Abstract
Neisseria meningitidis colonizes the nasopharynx and, unlike commensal Neisseria species, is capable of entering the bloodstream, crossing the blood-brain barrier, and invading the meninges. The other pathogenic Neisseria species, Neisseria gonorrhoeae, generally causes an infection which is localized to the genitourinary tract. In order to investigate the genetic basis of this difference in disease profiles, we used a strategy of genomic comparison. We used DNA arrays to compare the genome of N. meningitidis with those of N. gonorrhoeae and Neisseria lactamica, a commensal of the nasopharynx. We thus identified sequences conserved among a representative set of virulent strains which are either specific to N. meningitidis or shared with N. gonorrhoeae but absent from N. lactamica. Though these bacteria express dramatically different pathogenicities, these meningococcal sequences were limited and, in contrast to what has been found in other pathogenic bacterial species, they are not organized in large chromosomal islands.
Cerebrospinal meningitis remains a devastating disease worldwide, with a morbidity of 1 to 3 per 100,000 in North America and Europe and considerably higher in poorer regions, such as Africa (4). The mechanisms by which the pathogen Neisseria meningitidis passes from the site of initial colonization, the nasopharyngeal mucosa, to invade the cerebrospinal fluid are not well understood. Two bacterial attributes are constantly present in clinical isolates and thus are identified as being essential in meningococcal pathogenesis, the capsular polysaccharide and type IV pili. The former is important for extracellular growth (39), while the latter have been shown to mediate interaction both with the nasopharyngeal epithelium and with the components of the blood-brain barrier (23, 30).
One approach to the search for new determinants of bacterial pathogenesis is comparison of the genetic contents of closely related species expressing different pathogenicities. N. meningitidis is very closely related to several other Neisseria species, such as Neisseria gonorrhoeae and Neisseria lactamica (11, 16). Despite this genetic similarity, the characteristic disease profiles of these bacteria are very different. N. meningitidis, a normal inhabitant of the human nasopharynx (29), has the ability, in a proportion of those colonized, to invade the epithelium, to disseminate within the bloodstream, and to cross the blood-brain barrier. N. gonorrhoeae, the gonococcus, colonizes and invades the urogenital epithelium to cause a localized inflammation, gonorrhea. Though it is able in some cases to invade the bloodstream, disseminated disease is unusual and meningitis is extremely rare, with 20 cases reported in the United States between 1922 and 1972 (12, 28). N. lactamica is one of the several Neisseria species which are harmless commensals of the nasopharynx and are not associated with invasive disease.
Though different pathogenic potentials may be due to subtle genetic or transcriptional differences, the determinants of pathogenicity in many medically important bacteria consist of strain-specific genes which are often grouped in large chromosomal regions, or pathogenicity islands (9, 10). These differences will therefore be identified by chromosome comparison techniques, such as subtractive hybridization and DNA array technology. Genes present only in N. meningitidis would be important for the specific aspects of meningococcal disease, i.e., bloodstream dissemination and crossing of the blood-brain barrier. Again, one would expect that regions of the meningococcal chromosome shared with N. gonorrhoeae but not with N. lactamica will be important in the stages of the diseases caused by the two bacteria which are similar, that is, primary colonization and invasion of the mucosa at the port of entry—the nasopharynx for the meningococcus and the urogenital tract for the gonococcus.
Preliminary work using subtractive hybridization (17, 25, 37) had identified several species-specific regions in the neisseriae. However, this technique is not easily applicable to extensive comparison of large sets of strains. The aim of this work was therefore to take advantage of DNA array technology to identify those species-specific regions conserved among a large set of representative virulent N. meningitidis strains.
MATERIALS AND METHODS
Bacterial strains.
The strain on which the fabrication of the DNA arrays was based was N. meningitidis Z2491 (1), a serogroup A subtype IV-1 meningococcus (sequence type ST4 [19]) isolated from an African epidemic. This and other neisserial strains are listed in Table 1.
TABLE 1.
Strains used in this study
| Strain | Serogroup | Epidemiological group | ST | Origin | Reference |
|---|---|---|---|---|---|
| N. meningitidis | |||||
| Z2491 | A | Subgroup IV-1 | ST4 | The Gambia | 1 |
| Z5463 | A | Subgroup IV-1 | ST4 | The Gambia | 1 |
| MC58 | B | ET5 complex | ST74 | United Kingdom | 38 |
| 94N369 | B | ET37 complex | ST11 | Australia | 5 |
| FAM18 | C | ET37 complex | ST11 | USA | 2 |
| ROU | W135 | ET37 complex | ST11 | France | 26 |
| 98068 | C | ST41 complex | ST41 | France | Gift of P. Nicholas |
| Z4673 | B | ST41 complex | ST41 | The Netherlands | Gift of M. Achtman |
| N. gonorrhoeae | |||||
| F62 | United States | 15 | |||
| FA1090 | United States | 3 | |||
| Ng MS11 | United States | 33 | |||
| N. lactamica | |||||
| 8064 | France | Collection of X. Nassif | |||
| 9764 | France | Collection of X. Nassif |
Computer-assisted genomic analysis.
Chromosome sequence data were obtained from the following sites on the World Wide Web: http://www.sanger.ac.uk/Projects/N_meningitidis/(N. meningitidis Z2491), http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=gnm (N. meningitidis MC58), http://www.sanger.ac.uk/Projects/N_meningitidis/seroC.shtml (N. meningitidis FAM18), and http://www.genome.ou.edu/gono.html (N. gonorrhoeae FA1090).
Production and use of DNA arrays.
