2 TTSS are essential for Salmonella-induced delayed cytotoxicity of human macrophages.
Salmonella enterica subspecies I strains are responsible for enteric fever, gastroenteritis, and bacteremia in humans and a broad array of illnesses in animals, including enteritis and septicemia in livestock. A number of subspecies I serovars carry virulence plasmids encoding the highly conserved spv locus associated with disease syndromes of greater severity (13). The spv genes are found more frequently in human extraintestinal isolates than in environmental or fecal isolates, and most cases of human nontyphoidal bacteremia are caused by serovars that contain the spv genes (12, 13).
The plasmid-encoded spv locus consists of the spvR regulatory gene and four structural genes, spvABCD (26). spvR encodes a transcriptional regulator activating its own expression and the spvABCD operon (9, 27). Transcription of the spv operon also requires the stationary-phase alternate σ factor RpoS (5, 10). Since both rpoS and spv are upregulated after phagocytosis by macrophages (4), expression of the spv genes appears to be strongly induced by the intracellular environment of the host cell. The spv genes appear to promote the macrophage phase of the disease process, avoiding destruction by neutrophils and facilitating proliferation of Salmonella strains at extraintestinal sites of infection (13).
In vivo experiments suggest that the spv genes enhance bacterial replication in macrophages (18). In human macrophages, spv gene expression is required for induction of cytotoxicity characterized by cell detachment and eventual apoptosis (29). The spvB gene within the spv operon is essential for the spv virulence phenotype, as shown by mutational analysis (35). The spvB gene encodes a 65.6-kDa protein (recently characterized as an ADP-ribosyl transferase) that modifies actin and blocks polymerization to F-actin filaments (28, 38). Site-directed mutagenesis of the conserved NAD-binding site sequence demonstrates that this ADP-ribosylation activity is essential for virulence in mice (28).
Macrophages infected with Salmonella show complex patterns of cytotoxicity, with early and late phases that depend on different sets of virulence genes (2, 6, 22, 24, 30, 32, 33, 40). Early-phase cytotoxicity requires Salmonella pathogenicity island 1 (SPI1). SPI1 is known to encode a type III secretion system (TTSS) which exports proteins involved in host invasion (8, 16). The SPI1-encoded SipB protein activates caspase 1, which results in early cell death and lysis by mechanisms resembling necrosis (2, 22, 24). In contrast, Salmonella enterica serovar Dublin and Salmonella enterica serovar Typhimurium, when grown to stationary phase and phagocytosed by macrophages (conditions which do not activate SPI1 TTSS expression), have been shown to induce toxicity in murine, bovine, and human macrophages 18 to 24 h after infection (29, 30, 36, 40). The SPI2 TTSS, which is known to be required for systemic infection in mice (19, 20) and for the intracellular survival and proliferation of Salmonella strains in murine macrophages (7, 21, 34), has been shown to be important for this delayed killing in murine and bovine macrophages.
Human macrophages also exhibit cytotoxic changes after infection with serovar Typhimurium and serovar Dublin (29). In this paper, we examine genetic requirements for Salmonella-induced delayed cytotoxicity during infection of human monocyte-derived macrophages. We correlate previous measures of cytotoxicity, including numbers of intracellular bacteria and cell detachment, with loss of F-actin during infection. Specific mutants are used to evaluate the requirements for SpvB ADP-ribosylation activity and for functional SPI1 and SPI2 TTSSs.
Blood was collected in heparinized syringes from normal donors, and the monocytes were purified by density gradient centrifugation and differential adherence (14, 29). Monocytes were seeded into 24-well plates at a density of 4 × 105 to 5 × 105 cells/well and incubated in RPMI medium with 18% autologous serum for 7 days to allow differentiation into macrophages (14). Single colonies of the wild-type S. enterica serovar Dublin Lane strain (29) and serovar Typhimurium 14028s strains or mutants derived from these strains (spvR15-2 [29], spvBmut1 [28], sipB::aphT [25], invA::aphT [15], ssaV::mTn5 [strain P9B6], and ssaJ::mTn5 [strain P11D10] [20]) were grown to stationary phase in minimal essential medium (9). Bacteria were opsonized with 50% autologous serum for 15 min at 37°C, and the mixture was diluted and used to inoculate macrophages at a multiplicity of infection (MOI) of approximately 1. Culture plates were centrifuged at 200 × g and incubated for 1 h at 37°C to allow phagocytosis to occur. The cells were washed once with RPMI medium, and fresh medium containing gentamicin (20 μg/ml) was then added to kill the remaining extracellular bacteria. Cultures were harvested at 18 to 23 h after infection. The culture supernatants of three wells were combined, centrifuged, and resuspended in RPMI medium. A portion of these cells was added at a 1:1 ratio to 0.5% trypan blue stain (Gibco BRL, Rockville, Md.) and counted using a Fisher Scientific hemocytometer (Pittsburgh, Pa.). The remainder of these resuspended cells was added to 0.5% deoxycholate in phosphate-buffered saline (PBS) to lyse macrophages. Dilutions of this solution, representing the lysate of detached cells, were plated on Luria-Bertani agar. Adherent cells from the corresponding wells were lysed with 1 ml of deoxycholate in PBS, the products of three wells were combined as described above, and serial dilutions of this lysate from attached macrophages were plated on Luria-Bertani agar.
