Abstract
While comparing gene expression in the pathogenic organism Entamoeba histolytica and the closely related but nonpathogenic species Entamoeba dispar, we discovered that the E. histolytica abundant polyadenylated transcript 2 (ehapt2) and corresponding genomic copies are absent in E. dispar. Although polyadenylated, ehapt2 does not contain any overt open reading frame. Southern blot and sequence analyses revealed that about 500 copies of ehapt2 genomic elements were present in each cell and that the copies were distributed throughout the ameba genome. The various ehapt2 elements are regularly located in the vicinity of protein-encoding genes, downstream of pyrimidine-rich sequence stretches (40 to 125 bp; CT content, 79.2 to 85.5%), and are flanked by duplicated target sites of variable length. Target site duplications were obviously generated during integration of ehapt2 into the E. histolytica genome as one copy of the flanking repeat and the complete ehapt2 element are specifically absent in orthologous E. dispar genomic sequences. ehapt2 shares 3′ sequences with EhRLE, a recently identified non-long-terminal-repeat (non-LTR) retrotransposon-like element of E. histolytica, which contains a conceptual open reading frame for reverse transcriptase. Thus, ehapt2 has all of the properties of nonautonomous non-LTR retrotransposons. A comparison of various E. histolytica isolates suggested that transposition of ehapt2 takes place at a very low frequency as the genomic localization of ehapt2 elements was found to be well conserved. A mobile element such as ehapt2 could be a suitable mechanism to explain the infrequent and late transition of E. histolytica from a harmless gut commensal to an invasive pathogen.
The intestinal protozoan parasite Entamoeba histolytica, the causative agent of human amebiasis, is responsible for millions of cases of ulcerative colitis or extraintestinal abscesses annually (25). In addition to E. histolytica, there is a second, morphologically indistinghuishable Entamoeba species, Entamoeba dispar (10). Genetically, E. histolytica and E. dispar are the closest relatives among all Entamoeba species known so far (4), and these two species are highly similar in terms of cell biology and host range (both can infect only humans and a few Old World monkey species). However, after colonization of the human gut, only E. histolytica is able to invade tissues and cause disease, whereas E. dispar obviously is a harmless gut commensal as infection with this parasite has never been found to be associated with any form of amebic disease.
The exceptionally close phylogenetic relationship between E. histolytica and E. dispar has been documented by DNA comparisons. The rRNA genes are 98.4% identical (5), and on average, the nucleotide sequences of the protein-encoding and noncoding integenic regions differ by only 6.5 and 13.1%, respectively (27). In addition, mapping of chromosomes with a large number of cDNA probes has revealed that gene linkage groups are highly conserved in the two species. Moreover, sequencing of a small number of orthologous genomic regions has suggested that the two organisms are fully synthenic (27, 28).
In recent years, a number of E. histolytica molecules considered to be important for host tissue destruction have been identified and characterized on the molecular level; these molecules include the galactose-inhibitable surface lectin (15), pore-forming peptides (14), and cysteine proteinases (20). Although quantitative differences have been observed, qualitatively the various classes of molecules are found to be present in both ameba species (22). Thus, it appears that the key for E. histolytica pathogenicity has not become evident yet.
Recently, we reported the identification of two abundant polyadenylated transcripts (0.5 and 0.6 kb; designated E. histolytica abundant polyadenylated transcript 1 [ehapt1] and ehapt2, respectively), which together represent about 19% of the total E. histolytica poly(A)+ RNA but do not contain any overt open reading frame (ORF) (29). The corresponding genomic sequences for the two transcripts were found to be present at high copy numbers and to be distributed throughout the ameba genome. While the region coding for ehapt1 is part of a larger >3.3-kb repetitive unit (Willhoeft and Tannich, unpublished data), ehapt2 appears to represent the full-size element as two genomic clones were found to contain the entire sequence but no additional conserved up- or downstream regions (1, 7).