Primers for PCR were designed according to the chromosomal sequence of N. meningitidis Z2491 (http://www.sanger.ac.uk/Projects/N_meningitidis/), published on the World Wide Web before annotation, to amplify contiguous stretches of DNA about 1 kb long covering the chromosome. Oligonucleotide primers were designed, avoiding previously described repetitive sequences (Correia and highly represented insertion sequences [24]). The PCR products produced from Z2491 chromosomal DNA (2,045 amplicons) were spotted robotically (MicroGrid; Biorobotics Ltd., Cambridge, United Kingdom) in duplicate onto nylon membranes and fixed by treatment with alkali. The DNA arrays were designed to search for genetic islands which distinguish the meningococcus from the gonococcus and commensal Neisseria species. The use of amplicons of a similar size (1 kb), rather than predicted genes, avoids the problem of comparison of very low hybridization intensities associated with short genes (16% of the open reading frames [ORFs] in the annotation of the Z2491 chromosome are <300 bases long, and 8% are <200 bases; the signal from the DNA arrays is roughly proportional to the length of the PCR product spotted [data not shown]). In addition, it avoids possible inaccuracies in gene prediction and is a better strategy for comparisons of genomic content between strains.
Membranes were hybridized with 33P-labeled chromosomal DNAs of various Neisseria species and washed under standard Southern blotting conditions (6). Images were revealed with a STORM PhosphorImager (Molecular Dynamics) and interpreted with the software XDotsReader (COSE, Dugny, France), which quantitated the intensity of the signal associated with each spot.
The extents of the strain- and species-specific regions were derived from the presence or absence of the amplicon sequences in each strain. In order to identify the genes included within these regions, the corresponding amplicon coordinates were compared with the gene coordinates (base numbers) given by the Z2491 chromosome annotation (http://www.sanger.ac.uk/Projects/N_meningitidis/). All results are available at Supplementary Material, together with details of the amplicons, PCR, and Southern hybridization conditions.
Performance of DNA arrays.
The efficacy of the DNA arrays was assessed by comparison of their results with those of computer-assisted comparisons between the genome of the reference strain N. meningitidis Z2491 and those of the two other sequenced Neisseria strains, N. gonorrhoeae FA1090 (http://www.genome.ou.edu/gono.html) and N. meningitidis MC58 (34), as described in Supplementary Material. Comparison of these computer-assisted analyses with the DNA array results showed that 97% of the amplicons gave correct predictions of the presence or absence of the corresponding DNA sequence in the heterologous strains and of the presence or absence of the corresponding ORFs (3% [63 amplicons] gave false predictions and were rejected from the analysis). With respect to the ability of the DNA arrays to detect potential pathogenicity islands, further analysis of these results showed that the membranes identified 90% of strain-specific genes or regions >1.5 kb long, 96% of those >2 kb long, and all of those >3 kb long and therefore would identify genetic islands which might determine different pathogenicities.
RESULTS
In silico comparisons of pathogenic Neisseria genomes.
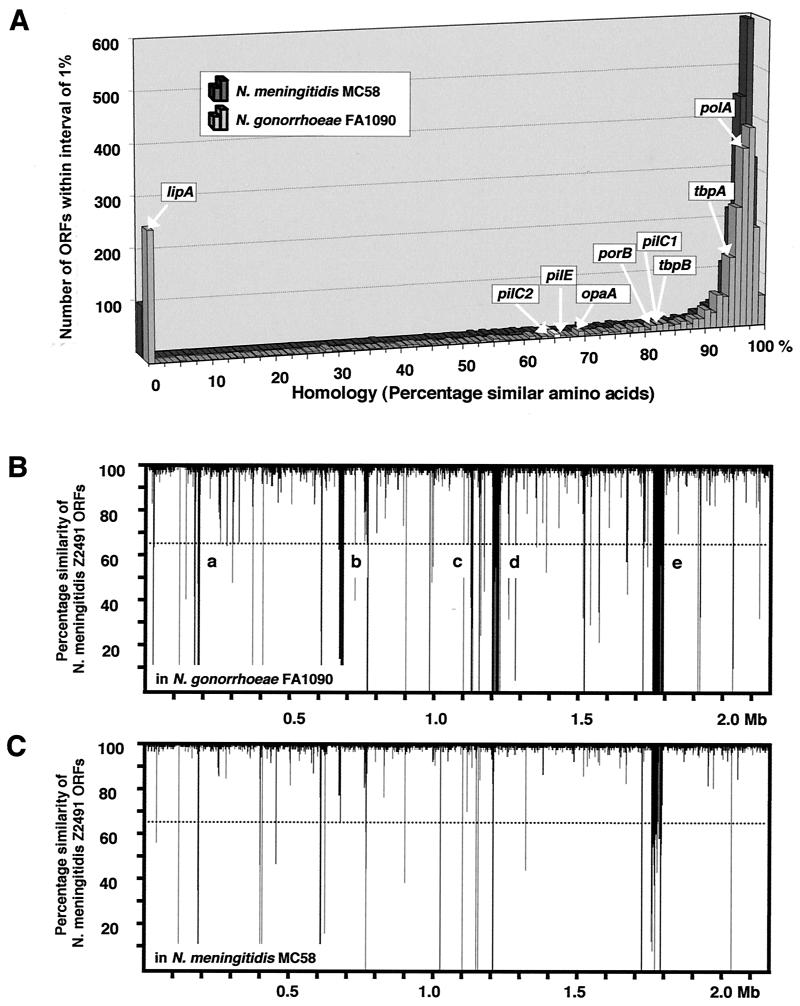
In order to identify sequences specific for N. meningitidis, we first took advantage of the completion of three genomic sequences of pathogenic Neisseria strains: N. gonorrhoeae strain FA1090 (http://www.genome.ou.edu/gono.html), N. meningitidis strain MC58 (34), and N. meningitidis strain Z2491 (1, 24). Based on the annotation of the Z2491 genome, we performed TBlastN comparisons (protein versus translated DNA) against the chromosome sequences of the other two strains. This computer-assisted comparison demonstrated that in general the genes were either very homologous among the strains or completely absent (Fig. 1A). Seventy-five percent of the N. meningitidis Z2491 genes had homologues with >80% amino acid similarity in N. gonorrhoeae FA1090, and 12% of the genes showed no significant homology. Ninety-four percent of the Z2491 ORFs had >80% homology in the meningococcus MC58, while 5% had no homologue. The chromosomal distribution of these genetic differences is shown in Fig. 1B and C. Though some large regions of the Z2491 chromosomal sequence (5 to 40 kb) were absent from the gonococcus FA1090 and could therefore represent meningococcus-specific islands, a significant proportion of this DNA is also absent from N. meningitidis MC58. Full data from these comparisons are available in the Supplementary Material.