At the outset, the amount of detachment was measured for human monocyte-derived macrophages infected with the wild-type serovar Dublin strain, the spvBmut1 mutant strain, which was constructed by substituting aspartates for glutamates (E538D and E540D) at the ADP-ribosylation active site (28), or with an spvR mutant strain that abolishes expression of the entire spv locus. Table 1 shows that after 20 h, the number of detached cells in cultures infected with the wild-type serovar Dublin strain is more than 20-fold greater than that in macrophage cultures infected with mutant strains. Infection with the spvB mutant strain showed no significant difference from that with the spvR mutant strain (Table 1). Thus, detachment of human macrophages infected with serovar Dublin required expression of the SpvB protein with intact ADP-ribosylation activity.
TABLE 1.
Numbers of detached macrophages in the culture fluid of wells infected at an MOI of 1.0 with a serovar Dublin wild-type or spvB or spvR mutant straina
| Strain | No. (± SE) of:
|
||
|---|---|---|---|
| Detached cells | Detached bacteria | Attached bacteria | |
| Wild type | 10,650 (1,417) | 39,000 (11,676) | 76,666 (10,349) |
| spvB | 641 (182) | 2,033 (635) | 151,000 (9,712) |
| spvR | 122 (27) | 776 (311) | 127,000 (3,511) |
Only intact cells were counted, and values represent the means of the results for three wells. Also shown are the results for the viable bacteria recovered from the detached and attached macrophage fractions. Aliquots of the concentrated detached cells were diluted and plated for determination of numbers of CFU. After removal of culture fluid, attached cells were lysed as described in the text and the numbers of viable bacteria were determined by plating. Values represent the means of the results for four wells. The entire experiment was repeated, with similar results.
The number of bacterial CFU in each detached macrophage fraction was determined by dilution and plating (Table 1). The results demonstrate considerably higher levels of wild-type bacteria in the detached cell population after 20 h than in cells infected with the spvB or spvR mutant strain. The bacterial CFU levels in the attached macrophage fraction show no significant difference between strains (Table 1). Combining results in Table 1, the fraction of wild-type bacteria in the detached macrophage population was 34% of the total CFU per well, in contrast to 1% and 0.6% of total viable bacteria per well present in the detached macrophage populations infected by spvB and spvR mutants, respectively. Viable bacteria in the detached cell fraction correlated with the cell numbers (Table 1).
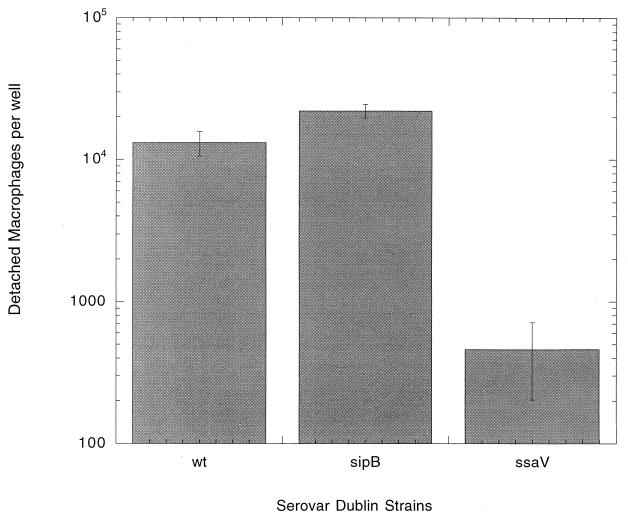
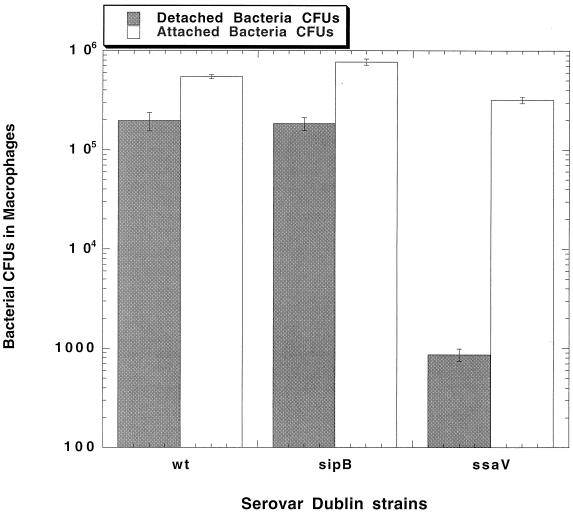
Next, the roles of Salmonella SPI1 and SPI2 loci in human monocyte-derived macrophage cytopathology were examined in the cell detachment assay. Figure 1 shows that detachment of cells infected with a sipB (SPI1) mutant strain of serovar Dublin reaches levels comparable to that of wild-type infection. In contrast, infection of human macrophages with serovar Dublin mutated at the ssaV gene locus within SPI2 produces a phenotype that virtually abolishes macrophage detachment. This effect is comparable to that of mutating the spvB gene, as seen for the previously described experiment. The number of CFU in these detached cell fractions, as determined by dilution and plating (Fig. 2), demonstrates much higher numbers of wild-type and sipB mutant bacteria than of the ssaV mutant strain. At 18 h, the fractions of wild-type and sipB mutant bacteria in detached cells represented 24% and 20%, respectively, of the total CFU in the well. The number of ssaV mutant strain bacteria in detached cells represents 0.2% of the viable bacteria per well. The bacterial CFU value for the attached cell fraction (Fig. 2) shows that the lack of detached cells in the ssaV mutant-infected cultures is not simply due to a difference in the numbers of bacteria in the attached cells.