Here we describe a detailed analysis of ehapt2 sequences performed within the genomic context and provide evidence that ehapt2 is a nonautonomous non-long-terminal-repeat (non-LTR) retrotransposon-like element. Interestingly, sequences corresponding to ehapt2 were found to be absent in E. dispar.
MATERIALS AND METHODS
Entamoeba isolates and culture conditions.
The E. histolytica and E. dispar isolates used were obtained from the following sources: HM:1-IMSS, HK-9, and NIH:200, provided by the American Type Culture Collection; SAW142, SAW760, and SAW 1798, provided by Peter Sargeaunt, London School of Hygiene and Tropical Medicine; ERI27, ERI1007, and ERI1769, provided by Rolf Michel, Ernst Rodenwald Institute, Koblenz, Germany; and BNI0496:4, BNI1196:1, BNI1296:1, BNI0297:1, BNI0397:2, BNI0497:1, and BNI0398:2, isolated at the Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany. All ameba isolates were cultured under axenic or xenic conditions in TYI-S33 or TYSGM-9 medium, as previously described (8, 9). E. histolytica isolates HM:1-IMSS, HK9, NIH:200, and BNI0398:2 were grown axenically, whereas E. histolytica isolates ERI27, ERI1796, BNI0496:4, BNI1296:1, and BNI0497:1, as well as all E. dispar isolates (namely, SAW142, SAW760, SAW1798, ERI1007, BNI1196:1, BNI0297:1, and BNI0397:2) were grown xenically. All isolates were classified as E. histolytica or E. dispar on the basis of zymodeme and DNA analyses (19, 23).
Northern and Southern blot analyses.
Ameba DNA or RNA were isolated by standard procedures (18). For Northern blot analysis, 10 μg of total RNA was separated on a 1% agarose-formaldehyde gel and transferred to nylon membranes. For Southern blot analysis 3 μg of genomic DNA was digested with restriction endonuclease EcoRI, separated on a 1% agarose gel, and transferred to nylon membranes. Field inversion gel electrophoresis (FIGE) was carried out at 200 V for 8 h with reverse-mode intervals of 1 to 20 s and then for another 8 h with reverse-mode intervals of 0.8 to 1.5 s by using an A5 electrophoresis chamber obtained from AGS, Heidelberg, Germany, and a Switch Back pulse controller obtained from Hoefer Pharmacia Biotech Inc., San Francisco, Calif. All blots were hybridized with random primed EhEST44, an ehapt2 cDNA probe previously cloned in our laboratory (29).
Molecular cloning of larger ehapt2-containing genomic E. histolytica DNA sequences.
A genomic E. histolytica library derived from isolate HM:1-IMSS, a generous gift from John Samuelson (Harvard School of Public Health, Boston, Mass.), was screened with the ehapt2 cDNA probe. Five of the various clones identified and purified were sequenced entirely by the dideoxy chain termination method with an ABI 377 sequencer.
PCR analysis.
A number of different genomic DNA fragments of the various E. histolytica and E. dispar isolates were amplified by PCR and subjected to DNA sequencing. PCR was performed under standard conditions (18) with template DNA isolated from cultured ameba trophozoites. DNA fragments from the genomic regions were amplified with the following primer pairs: p1/np1, 5′-GCA ACT AGT GTT AGT TA and 5′-CCT CCA AGA TAT GTT TTA AC; ehapt2 internal fragment, 5′-CGT CTG AAA CAC CAC ACA CAA CCC C and 5′-CCT AGC TCA GGG GAG ACT AAT CCC; genomic fragment comprising ehapt2 plus up- and downstream sequences corresponding to clone gEh-AP, 5′-CAC CTC TCA CTC TTA CTC C and 5′-CGA AGA TTA AAA CAA AGA C; genomic fragment comprising ehapt2 plus up- and downstream sequences corresponding to clone gEh-V.1, 5′-GCT TTG AGG AGG TAA AGG and 5′-GAA GTA GAG ATG ATA AGG; genomic fragment comprising ehapt2 plus up- and downstream sequences corresponding to clone gEh-V.2, 5′-CAA ACC ATT TCA AAT GAT CC and 5′-CTT TGC TCA TTG AAC TTA GC; and genomic fragment comprising ehapt2 plus up- and downstream sequences corresponding to clone gEh-VIII, 5′-CGT CGT ATT TTT TAT CAA TTC ATT TC and 5′-GCC CTA TTT TCA AGT TTG CAG.