FIG. 1.
Computer-assisted comparison of publicly available neisserial genome sequences. (A) TBlastN comparison of ORFs in Z2491 against the genomes of N. gonorrhoeae FA1090 and N. meningitidis MC58. The comparison used a minimum E value of 10−4 as the cutoff for reporting hits, which corresponds to ∼20% amino acid similarity for an ORF of 100 bases and <10% for an ORF of 300 bases or larger. The percentage of homologous amino acids in a Z2491 ORF (abscissa) is plotted against the number of ORFs (ordinate) presenting that percentage of homology. The positions of selected genes are shown. lipA is a meningococcus-specific capsular biosynthesis gene. PolA is a DNA polymerase involved in chromosome replication. The protein PilC1 is an adhesin, PilE is the pilin subunit, and OpaA and PorB are surface antigens. TbpA and TbpB are the membrane transport and surface-exposed components, respectively, of a human transferrin binding and iron acquisition system. Hence, proteins known experimentally to have similar functions but varying sequences in strains of pathogenic neisseriae are found to have between 65 and 80% amino acid similarity and allow a choice of 65% homology as a cutoff to define the presence or absence of a functional homologue in a test strain. (B) Chromosomal distribution of genetic differences. The degree of homology of the N. meningitidis Z2491 ORFs to sequences in the FA1090 genome (percentage amino acid similarity of predicted proteins) is plotted along the length of the Z2491 chromosome. The larger islands of strain-specific DNA are as follows: a, part of the capsule locus NMA0184-NMA0185 and NMA0195-NMA0202; b, two-partner secretion family proteins NMA0687-NMA0698; c and d, phage-related proteins NMA1183-NMA1200 and NMA1298-NMA1324; and e, prophage NMA1820-NMA1883. (C) Chromosomal distribution of differences between N. meningitidis Z2491 and MC58. The homology of ORFs to sequences in the MC58 genome is plotted along the length of the Z2491 chromosome. Note that many of these differences correspond to those between Z2491 and N. gonorrhoeae FA1090 and are hence strain and not species specific.
This analysis demonstrates chromosomal differences between the meningococcus and the gonococcus and also between the two meningococcal strains investigated. Therefore, in order to identify regions which are both specific to N. meningitidis and conserved among disease isolates, we undertook a larger-scale comparison, using DNA arrays and a defined panel of virulent strains of meningococcus. In addition, such a strategy permits a comparison of the meningococcal genome with that of the nonpathogenic N. lactamica, whose genomic sequence is not available.
Dramatic differences in pathogenic potential result from small genetic changes.
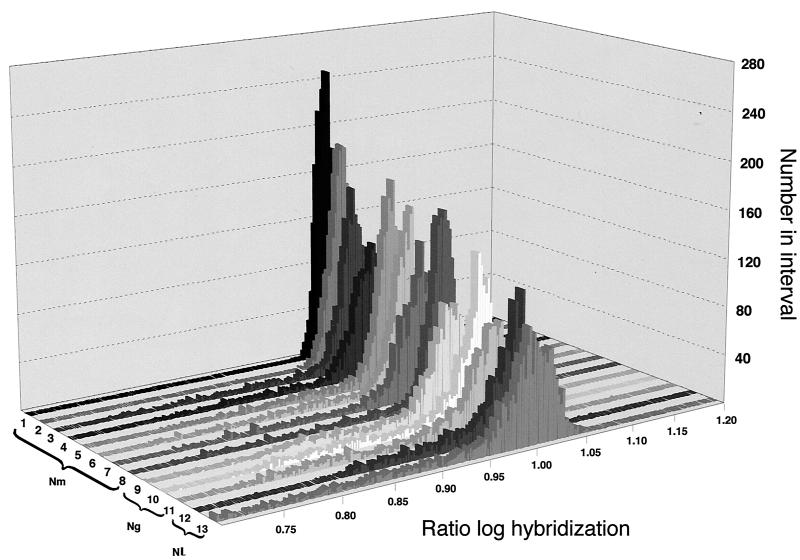
DNA arrays were produced as described in Materials and Methods. A panel of virulent meningococci was chosen to represent the major phylogenetic groups presently causing disease. Epidemiological studies have shown that the majority of meningococcal diseases worldwide are caused by a relatively restricted number of clonal groups (4), now defined as sequence types (STs) (19). Thus, most meningococcal disease in Europe is caused by meningococci of STs 11, 32, and 41 (ET37, ET5, and ST41 complexes, respectively), while ST4 (serogroup A) causes severe epidemics in Africa. We used disease isolates from each of these clonal groups, so that genes common to and specific for these strains would be likely to be involved in properties important for the lifestyle of the meningococcus and, hence, the differential pathogenesis of meningococcal disease. The DNA arrays were therefore reacted with genomic DNAs extracted from these meningococcal strains, from three strains of N. gonorrhoeae, and from two strains of N. lactamica (Table 1). The majority of the amplicons on the DNA array reacted with sequences in each of the chromosomes tested (Fig. 2; note the large peak around unity). In addition to this peak of homologous sequences, in each case the tested strains showed genomic differences. These correspond to the amplicons of lower hybridization ratios, hence sequences absent from the strains, and represent between 3 and 8% of the total for N. meningitidis, about 10% for N. gonorrhoeae, and between 15 and 20% for N. lactamica, in agreement with earlier analyses (16). It is notable, in addition, that these values are very similar to those obtained from the above-mentioned in silico comparison between N. meningitidis strain Z2491 and N. meningitidis strain MC58 or N. gonorrhoeae strain FA1090.