FIG. 1.
Numbers of detached macrophages in the culture fluid of wells infected at an MOI of 1.0 with either the serovar Dublin wild-type, ssaV mutant, or sipB mutant strain. Only intact cells were counted, and the values represent the means of the results for four wells. The entire experiment was repeated, with similar results.
FIG. 2.
Viable bacteria recovered from the detached and attached macrophage fractions from the experiment whose results are shown in Fig. 1. Aliquots of the concentrated detached cells were diluted and plated for CFU counts. After removal of the culture fluid, attached cells were lysed as described in the text and the numbers of viable bacteria were determined by plating. Values represent the means of the results for four wells. The entire experiment was repeated, with similar results.
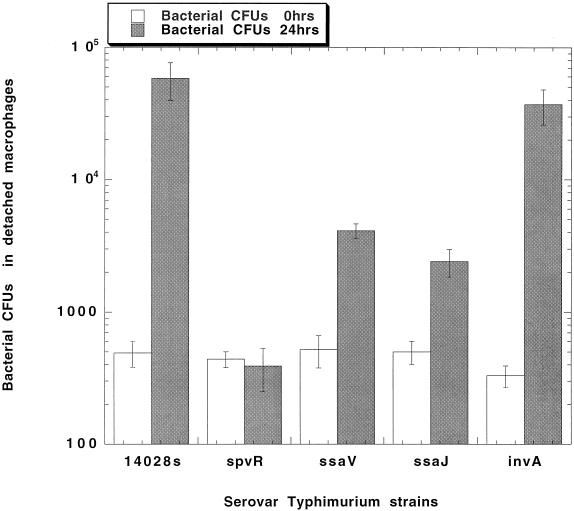
Human macrophages infected by the wild-type serovar Typhimurium strain or strains with mutations in SPI1, SPI2, or spv showed a cytotoxic phenotype similar to that of serovar Dublin. Human macrophages were infected with the wild-type serovar Typhimurium strain or one of the following mutant strains: spvR, SPI1 loci (invA and sipB::aph), and SPI2 loci (ssaV and ssaJ). The numbers of bacterial CFU in the detached cell fractions of these infections were determined at 0 and 24 h. The results shown in Fig. 3 demonstrate a large increase in viable wild-type serovar Typhimurium bacteria in the detached cell fraction recovered in the culture fluid after 24 h. A similar increase was observed with the invA (SPI1) mutant. In additional experiments, other SPI1 mutations (sipB::mudJ and sipC::aph) demonstrated a detached cell fraction result similar to that for wild-type infection (data not shown). No increase in bacterial numbers was seen with the spvR and SPI2 loci mutants, ssaV and ssaJ, after 24 h (Fig. 3). Since previous results have shown a close correlation between bacterial counts in detached cells and cytopathology (29), these data indicate that serovar Typhimurium also requires the presence of an intact SPI2 locus for the induction of cytopathic effects in human monocyte-derived macrophages.
FIG. 3.
Viable bacteria recovered from the detached macrophage fraction of human monocyte-derived macrophages infected with the serovar Typhimurium wild-type 14028s strain or a serovar Typhimurium strain with mutations in spv or the in SPI1 (invA) or SPI2 (ssaJ or ssaV) locus. Aliquots of the concentrated detached cells were diluted and plated for CFU counts. Values represent the means of the results for three wells. The entire experiment was repeated, with similar results.
Our results confirm that the cytopathology of human macrophage detachment depends on the NAD-binding sequence conferring ADP-ribosylation activity on SpvB. The substrate of this enzyme has been identified as actin, and the enzyme depolymerizes F-actin to G-actin filaments in CHO cells (28) and MDCK cells (38). Therefore, we investigated F-actin depolymerization in human macrophages 20 h after infection with a wild-type serovar Dublin strain or a serovar Dublin spvB, spvR, sipB, or ssaV mutant strain.
