Abstract
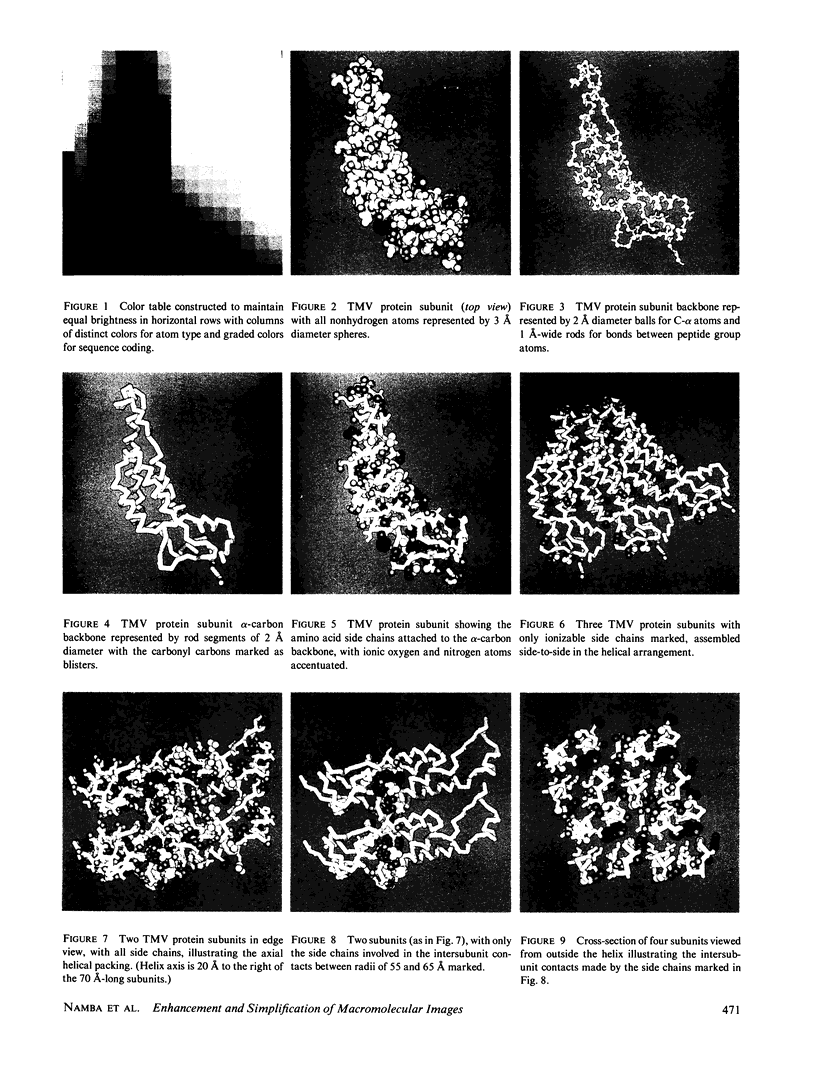
Computer graphics programs have been devised to display selected atomic features and to simplify images of complex macromolecular structures. By using boundary outlines, adjustment of size and shape of the molecular components, color coding, shading, and selective omission of obscuring detail, attention can be focused on specific interactions which determine higher levels of organization. A balanced color table has been constructed in which different hues have equal steps in brightness; this table has facilitated distinction of atom types and sequence coding together with representation of an optimum range of depth cueing and surface shading. The graphics system has been used with the atomic coordinates of the tobacco mosaic virus structure to simplify images of the protein subunit, to illustrate intermolecular interactions, and to relate subunit packing arrangements in different assemblies to the underlying atomic structure. The system has also been used to construct a schematic representation of the polyomavirus capsid, based on low resolution data. Application of artistic methods contributes to the effective presentation and interpretation of detailed scientific information about complex macromolecular structures.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bash P. A., Pattabiraman N., Huang C., Ferrin T. E., Langridge R. Van der waals surfaces in molecular modeling: implementation with real-time computer graphics. Science. 1983 Dec 23;222(4630):1325–1327. doi: 10.1126/science.222.4630.1325. [DOI] [PubMed] [Google Scholar]
- CASPAR D. L. ASSEMBLY AND STABILITY OF THE TOBACCO MOSAIC VIRUS PARTICLE. Adv Protein Chem. 1963;18:37–121. doi: 10.1016/s0065-3233(08)60268-5. [DOI] [PubMed] [Google Scholar]
- Feldmann R. J. The design of computing systems for molecular modeling. Annu Rev Biophys Bioeng. 1976;5:477–510. doi: 10.1146/annurev.bb.05.060176.002401. [DOI] [PubMed] [Google Scholar]
- Langridge R., Ferrin T. E., Kuntz I. D., Connolly M. L. Real-time color graphics in studies of molecular interactions. Science. 1981 Feb 13;211(4483):661–666. doi: 10.1126/science.7455704. [DOI] [PubMed] [Google Scholar]
- Namba K., Caspar D. L., Stubbs G. J. Computer graphics representation of levels of organization in tobacco mosaic virus structure. Science. 1985 Feb 15;227(4688):773–776. doi: 10.1126/science.3994790. [DOI] [PubMed] [Google Scholar]
- Namba K., Stubbs G. Structure of tobacco mosaic virus at 3.6 A resolution: implications for assembly. Science. 1986 Mar 21;231(4744):1401–1406. doi: 10.1126/science.3952490. [DOI] [PubMed] [Google Scholar]
- Rayment I., Baker T. S., Caspar D. L., Murakami W. T. Polyoma virus capsid structure at 22.5 A resolution. Nature. 1982 Jan 14;295(5845):110–115. doi: 10.1038/295110a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salunke D. M., Caspar D. L., Garcea R. L. Self-assembly of purified polyomavirus capsid protein VP1. Cell. 1986 Sep 12;46(6):895–904. doi: 10.1016/0092-8674(86)90071-1. [DOI] [PubMed] [Google Scholar]