FIG. 2.
Distribution of reactivities with different Neisseria strains. The histogram shows the distribution of reactivities of amplicons for each of the strains. Related strains are grouped on the basis of previous epidemiological studies (Table 1). Lanes: 1, N. meningitidis Z2491 (serogroup A); 2, Z5463 (serogroup A); 3, MC58 (serogroup B); 4, 94N369 (serogroup B); 5, FAM18 (serogroup C); 6, ROU (serogroup W135); 7, 98068 (serogroup C); 8, Z4673 (serogroup B); 9, N. gonorrhoeae F62; 10, MS11; 11, FA1090; 12, N. lactamica 8064; 13, 9764.
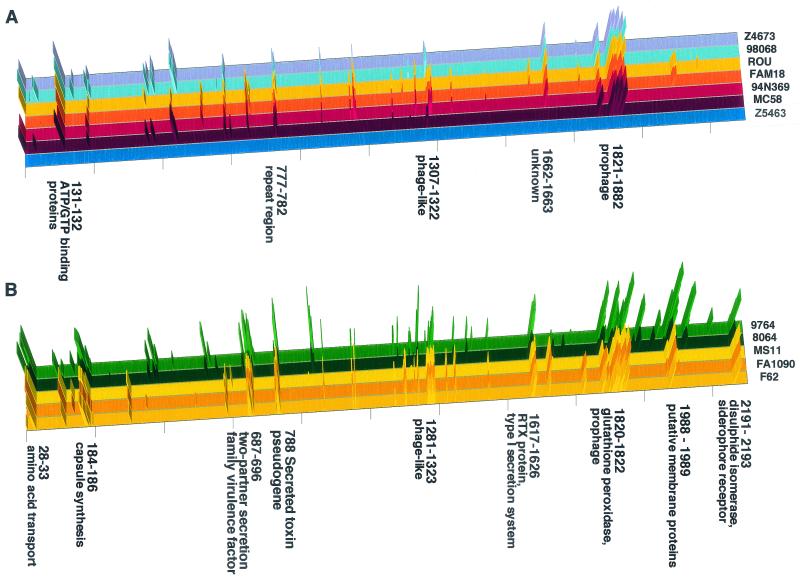
To determine the extent of the genetic differences between the species, these differences were mapped along a linear representation of the meningococcal (Z2491) chromosome (Fig. 3). Comparison of the meningococci alone showed that 89% of the chromosome of Z2491 was shared with all other strains, while strain-specific differences characterized the remaining 11% (Fig. 3A). In contrast, comparison with N. gonorrhoeae and N. lactamica (Fig. 3B) demonstrated species-specific chromosomal regions—most of the regions absent from one gonococcus were also absent from the others, and similarly for N. lactamica.
FIG. 3.
Distribution of strain-specific sequences. Distribution along the chromosome of Z2491 of amplicons absent from each of the strains of N. meningitidis (A) and from each of the strains of N. gonorrhoeae and N. lactamica (B). The strains are arranged as in Table 1 and Fig. 2. The peak height represents the difference between the value for an amplicon and the cutoff value, where the sequence is absent from the strain. Only groups of at least two contiguous nonreacting amplicons are shown. Selected genes or groups of genes, meningococcus serogroup A specific (A) and meningococcus specific (B), are named. Strain Z5463 (A) is an isolate from the same epidemic as the reference strain, Z2491, and bears witness to their high sequence homology.
In order to obtain new insights into meningococcal pathogenesis, our analysis focused on the large majority of the genome of Z2491 that was shared by all virulent strains of N. meningitidis. Most of these sequences (78% of the chromosome; 1.7 Mb) were present in all isolates of the three species N. meningitidis, N. gonorrhoeae, and N. lactamica and thus correspond to the core neisserial genome. Of the rest, 46 kb (2.2%) are strictly meningococcus specific, that is, present in all strains of invasive meningococci and absent from all the N. gonorrhoeae and N. lactamica strains. Seventy-three kilobases (3.3%) of the Z2491 genome is pathogen specific, i.e., shared with the gonococcus and absent from N. lactamica. Twelve kilobases (0.6%) is shared with N. lactamica but absent from the gonococcus. The genes corresponding to these sequences are reported in Table 2. Their analysis reveals that they correspond in general to single genes or small groups of genes scattered around the genome, since only one genetic island >10 kb in length was found (NMA0687-NMA0698) (Table 2). It should be noted that the capsular polysaccharide synthesis gene cluster (∼20 kb) is known to be meningococcus specific; the technique identified the genes encoding the conserved enzymes which attach the lipid anchor to the polysaccharide chain, while genes encoding polysaccharide biosynthesis and secretion show interstrain variation.
TABLE 2.