Intracellular actin was detected using phalloidin-rhodamine staining. Human macrophages were prepared, cultured, and infected as described above. After 24 h, each well was carefully scraped using a Costar cell scraper 3010 (Corning, N.Y.). The entire content of each well was aspirated, and the suspended cells were centrifuged at 300 × g for 15 min and then fixed with 3.7% formalin in PBS and spun onto microscope slides by using a Cytospin apparatus. Cells on slides were washed in PBS, rehydrated, and exposed to 0.1% Triton X-100 for 5 min. Following two washes with PBS, slides were preincubated with 1% bovine serum albumin in PBS for 30 min and then incubated with phalloidin-rhodamine (catalog no. R415; Molecular Probes, Eugene, Oreg.) for 20 min to stain F-actin filaments. After washing with PBS, the slides were counterstained for nucleic acid with 4′,6′-diamidino-2-phenylindole (DAPI) (1 μg/ml) in PBS for 15 min. Slides were visualized by fluorescent microscopy, and cells staining with phalloidin-rhodamine and/or DAPI were counted. Upon staining with DAPI, macrophages were identified by the presence of a bright nucleus and an abundant pale-blue cytoplasm. A total of 300 to 600 cells for each infecting Salmonella strain were identified, and the proportions of cells costaining for any detectable F-actin were determined. Only macrophages without any detectable actin staining were counted as negative.
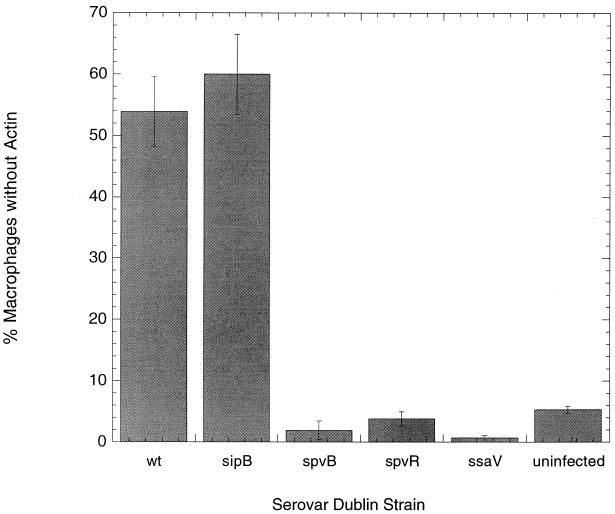
Figure 4 shows the percentages of human macrophages with F-actin depolymerization observed 20 h after infection with wild-type serovar Dublin or the spvR, spvB, sipB, or ssaV mutant strain. Only 2 and 3% of macrophages produced with the spvB and spvR mutants, respectively, were without actin staining. This proportion was essentially identical to that of uninfected human macrophages (1%). In contrast, examination of macrophages from cultures infected by wild-type bacteria showed 54% of macrophages without F-actin. The roles of SPI1 and SPI2 in actin cytoskeletal rearrangements were also examined. Figure 4 shows that 60% of macrophages from sipB mutant-infected cultures had total loss of F-actin, which is comparable to the result with wild-type serovar Dublin strain infection. In contrast, only 1% of cells from cultures infected with the ssaV mutant had F-actin loss, a proportion identical to that with the uninfected group. Thus, we demonstrate that cytopathology in human macrophages infected with serovar Dublin is associated with actin depolymerization requiring the spv and SPI2 loci.
FIG. 4.
The percentages of the total amount of human monocyte-derived macrophages in the culture well with complete actin depolymerization, as determined via F-actin staining with phalloidin-rhodamine 20 h after infection. Values represent the means of the results for three wells. Uninfected macrophages are compared with those infected with a serovar Dublin wild-type or spvB, spvR, ssaV, or sipB mutant strain.
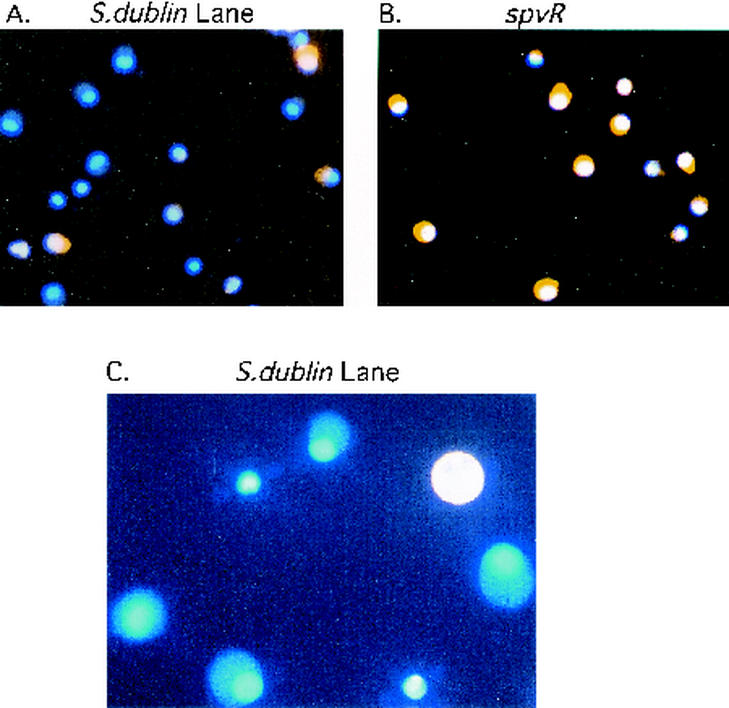
The appearance of macrophages from these cultures is presented in Fig. 5 and 6. In Fig. 5, human monocyte-derived macrophages were simultaneously stained with phalloidin-rhodamine (for polymerized actin) and DAPI. The results for many human monocyte-derived macrophages from cultures infected with the wild-type strain (Fig. 5A) demonstrate the absence of polymerized actin, while F-actin is intact in nearly all of the cells from the culture infected with the spvR mutant strain (Fig. 5B). Figure 5C shows a closer view of a wild-type strain-infected macrophage with a complete absence of polymerized actin. The “ghost” cells visible in the figure were usually associated with the presence of several bacteria. Gradations of actin loss could be observed, with some wild-type strain-infected cells showing a thin ring of periplasmic actin. The latter probably represents a partial stage of polymerized actin degradation. However, only macrophages without any detectable actin staining were counted as representing a negative result.