DNA sequence analysis.
Sequences were edited by using the DNASIS-MACv2.0 software package. Two sequences were compared by using the BLAST-2-SEQUENCES program from the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html), and multiple-sequence alignments were constructed by using ClustalW (www2.ebi.ac.uk/clustalw/). Genome sequence searches were done by using BLASTN and the NCBI GenBank database (www.ncbi.nlm.nih.gov/BLAST), as well as the Entamoeba genome databases of The Institute for Genomic Research (www.tigr.org/tdb/e2k1/eha1) and the Sanger Center (www.sanger.ac.uk/Projects/E_histolytica).
Nucleotide sequence accession numbers.
The nucleotide sequences for the E. histolytica genomic regions described in this study have been submitted to GenBank under accession numbers X70851 for gEh-AP, AY141198 for gEh-111, AY141199 for gEh-V.1, AY141200 for gEh-V.2, AY141201 for gEh-VIII, and AY141202 for gEh-X.
RESULTS
Sequences corresponding to ehapt2 are absent in E. dispar.
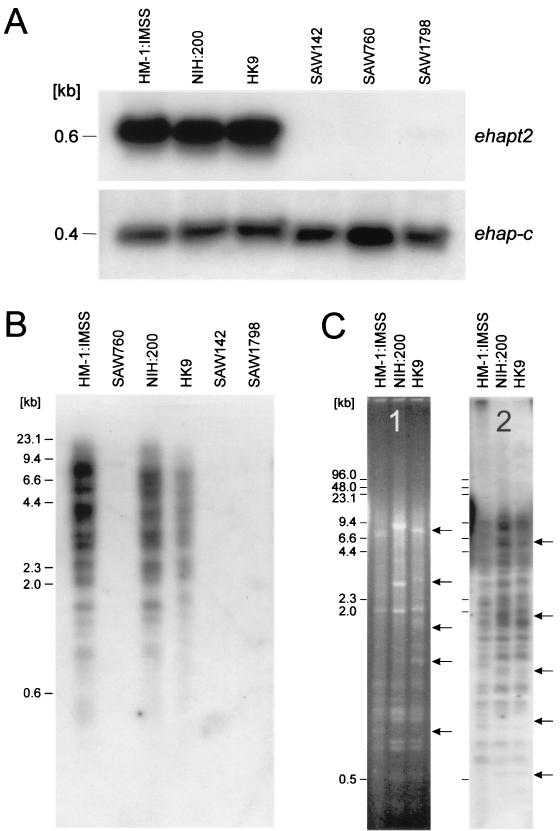
To compare the expression of various genes in E. histolytica and E. dispar, a large number of independent cDNA probes, including a probe for ehapt2, were hybridized to Northern blots prepared from total RNA of three E. histolytica and three E. dispar isolates. The results indicated that with the exception of the ehapt2 probe, all of the probes hybridized to RNA of both ameba species. The ehapt2 probe reacted strongly only with RNA of the various E. histolytica isolates, and no signal was obtained with RNA of the various E. dispar isolates (Fig. 1A). In order to examine whether corresponding ehapt2 genomic copies are present in E. dispar, Southern blot analysis of restriction enzyme-digested ameba DNA was performed. As shown in Fig. 1B, no hybridization of ehapt2 was obtained with any of the DNA extracted from the various E. dispar isolates, whereas, consistent with previous findings, ehapt2 hybridized to a large number of DNA fragments in all of the E. histolytica isolates examined. The complex band patterns obtained appeared to be rather similar, if not identical, for the various isolates, suggesting that the location of ehapt2 sequences within the genome is conserved in these isolates. This suggestion was supported by Southern blot analysis of high-resolution FIGE-separated DNA (Fig. 1C). Hybridization of the ehapt2 probe to FIGE blots resulted in identification of about 50 ehapt2-containing genomic fragments (Fig. 1C, gel 2). However, despite the multitude of hybridizing bands, only a few restriction fragment length polymorphisms were observed for the three E. histolytica isolates, which resembled the results obtained for restriction fragment length polymorphisms obtained by comparison of the highly conserved ribosomal DNA-containing E. histolytica episome (Fig. 1C, gel 1).