Genes present within the regions defined as N. meningitidis or pathogen specific or common to N. meningitidis and N. lactamicaa
| Gene(s)b | Description | No. of ORFs | No. of kb specificc | % GC | No. of subtractive clonesd |
|---|---|---|---|---|---|
| N. meningitidis specific | |||||
| Virulence associated | |||||
| NMA0184 to NMA0186 | Capsule synthesis | 3 | 5.3 | 55 | 2 |
| NMA0788 | Secreted toxin (Frp) | 1 | 2.1 | 46 | 1 |
| NAM1617 to NMA1626 | Superoxide dismutase, probable RTX family exotoxin (Frp), and others | 10 | 6.9 | 44 | |
| NMA2191 and NMA2193 | DsbA, disulfide isomerase; TonB-dependent receptor protein | 2 | 2.8 | 49, 54 | 2 |
| Possible virulence associated | |||||
| NMA0687-NMA0696 | Filamentous hemagglutinin FhaB homologue (B. pertussis) | 13 | 17.3 | 45 | 9 |
| NMA1994 to NMA1996 | TolC and Hly4; possible hemolysin secretion system (Escherichia coli) | 3 | 3.0 | 54 | 1 |
| Metabolic | |||||
| NMA0028 to NMA0033 | Amino acid metabolism and transposase | 6 | 5.6 | 55 | 1 |
| NMA1820 (to NMA1884) | GpxA; glutathione peroxidasee | 1 (53) | 0.5 (40) | 48 (53) | 9 |
| Others | |||||
| NMA0093 | Probable integral membrane protein | 1 | 0.8 | 54 | |
| NMA1636 to NMA1637 | No homology | 2 | 1.1 | 53 | |
| NMA1988 to NMA1989 | Possible integral membrane proteins | 2 | 1.7 | 34 | |
| NMA2035 | Conserved hypothetical protein | 1 | 0.8 | 35 | |
| Insertion sequences; bacteriophages | |||||
| NMA1814 | Insertion element IS1655 transposase | 1 | 1.5 | 51 | |
| Pathogen specific | |||||
| Virulence associated | |||||
| NMA0609 | PilC; pilin-associated adhesinf | 1/2 (3′) | 1.5 | 49 | 4 |
| NMA0905 | Immunoglobulin A1 protease | 1 | 5.3 | 47 | 3 |
| NMA1925 | HmbR; hemoglobin receptor | 2 | 3.2 | 52 | |
| Possible virulence associated | |||||
| NMA0478 | Possible outer membrane peptidase; Ssa1 (Pasteurella haemolytica)f | 1/2 (5′) | 1.7 | 60 | 2 |
| NMA0575 to NMA0578 | Probable ferric siderophore receptors; ORF without homology | 2 | 5.7 | 61 | |
| NMA1642 | PorA; class I outer membrane poring | 1 | 1.2 | 52 | 2 |
| NMA1676 | Variant of opacity protein | 1 | 0.6 | 48 | |
| NMA1725 | Possible virulence-associated protein; VirG (Shigella flexneri) | 1 | 1.9 | 41 | 1 |
| NMA1900 | Possible hemolysin Hly3 (Bacillus cereus) | 1 | 0.6 | 51 | 1 |
| Metabolic | |||||
| NMA1255 | Ggt; probable gamma-glutamyltranspeptidaseg | 1 | 1.8 | 47 | |
| NMA2011, NMA2013 | BioF; biotin synthesis | 2 | 1.8 | 42, 44, 51 | |
| NMA2015 to NMA2017 | SpeA and -B; polyamine metabolism | 3 | 4.5 | 4 | |
| NMA2050, NMA2052, and NMA2054 to NMA2055 | AckA2, acetate kinase, and AcnA, aconitase; PrpC, citrate synthase, and PrpB, possible carboxyphosphoenolpyruvate phosphomutase | 4 | 4.6 | 3 | |
| Others | |||||
| NMA0020 to NMA0021 | Probable integral membrane protein | 2 | 2.1 | 48 | |
| NMA0091 | Probable amino acid transporter | 1 | 1.4 | 54 | |
| NMA0109 to NMA0114 | Preprotein translocase; ribosomal proteins | 6 | 3.4 | 37, 42 | |
| NMA0305A | Probable transposase | 1 | 0.6 | 47 | |
| NMA0431 | Possible inner membrane protein | 1 | 1.3 | 49 | |
| NMA0586 | Possible lipoprotein | 1 | 0.8 | 54 | |
| NMA0668 to NMA0670 | Two-component sensor/kinase and transporter pseudogene | 1 | 3.0 | 44 | 1 |
| NMA0678 to NMA0683 | Possible lipoprotein, secretion protein pseudogene, and ORFs without homology | 5 | 3.0 | 44 | 1 |
| NMA0700 | Possible ribonuclease | 1 | 1.2 | 57 | |
| NMA1101 to NMA1102 | Possible integral membrane protein and ORF without homology | 2 | 1.0 | 43 | |
| NMA1777 | Possible integral membrane protein | 1 | 0.9 | 54 | |
| NMA1797 to NMA1799 | TspB; Neisseria-specific antigen and other ORFs without homology | 3 | 3.4 | 47 | 2 |
| NMA2069 | Probable integral membrane protein | 1 | 1.6 | 48 | |
| Insertion sequences; bacterio- phages | |||||
| NMA1167 to NMA1171 | Possible phage-related proteins | 5 | 1.6 | 45 | |
| NMA1212 | Transposase | 1 | 1.1 | 44 | |
| NMA1600 | Possible transposase | 1 | 0.4 | 37/PICK> | |
| No homology | |||||
| NMA0171A to NMA0173 | No homology | 3 | 1.1 | 37 | |
| NMA0565 to NMA0568 | No homology | 3 | 1.1 | 46 | |
| NMA1067 to NMA1068 | No homology | 2 | 1.0 | 33 | |
| NMA1218 to NMA1220 | No homology | 3 | 0.8 | 53 | |
| NMA1424 | No homology | 1 | 1 | 52 | |
| NMA1438 | No homology | 1 | 1.4 | 52 | |
| NMA1913 | No homology | 1 | 0.8 | 48 | |
| NMA1997 to NMA1999 | No homology | 2 | 1.5 | 46 | |
| Common to N. meningitidis and N. lectamica | |||||
| NMA1035 to NMA1036 | Restriction modification system | 2 | 2.1 | 33 | 1 |
| NMA1200 | Probable surface fibril protein; Hsf | 1 | 1.8 | 49 | |
| NMA1283 to NMA1286 | Phage-related protein | 4 | 3.4 | 51 | |
| NMA1365 to NMA1366 | Sulfate metabolism | 2 | 2.5 | 56 |
Comparisons were made between the genetic complement of the eight virulent meningococcal strains and three N. gonorrhoeae and two N. lactamica strains. The presence or absence of amplicons in a test strain was translated into that of the corresponding genes, based on the coordinates (base numbers) of the amplicons and genes on the meningococcal chromosome (Z2491). Genes corresponding to one or more amplicons absent in the test strain were considered to be absent; genes which overlapped two amplicons one of which was present and one absent could not be designated clearly and are not tabulated.