FIG. 5.
The appearance of human monocyte-derived macrophages after 20 h of infection with a wild-type or spvR mutant serovar Dublin strain. These cells were simultaneously stained with phalloidin-rhodamine (for polymerized actin) and DAPI. (A) spvR mutant-infected macrophages (×400 field); all cells have fluorescent orange labeling of cytoplasm F-actin. (B) Wild-type serovar Dublin-infected human monocyte-derived macrophages (×40 field); a number of these show an absence of polymerized actin and appear as ghost cells. (C) A view at a magnification of ×1,000 of a wild-type-infected macrophage with a complete absence of polymerized actin. Only macrophages without any actin staining were counted as representing negative results. These findings were duplicated in additional experiments not shown here.
FIG. 6.
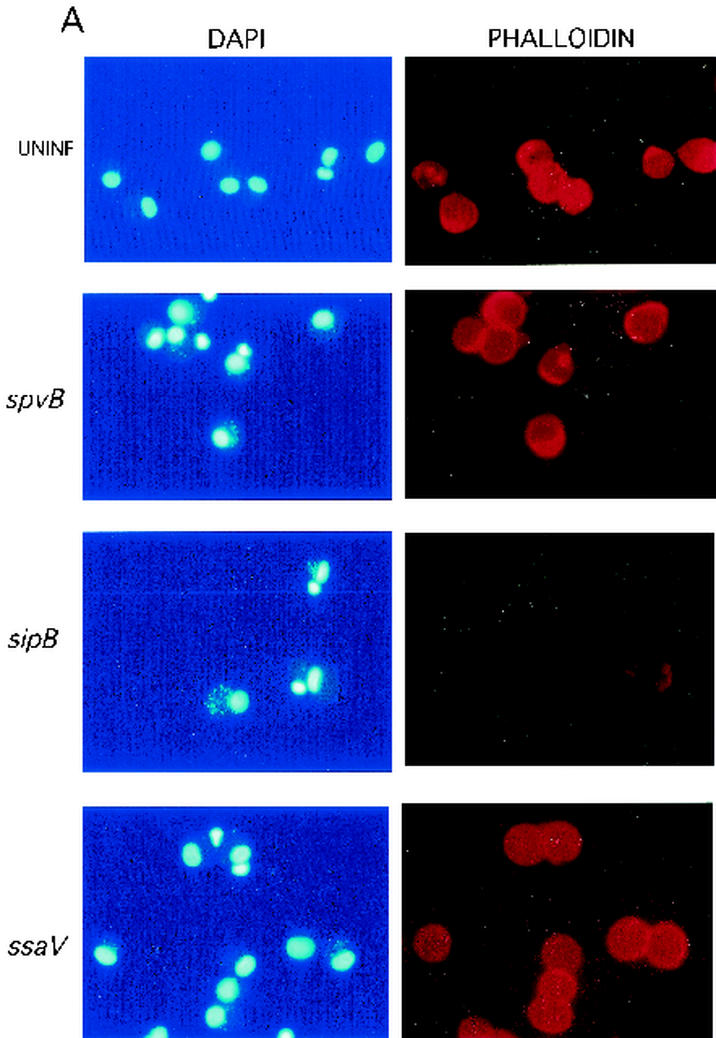
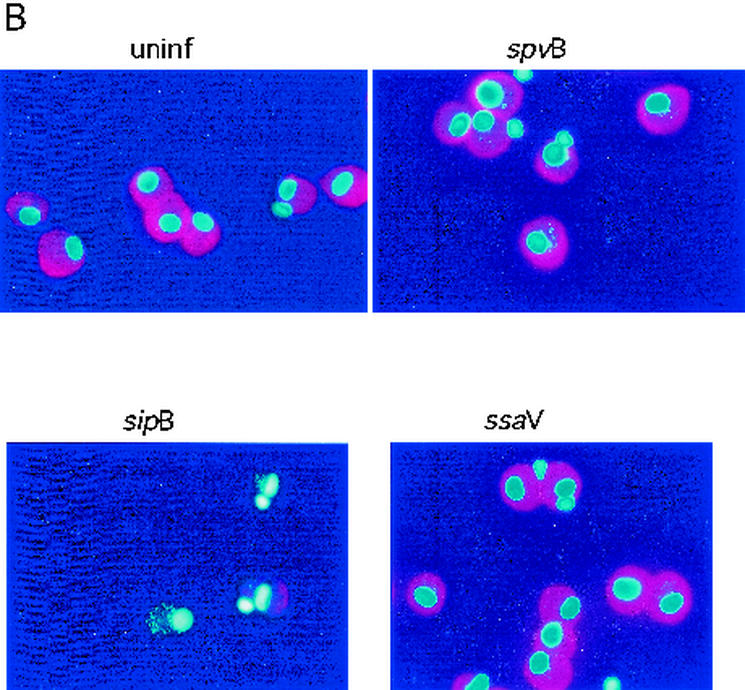
The appearance of uninfected human monocyte-derived macrophages compared with that of macrophages after 20 h of infection with a serovar Dublin wild-type or spvB, sipB, or ssaV mutant strain. (A) Fields examined separately with DAPI to stain the macrophages and associated bacteria and with phalloidin-rhodamine to stain F-actin; (B) merged image to demonstrate differences between cells with and without F-actin. Only macrophages without any actin staining were counted as representing negative results.