FIG. 1.
Northern and Southern blot analyses of E. histolytica isolates HM-1:IMSS, NIH:200, and HK9 and E. dispar isolates SAW142, SAW760, and SAW1798. (A) Autoradiograms of total RNA of the various Entamoeba isolates separated on an agarose gel, blotted onto a nylon membrane, and sequentially hybridized with radiolabeled ehapt2 and ehap-c probes. The ehap-c probe is a cDNA probe for E. histolytica amebapore C. (B) Autoradiogram of a Southern blot containing gel-separated EcoRI-digested genomic DNA of the various Entamoeba isolates, which were hybridized with ehapt2. (C) EcoRI-digested genomic DNA of the three E. histolytica isolates separated by FIGE. Gel 1 is an ethidium bromide-stained FIGE gel to visualize fragments of the multicopy ribosomal DNA-containing circular episome, and gel 2 is an autoradiogram of the same FIGE gel transferred to a nylon membrane and hybridized with ehapt2. The arrows indicate differences among the three isolates. The positions of size markers are indicated on the left.
ehapt2 elements are flanked by direct repeats and are located in the vicinity of protein-encoding regions, immediately downstream of pyrimidine-rich sequences.
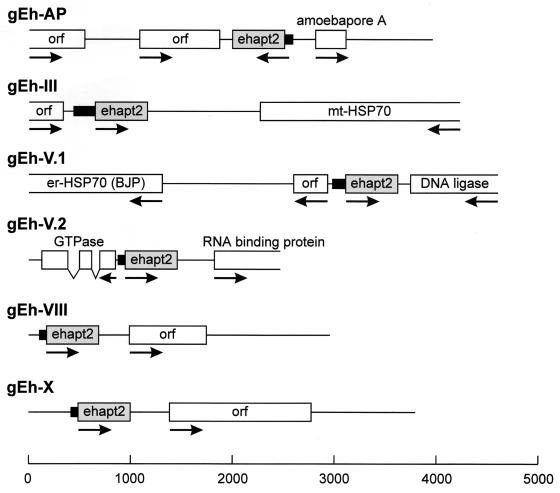
Two ehapt2-containing genomic sequences (gEh-AP [accession no. X70851] and IE-gen1 [accession no. M94257]) have been described previously (1, 7). However, this small number of sequences did not allow any assumption about possible preferences for the location of ehapt2 sequences within the E. histolytica genome. Therefore, additional ehapt2-containing clones were isolated from a genomic E. histolytica library. Five of these clones, comprising between 2.8 and 4.6 kb of amebic DNA, were sequenced entirely (Fig. 2), whereas for the remaining clones only about 500 bp of adjacent DNA upstream and downstream of the ehapt2 elements was analyzed (data not shown). As shown in Fig. 2, each of the fully sequenced genomic clones contained one copy of ehapt2, and all of the various copies were located in the vicinity of ORFs. Some of the ORFs were found to encode proteins with significant similarity to proteins in public databases. A careful analysis of the ehapt2 flanking regions did not reveal any consensus sequence that might predict the location of ehapt2 elements within the ameba genome. However, two sequence peculiarities were identified, which were present in all ehapt2 flanking regions. Upstream of the various ehapt2 elements, the genomic sequences contain a stretch of at least 40 nucleotides and up to 125 nucleotides which is extraordinary rich in pyrimidine residues (79.2 to 85.5%). In addition, the various ehapt2 elements are located between individual stretches of identical sequences (direct repeats) comprising at least 8 nucleotides and up to 26 nucleotides. This was confirmed by mining the NCBI GenBank database, as well as The Institute for Genomic Research and Sanger Center E. histolytica genome databases. BLASTN searches of these databases identified about 500 sequences with significant similarity to ehapt2, all of which were from E. histolytica. There was no apparent homology to sequences from any other organism. The majority of the 500 sequences identified contained only partial ehapt2 sequences. Detailed analysis of the 30 sequences with the highest identity scores revealed that they all represent full-length ehapt2 elements, and 26 (87%) of them were found to be flanked by a direct repeat (data not shown). In contrast, sequences containing only portions of ehapt2 usually did not have flanking repeats. A large number of these sequences were homologous to EhRLE, a recently identified retrotransposon-like element of E. histolytica (21).