ORF nomenclature of Parkhill and colleagues (24).
Regions over 2 kb are in boldface type.
Regions previously brought to light by subtractive hybridization methods and the number of corresponding subtractive clones (1).
The genes pilC and ssa1 are both represented by two amplicons which divide them into a 5′ half and a 3′ half.
Both PorA and Ggt, previously described as N. meningitidis-specific attributes, are represented as pseudogenes in N. gonorrhoeae FA1090.
Among the N. meningitidis-specific sequences, only seven were >2 kb long. As mentioned above and as expected, one of these regions corresponds to the cps locus specifying the production of capsular polysaccharide, which is known to be necessary for meningococcal virulence (27). Two other meningococcus-specific regions (Table 2) encode the production and secretion of the protein Frp of the RTX toxin family (35, 36). The largest of the five regions showed homology to proteins of the newly described family of two-partner virulence factor secretion systems (14) and would elaborate an ∼200-kDa protein with homology to the filamentous hemagglutinin of Bordetella pertussis. This region, along with three others encoding a group of metabolic enzymes, a putative type I secretion system, and a disulfide oxidoreductase involved in the correct folding of secreted proteins, respectively, has been described previously (17)
As for meningococcus-specific regions, pathogen-specific sequences (i.e., common to N. meningitidis and N. gonorrhoeae but absent from N. lactamica) are scattered as small islands around the chromosome. They were present in larger numbers than the meningococcus-specific regions, but nevertheless, only three of these genes have been shown to play roles in pathogenesis: those for the immunoglobulin A protease (13), the PilC adhesin (20, 26), and the hemoglobin receptor (31). As expected, there were also a more limited number of sequences which were shared between N. meningitidis and N. lactamica. Since these two species inhabit the same anatomical site, some of these genes (e.g., that for the fibrilar protein NMA1200) may play a role in the initial colonization of the nasopharyngeal mucosa.
These data demonstrate that, despite the dramatically different pathogenic potentials of the bacteria within the genus Neisseria, the chromosomal differences which could be responsible for these differences remain small. In addition the small sizes of these differential regions are in contrast with what is observed in other closely related bacteria expressing different pathogenicities, in which large pathogen-specific chromosomal regions, or pathogenicity islands, between 20 and 200 kb in extent (10) are a characteristic feature.
DISCUSSION
We have compared the genomic contents of three species of the genus Neisseria: the meningococcus N. meningitidis, the gonococcus N. gonorrhoeae, and a commensal of the nasopharynx, N. lactamica. Our analysis demonstrates the presence of species- and strain-specific differences corresponding to sequences of about 1 to 40 kb of chromosomal DNA. The association of these sequences with a range of virulent meningococcal, or gonococcal, isolates of diverse epidemiological groups isolated at different times in different countries suggests that the products of the species-specific genes play roles in the different lifestyles of the bacteria.
Interpreting the roles of these sequences in light of the differences in pathogenesis would imply that those present only in virulent meningococci are responsible for specific aspects of meningococcal pathogenesis. It is therefore possible that a specific interaction of N. meningitidis with the blood-brain barrier is mediated by one of the meningococcus-specific genes, and in fact, some of these genes show homology to known bacterial virulence factors, e.g., NMA0688, a filamentous hemagglutinin homologue (Table 2). However, to date, no phenotype has been associated with this gene (17), underlining our incomplete understanding of meningococcal genetics and physiology. Indeed, all of the larger N. meningitidis-specific regions have previously been investigated, and only two (the capsule locus [32] and the DsbA region [17]) have been implicated in pathogenesis. Mutations in these regions produce defects associated with the level of bacteremia in an infant rat model, the most dramatic effect being seen after deletion of the capsule locus. However, to invade the meninges from the bloodstream, N. meningitidis must cross the blood-brain barrier, probably by a transcellular route through brain endothelial cells. Surprisingly, none of the meningococcus-specific sequences have been shown to be involved in the interaction of the bacteria with endothelial cells. Though the interactions of Neisseria with human cellular barriers are complex processes, and present models of the process may not reveal some of their more subtle facets, these data suggest that the specificity of meningococcal pathogenesis depends on the ability of the bacteria to survive in the bloodstream, as has been shown for another cause of meningitis, Haemophilus influenzae (22). Moreover, this is in agreement with studies of human susceptibility to meningococcal infection (7), which demonstrate a correlation between serum bactericidal activity and resistance to disease. The ability to adhere to and invade endothelial cells is a property of both N. meningitidis and N. gonorrhoeae, whereas N. lactamica interacts inefficiently with human cells, does not invade, and induces no intracellular cytoskeletal rearrangements as do the pathogenic species. This suggests that in N. meningitidis the genes important for the crossing of the blood-brain barrier are shared with N. gonorrhoeae but absent from N. lactamica. In support of this hypothesis, the only gene which has so far been associated with the crossing of the blood-brain barrier in vivo is that for PilC1 (26). This protein, which transforms the type IV pili into an adhesive structure, is found in both of the pathogenic Neisseria species but not in N. lactamica.