Figure 6 shows the appearances of human monocyte-derived macrophages from various infection conditions; the figure shows separate (Fig. 6A) and superimposed (Fig. 6B) images of bacteria examined separately for DAPI fluorescence and phalloidin-rhodamine. Despite the presence of multiple bacteria (stained with DAPI), polymerized actin stained with phalloidin-rhodamine appears intact in spvB mutant bacterial infections and identical to actin staining in uninfected cells. Figure 6 also shows the appearance of human monocyte-derived macrophages with a sipB mutant infection compared to those of uninfected and ssaV mutant-infected cells. sipB mutant-infected macrophages showed a complete loss of polymerized actin, as was the case for wild-type-infected macrophages. In contrast, macrophages in the ssaV mutant-infected culture showed the preservation of polymerized actin, despite similar numbers of cell-associated bacteria, which resembles the results for the spvB and spvR mutant infections and uninfected controls.
These results demonstrate by site-specific mutational analysis that the spv cytotoxic effect (29) in human monocyte-derived macrophages is dependent on the ADP-ribosylation activity of the SpvB protein. Furthermore, our studies show that the SPI1 locus has no role in this delayed cytotoxicity seen in human monocyte-derived macrophages. Cell detachment and proliferating bacteria were observed in human monocyte-derived macrophages infected with mutants deficient in SipB and InvA, which are encoded within SPI1. SipB is both a component of the TTSS and an effector protein, is involved in activation of caspase 1 and caspase 2 in murine macrophages (22, 24, 32), and is required by Salmonella strains for induction of murine macrophage cytotoxicity shortly after invasion (4, 22, 33). InvA is a component of the SPI1 TTSS required for translocation and delivery of type III-secreted effectors and is critical to host cell invasion (8, 15, 16). In contrast to the results with SPI1 mutants, the detachment of Salmonella-infected human monocyte-derived macrophages was virtually abolished in infections by strains with null mutations of the ssaV and ssaJ genes of the SPI2 locus. SsaV and SsaJ are essential components of the secretion system apparatus involved in delivery of a variety of type III-secreted effectors. The mechanism used by these effectors to promote virulence is unknown, although effects on vacuole morphology (1, 3), actin structure (31), vacuolar traffic (39), and assembly of vacuolar NADPH oxidase (17, 41) have been reported in murine macrophages.
SpvB modifies actin monomers in vitro and produces loss of F-actin filaments in cells (28, 38), and our experiments show that F-actin is a substrate for the SpvB protein in human macrophages. The macrophage populations studied in the actin experiments included both detached macrophages and those which remained attached (total culture population). The proportion of macrophages devoid of F-actin at 18 to 24 h after infection (approximately 50 to 60%) was much greater than the proportion of detached cells with proliferating bacteria observed in Salmonella-infected macrophage cultures in this and previous experiments (29). Thus, the cytotoxic effect of the SpvB protein is more extensive at this time point of infection than is indicated by measurements of the detached cell population alone.
Our experiments examining F-actin polymerization in human macrophages further demonstrate that SPI2 locus genes are required for expression of SpvB cytotoxicity. Macrophages infected with the ssaV mutant were indistinguishable from uninfected cells, whereas macrophages infected with SipB null mutants had the same level of F-actin loss as wild-type-infected cells. The finding that interruption of the SPI2 locus also affects actin depolymerization must mean either that SpvB is translocated by the SPI2 TTSS or that its intracellular action requires the SPI2 effector function. The present studies do not distinguish between these possibilities. Reports of prior studies relevant to the relationship between SpvB and SPI2 genes are sparse. Shea et al. (37) found that the presence of an spvA mutation appeared to further attenuate the virulence of an ssaV mutant, as assayed by the competitive index method in mixed infections of mice. These investigators concluded that spv and SPI2 have independent virulence effects. The SpvA mutation is likely to be polar on the downstream spv genes (27); thus, transcription of not only SpvB but also SpvC and -D proteins would be affected. Our study was performed in vitro on human macrophages and focused on cytopathology dependent on the ADP-ribosylation enzyme activity specific to the SpvB protein. The SpvC and SpvD proteins could have an accessory virulence role in vivo, as suggested in an earlier mutational analysis (35). Alternatively, SpvB may have an additional virulence function in vivo that is independent of ADP-ribosylation activity and that is detected when the SPI2 TTSS secretion system is inactive.