FIG. 2.
Schematic diagrams of six E. histolytica ehapt2-containing genomic regions. The various ORFs or regions coding for proteins with significant similarity to proteins in public databases are represented by boxes, and their orientations are indicated by arrows. Connected gaps represent intron positions. The various ehapt2 elements are indicated by grey boxes, and the adjacent 5′ pyrimidine-rich sequence stretches are indicated by solid bars. A size marker (in base pairs) is shown at the bottom.
The direct repeats flanking the ehapt2 elements are a consequence of target site duplications.
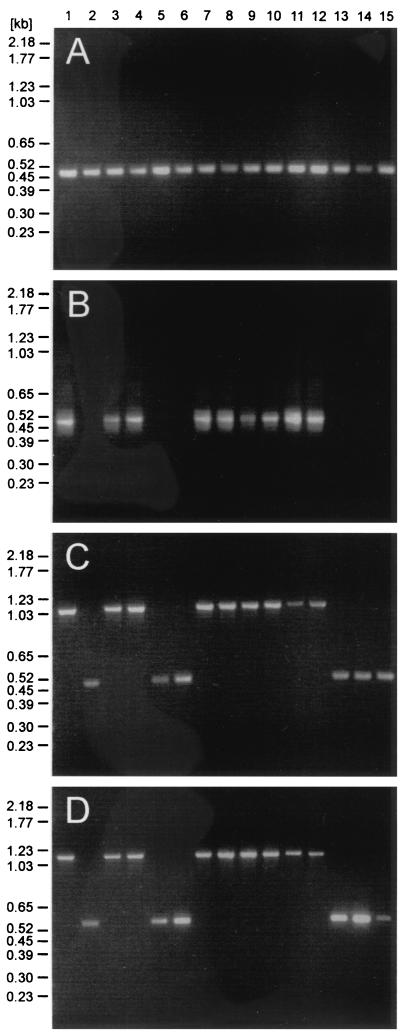
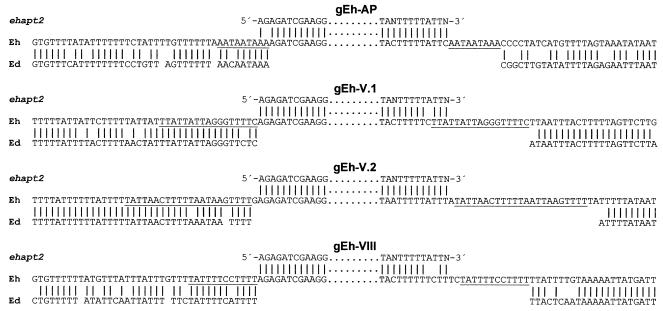
To determine the breakpoints of ehapt2 deletions in E. dispar, we used previous genetic comparisons of E. histolytica and E. dispar, which revealed that these two ameba species are syntenic and that the level of identity of orthologous sequences usually exceeds 80% (27). Accordingly, oligonucleotide primers derived from flanking sequences of four ehapt2 elements were used for PCR to amplify the corresponding genomic regions from various E. histolytica and E. dispar isolates. In addition, primers specific for ehapt2 and for p1/np1 were used as controls. The latter primers are specific for E. histolytica and E. dispar and amplify a 482-bp fragment, but they do not discriminate between the two species (23). As expected, PCR with primers specific for p1/np1 resulted in the corresponding amplification product with template DNA from all ameba isolates examined, whereas ehapt2-specific primers exclusively amplified DNA of the various E. histolytica isolates (Fig. 3A and B). Consistent with previous findings, which revealed minor length polymorphisms in different ehapt2 transcripts (29), the ehapt2-specific amplification products showed broader, less sharp bands after separation in ethidium bromide-stained agarose gels (Fig. 3B). PCR analysis with pairs of primers derived from the ehapt2 flanking regions revealed specific amplification products from all isolates of both ameba species. However, compared to the E. histolytica fragments, the amplified fragments from E. dispar were consistently smaller by about 550 bp, which corresponds to the length of ehapt2 (Fig. 3C and D). Sequence analysis and alignment of the corresponding amplified fragments revealed virtually no differences among the various isolates within each group of amebae. However, when the two groups were compared, the sequences flanking ehapt2 differed by about 17%. In addition, the complete ehapt2 elements, as well as one copy of the direct flanking repeats, were found to be absent in E. dispar (Fig. 4), which strongly suggests that insertion of ehapt2 sequences into the E. histolytica genome took place by a mechanism which resulted in duplication of a short stretch of sequence at the various target sites (target site duplication [TSD]).