Usually, the pathogenesis of bacteria belonging to related species and responsible for different diseases is determined by large (20- to 200-kb), horizontally acquired pathogenicity islands (10) inserted into the core genome which may specify successive steps in infection (8). Our comparative genomic analysis did not reveal any such pathogenicity islands responsible for the dramatic difference in pathogenesis between N. meningitidis and the other closely related members of the genus, but rather sequences of relatively small extent scattered about the genome. Physical explanations (the size of transforming DNA [21] and frequent genomic rearrangements) may not be sufficient to explain this difference, since large islands (NMA1820 to NMA1884; apparently a prophage [Table 2]) do exist, and cotranscribed or corregulated genes (e.g., the capsular gene cluster) would tend to remain physically linked (18). The situation may be analyzed in terms of the lifestyles of the bacteria, and in this regard it is notable that the usual meningococcus-host interaction is one of asymptomatic carriage. Meningococcal infection is a deadly disease which moreover does not favor transmission. In this light, a pathogenicity island would provide no selective advantage to its host meningococcus, accounting for the absence of such large, complex islands. This genomic organization therefore strongly supports the idea that N. meningitidis is essentially a commensal species. Besides the anatomical site, the main difference between N. meningitidis and N. gonorrhoeae is the mode of transmission. N. gonorrhoeae is transmitted by direct contact, whereas N. meningitidis is spread from person to person by respiratory droplets, and some of the meningococcus-specific sequences presumably serve to optimize this mode of transmission. In addition, the meningococcal sequences important for interaction with the blood-brain barrier are likely to have been initially selected for to promote interaction with the nasopharyngeal cells, thus leading to the asymptomatic carriage which also involves an intracellular lifestyle (29). Meningococcal pathogenesis may therefore result from the expression of sequences necessary for bacterial transmission and pharyngeal colonization.
Acknowledgments
We thank Fred Heffron for careful reading of the manuscript and helpful suggestions. Julian Parkhill from the Sanger Centre, Hinxton, United Kingdom, provided much help with interpretation of the unannotated genome sequence. Some of the strains used in this study were the kind gifts of M. Achtman of the Max-Planck Institut für Infektionsbiologie, Berlin, Germany, or of P. Nicolas of the Meningococcal Reference Centre, Marseilles, France.
This work was supported by the Université Paris 5 René Descartes, the INSERM, and special Apex grant 99-03.
Editor: A. D. O'Brien
REFERENCES
- 1.Achtman, M., R. A. Wall, M. Bopp, B. Kusecek, G. Morelli, E. Saken, and M. Hassan-King. 1991. Variation in class 5 protein expression by serogroup A meningococci during a meningitis epidemic. J. Infect. Dis. 164:375-382. [DOI] [PubMed] [Google Scholar]
- 2.Aho, E. L., J. A. Dempsey, M. M. Hobbs, D. G. Klapper, and J. G. Cannon. 1991. Characterization of the opa (class 5) gene family of Neisseria meningitidis. Mol. Microbiol. 5:1429-1437. [DOI] [PubMed] [Google Scholar]
- 3.Black, W. J., R. S. Schwalbe, I. Nachamkin, and J. G. Cannon. 1984. Characterization of Neisseria gonorrhoeae protein II phase variation by use of monoclonal antibodies. Infect. Immun. 45:453-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caugant, D. A. 1998. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS 106:505-525. [PubMed] [Google Scholar]
- 5.Caugant, D. A., P. Bol, E. A. Hoiby, H. C. Zanen, and L. O. Froholm. 1990. Clones of serogroup B Neisseria meningitidis causing systemic disease in The Netherlands, 1958-1986. J. Infect. Dis. 162:867-874. [DOI] [PubMed] [Google Scholar]
- 6.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldschneider, I., E. C. Gotschlich, and M. S. Artenstein. 1969. Human immunity to the meningococcus. I. The role of humoral antibodies. J. Exp. Med. 129:1307-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groisman, E. A., and H. Ochman. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87:791-794. [DOI] [PubMed] [Google Scholar]
- 9.Hacker, J., G. Blum-Oehler, I. Mühldorfer, and H. Tschäpe. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on virulence. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 10.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 11.Hoke, C., and N. A. Vedros. 1982. Taxonomy of the Neisseriae: deoxyribonucleic acid base composition, interspecific transformation, and deoxyribonucleic acid hybridization. Int. J. Syst. Bacteriol. 32:57-66. [Google Scholar]
- 12.Holmes, K. K., G. W. Counts, and H. N. Beaty. 1971. Disseminated gonococcal infection. Ann. Intern. Med. 74:979-993. [DOI] [PubMed] [Google Scholar]
- 13.Hopper, S., B. Vasquez, A. Merz, S. Clary, J. S. Wilbur, and M. So. 2000. Effects of the immunoglobulin A1 protease on Neisseria gonorrhoeae trafficking across polarized T84 epithelial monolayers. Infect. Immun. 68:906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob-Dubuisson, F., C. Loche, and R. Antoine. 2001. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40:306-313. [DOI] [PubMed] [Google Scholar]