Salmonella virulence genes have been shown to be involved in both the polymerization (18, 23, 31, 42, 43) and depolymerization (28, 38) of actin. SPI1-encoded proteins coordinate short-lived local plasma membrane cytoskeletal reorganization, enabling the initial Salmonella invasion of epithelial cells. Since Salmonella enters macrophages primarily by phagocytosis, the mechanism of cytoskeletal rearrangements mediating bacterial entry is likely to differ between macrophages and epithelial cells. After 3 to 4 h, as bacteria are beginning to proliferate, polymerized actin appears to condense around the Salmonella-containing vacuole in an SPI2-dependent process (31). Since these actin polymerization processes are induced by Salmonella strains that also carry spvB, the expression and function of the SpvB protein must be regulated so as not to interfere with these earlier events. Therefore, the time course of actin cytoskeletal rearrangements appears critical during intracellular infection. Although spvB expression is detected during the first few hours of intracellular infection, cytotoxicity manifested by cell detachment occurs between 10 and 24 h following the entry of the bacteria (4, 29). Our results indicate that at 18 to 24 h after infection, F-actin is depolymerized by the SpvB protein and that cell detachment takes place in a process which depends on SPI2 function. This cytopathology is associated with apoptosis (29). These dying cells or resulting apoptotic bodies may be phagocytosed by surrounding macrophages, facilitating cell-to-cell spread of the infection. The finding that gentamicin is ineffective in Salmonella infections implies that Salmonella can complete the infectious cycle without being exposed to the extracellular environment (11).
Acknowledgments
This work was supported by Public Health Service grants AI32178 and DK35108.
We thank David Holden, Brian Ahmer, and Jorge Galán for generously providing Salmonella strains.
Editor: D. L. Burns
REFERENCES
- 1.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan, M. A., and B. T. Cookson. 2000. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 38:31-40. [DOI] [PubMed] [Google Scholar]
- 3.Brumell, J. H., C. M. Rosenberger, G. T. Gotto, S. L. Marcus, and B. B. Finlay. 2001. SifA permits survival and replication of Salmonella typhimurium in murine macrophages. Cell. Microbiol. 3:75-84. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C.-Y., L. Eckmann, S. J. Libby, F. C. Fang, S. Okamoto, M. F. Kagnoff, J. Fierer, and D. G. Guiney. 1996. Expression of Salmonella typhimurium rpoS and rpoS-dependent genes in the intracellular environment of eukaryotic cells. Infect. Immun. 64:4739-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C.-Y., N. A. Buchmeier, S. Libby, F. C. Fang, M. Krause, and D. G. Guiney. 1995. Central regulatory role for the RpoS sigma factor in expression of Salmonella dublin plasmid virulence genes. J. Bacteriol. 177:5303-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, L. M., K. Kaniga, and J. E. Galan. 1996. Salmonella spp. are cytotoxic for cultured macrophages. Mol. Microbiol. 21:1101-1115. [DOI] [PubMed] [Google Scholar]
- 7.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 8.Darwin, K. H., and V. L. Miller. 1999. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12:405-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang, F. C., M. Krause, C. Roudier, J. Fierer, and D. G. Guiney. 1991. Growth regulation of a Salmonella plasmid gene essential for virulence. J. Bacteriol. 173:6783-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, F. C., S. J. Libby, N. A. Buchmeier, P. C. Loewen, J. Switala, J. Harwood, and D. G. Guiney. 1992. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fierer, J., L. Hatlen, J.-P. Lin, D. Estrella, P. Mihalko, and A. Yau-Young. 1990. Successful treatment using gentamicin liposomes of Salmonella dublin infections in mice. Antimicrob. Agents Chemother. 34:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fierer, J., M. Krause, R. Tauxe, and D. Guiney. 1992. Salmonella typhimurium bacteremia: association with the virulence plasmid. J. Infect. Dis. 166:639-642. [DOI] [PubMed] [Google Scholar]
- 13.Fierer, J., and D. Guiney. 2001. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J. Clin. Investig. 107:775-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freundlich, B., and N. Avadlovic. 1983. Use of gelatin/plasma coated flasks for isolating human peripheral blood monocytes. J. Immunol. Methods 62:31-37. [DOI] [PubMed] [Google Scholar]
- 15.Galan, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 16.Galan, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallois, A., J. R. Klein, L. A. Allen, B. D. Jones, and W. M. Nauseef. 2001. Salmonella pathogenicity island 2-encoded type III secretion system mediates exclusion of NADPH oxidase assembly from the phagosomal membrane. J. Immunol. 166:5741-5748. [DOI] [PubMed] [Google Scholar]