- 15.Kellogg, D. S., Jr., I. R. Cohen, L. C. Norrins, A. L. Schroeter, and G. Reising. 1968. Neisseria gonorrhoeae. II. Colonial variation and pathogenicity during 35 months in vitro. J. Bacteriol. 96:596-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kingsbury, D. T. 1967. Deoxyribonucleic acid homologies among species of the genus Neisseria. J. Bacteriol. 94:870-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klee, S. R., X. Nassif, B. Kusecek, P. Merker, J.-L. Berett, M. Achtman, and C. R. Tinsley. 2000. Molecular and biological analysis of eight genetic islands distinguishing Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect. Immun. 68:2082-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence, J. G., and J. R. Roth. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merz, A. J., C. A. Enns, and M. So. 1999. Type IV pili of pathogenic Neisseriae elicit cortical plaque formation in epithelial cells. Mol. Microbiol. 32:1316-1332. [DOI] [PubMed] [Google Scholar]
- 21.Morelli, G., B. Malorny, K. Muller, A. Seiler, J.-F. Wang, J. del Valle, and M. Achtman. 1997. Clonal descent and microevolution of Neisseria meningitidis during 30 years of epidemic spread. Mol. Microbiol. 25:1047-1064. [DOI] [PubMed] [Google Scholar]
- 22.Moxon, E. R., and P. T. Ostrow. 1977. Haemophilus influenzae meningitis in infant rats: role of bacteremia in pathogenesis of age-dependent inflammatory responses in cerebrospinal fluid. J. Infect. Dis. 135:303-307. [DOI] [PubMed] [Google Scholar]
- 23.Nassif, X., and M. So. 1995. Interaction of pathogenic Neisseriae with nonphagocytic cells. Clin. Microbiol. Rev. 8:376-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 25.Perrin, A., X. Nassif, and C. R. Tinsley. 1999. Identification of regions of the chromosome of Neisseria meningitidis and Neisseria gonorrhoeae which are specific to pathogenic neisseriae. Infect. Immun. 67:6119-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pron, B., M. K. Taha, C. Rambaud, J. C. Fournet, N. Pattey, J. P. Monnet, M. Musilek, J. L. Beretti, and X. Nassif. 1997. Interaction of Neisseria meningitidis with the components of the blood-brain barrier correlates with an increased expression of PilC. J. Infect. Dis. 176:1285-1292. [DOI] [PubMed] [Google Scholar]
- 27.Ram, S., F. G. Mackinnon, S. Gulati, D. P. McQuillen, U. Vogel, M. Frosch, C. Elkins, H. K. Guttormsen, L. M. Wetzler, M. Oppermann, M. K. Pangburn, and P. A. Rice. 1999. The contrasting mechanisms of serum resistance of Neisseria gonorrhoeae and group B Neisseria meningitidis. Mol. Immunol. 36:915-928. [DOI] [PubMed] [Google Scholar]
- 28.Sayeed, Z. A., U. Bhaduri, E. Howell, and H. L. Meyers, Jr. 1972. Gonococcal meningitis. A review. JAMA 219:1730-1731. [PubMed] [Google Scholar]
- 29.Sim, R. J., M. M. Harrison, E. R. Moxon, and C. M. Tang. 2000. Underestimation of meningococci in tonsillar tissue by nasopharyngeal swabbing. Lancet 356:1653-1654. [DOI] [PubMed] [Google Scholar]
- 30.Stephens, D. S., and Z. A. McGee. 1981. Attachment of Neisseria meningitidis to human mucosal surfaces: influence of pili and type of receptor cell. J. Infect. Dis. 143:525-532. [DOI] [PubMed] [Google Scholar]
- 31.Stojilkovic, I., V. Hwa, L. de Saint Martin, P. O'Gaora, X. Nassif, F. Heffron, and M. So. 1995. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15:531-541. [DOI] [PubMed] [Google Scholar]
- 32.Sun, Y. H., S. Bakshi, R. Chalmers, and C. M. Tang. 2000. Functional genomics of Neisseria meningitidis pathogenesis. Nat. Med. 6:1269-1273. [DOI] [PubMed] [Google Scholar]
- 33.Swanson, J., E. Sparks, B. Zeligs, M. A. Siam, and C. Parrott. 1974. Studies on gonococcus infection. V. Observations on in vitro interactions of gonococci and human neutrophils. Infect. Immun. 10:633-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, S. A., L. L. Wang, and P. F. Sparling. 1993. Cloning and nucleotide sequence of frpC, a second gene from Neisseria meningitidis encoding a protein similar to RTX cytotoxins. Mol. Microbiol. 9:85-96. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, S. A., L. L. Wang, A. West, and P. F. Sparling. 1993. Neisseria meningitidis produces iron-regulated proteins related to the RTX family of exoproteins. J. Bacteriol. 175:811-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tinsley, C. R., and X. Nassif. 1996. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae, two closely related bacteria expressing two different pathogenicities. Proc. Natl. Acad. Sci. USA 93:11109-11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Virji, M., K. Makepeace, I. R. Peak, D. J. Ferguson, M. P. Jennings, and E. R. Moxon. 1995. Opc- and pilus-dependent interactions of meningococci with human endothelial cells: molecular mechanisms and modulation by surface polysaccharides. Mol. Microbiol. 18:741-754. [DOI] [PubMed] [Google Scholar]
- 39.Vogel, U., and M. Frosch. 1999. Mechanisms of neisserial serum resistance. Mol. Microbiol. 32:1133-1139. [DOI] [PubMed] [Google Scholar]