- 18.Gulig, P. A., and T. J. Doyle. 1993. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect. Immun. 61:504-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 20.Hensel, M., J. E. Shea, B. Raupach, D. Monack, S. Falkow, C. Gleeson, T. Kubo, and D. W. Holden. 1997. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol. Microbiol. 24:155-167. [DOI] [PubMed] [Google Scholar]
- 21.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 22.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasion SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 96:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holden, D. W. 2002. Trafficking of the Salmonella vacuole in macrophages. Traffic 3:161-169. [DOI] [PubMed] [Google Scholar]
- 24.Jesenberger, V., K. J. Procyk, J. Yuan, S. Reipert, and M. Baccarini. 2000. Salmonella-induced caspase-2 activation in macrophages: a novel mechanism in pathogen-mediated apoptosis. J. Exp. Med. 192:1035-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaniga, K., S. Tucker, D. Trollinger, and J. E. Galán. 1995. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 177:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krause, M., C. Roudier, J. Fierer, J. Harwood, and D. Guiney. 1991. Molecular analysis of the virulence locus of the Salmonella dublin pSDL2. Mol. Microbiol. 5:307-316. [DOI] [PubMed] [Google Scholar]
- 27.Krause, M., F. C. Fang, and D. G. Guiney. 1992. Regulation of plasmid virulence gene expression in Salmonella dublin involves an unusual operon structure. J. Bacteriol. 174:4482-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lesnick, M. L., N. E. Reiner, J. Fierer, and D. G. Guiney. 2001. The Salmonella spvB virulence gene encodes an enzyme that ADP-ribosylates actin and destabilizes the cytoskeleton of eukaryotic cells. Mol. Microbiol. 39:1464-1470. [DOI] [PubMed] [Google Scholar]
- 29.Libby, S. J., M. Lesnick, P. Hasegawa, E. Weidenhammer, and D. G. Guiney. 2000. The Salmonella virulence plasmid spv genes are required for cytopathology in human monocyte-derived macrophages. Cell. Microbiol. 2:49-58. [DOI] [PubMed] [Google Scholar]
- 30.Lindgren, S. W., I. Stojiljkovic, and F. Heffron. 1996. Macrophage killing is an essential virulence mechanism of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:4197-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meresse, S., K. E. Unsworth, A. Habermann, G. Griffiths, F. Fang, M. J. Martinez-Lorenzo, S. R. Waterman, J. P. Gorvel, and D. W. Holden. 2001. Remodelling of the actin cytoskeleton is essential for replication of intravacuolar Salmonella. Cell. Microbiol. 3:567-577. [DOI] [PubMed] [Google Scholar]
- 32.Monack, D. M., C. S. Detweiler, and S. Falkow. 2001. Salmonella pathogenicity island 2-dependent macrophage death is mediated in part by the host cysteine protease caspase-1. Cell. Microbiol. 3:825-837. [DOI] [PubMed] [Google Scholar]
- 33.Monack, D. M., B. Raupach, A. E. Hromockyj, and S. Falkow. 1996. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. USA 93:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochman, H., F. C. Soncini, F. Solomon, and E. A. Groisman. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. USA 93:7800-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roudier, C., J. Fierer, and D. G. Guiney. 1992. Characterization of translation termination mutations in the spv operon of the Salmonella virulence plasmid pSDL2. J. Bacteriol. 174:6418-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos, R. L., R. M. Tsolis, A. J. Bäumler, R. Smith III, and L. G. Adams. 2001. Salmonella enterica serovar Typhimurium induces cell death in bovine monocyte-derived macrophages by early sipB-dependent and delayed sipB-independent mechanisms. Infect. Immun. 69:2293-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shea, J. E., C. R. Beuzon, C. Gleeson, R. Mundy, and D. W. Holden. 1999. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect. Immun. 67:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tezcan-Merdol, D., T. Nyman, U. Lindberg, F. Haag, F. Koch-Nolte, and M. Rhen. 2001. Actin is ADP-ribosylated by the Salmonella enterica virulence-associated protein SpvB. Mol. Microbiol. 39:606-619. [DOI] [PubMed] [Google Scholar]
- 39.Uchiya, K., M. A. Barbieri, K. Funato, A. H. Shah, P. D. Stahl, and E. A. Groisman. 1999. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18:3924-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Velden, A. W. M., S. W. Lindgren, M. J. Worley, and F. Heffron. 2000. Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype Typhimurium. Infect. Immun. 68:5702-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vazquez-Torres, A., Y. Xu, J. Jones-Carson, D. W. Holden, S. M. Lucia, M. C. Dinauer, P. Mastroeni, and F. C. Fang. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655-1658. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, D., L. M. Chen, L. Hernandez, S. B. Shears, and J. E. Galan. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39:248-259. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, D., M. S. Mooseker, and J. E. Galan. 1999. An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc. Natl. Acad. Sci. USA 96:10176-10181. [DOI] [PMC free article] [PubMed] [Google Scholar]