FIG. 3.
Comparison of PCR-amplified genomic DNA of E. histolytica and E. dispar: ethidium bromide-stained agarose gels containing PCR-amplified fragments of various E. histolytica and E. dispar isolates. Amplified DNA were applied to the gels as follows: lane 1, HM-1:IMSS; lane 2, SAW760; lane 3, NIH:200; lane 4, HK9; lane 5, SAW142; lane 6, ERI1007; lane 7, BNI0398:2; lane 8, BNI0496:4; lane 9, BNI1296:1; lane 10, ERI27; lane 11, BNI0497:1; lane 12, ERI1769; lane 13, BNI0397:2; lane 14, BNI1196:1; and lane 15, BNI0297:1. (A) Amplified DNA obtained with primers for p1/np1. (B) Amplified DNA obtained with primers for an ehapt2 internal fragment. (C and D) Amplified DNA obtained with primers for genomic fragments comprising ehapt2 plus up- and downstream sequences corresponding to genomic clones gEh-AP (C) and gEh-V.1 (D).
FIG. 4.
Comparison of ehapt2-containing genomic regions of E. histolytica and orthologous sequences from E. dispar. Four E. histolytica sequences (Eh) corresponding to the genomic clones indicated were aligned with the ehapt2 consensus sequence (ehapt2), as well as with the corresponding orthologous sequences from E. dispar (Ed). Only the extreme 5′ and 3′ ends of ehapt2 sequences are shown, and the direct repeats flanking the ehapt2 elements in E. histolytica are underlined. Vertical lines indicate nucleotide identity. For optimal alignment gaps were introduced into the E. dispar sequences. Note that in all four examples the complete ehapt2 element and one copy of the flanking repeats are absent in E. dispar.
DISCUSSION
Better knowledge of the genetic differences between E. histolytica and E. dispar is considered to be important for understanding why only E. histolytica is able to cause disease during infection of the human host. In this study we showed that genomic copies corresponding to E. histolytica ehapt2 are absent in E. dispar. Southern blot and PCR analyses indicated that the presence of ehapt2 elements is a species-specific property as such elements are present in all nine E. histolytica isolates examined and absent in all seven E. dispar isolates examined. As these isolates were obtained from various geographic areas, ehapt2 appears to be a reliable marker for differentiating between the two ameba species. Previous studies of the E. histolytica IE element (which is identical to ehapt2) in which dot blot hybridization was used suggested that the copy number is about 500 copies per cell (1). This value is in agreement with our results from the database BLAST search analysis, although the number of full-length ehapt2 element copies appears to be smaller. Southern blot and sequence analyses of larger ehapt2-containing genomic sequences indicated that the various elements are distributed throughout the genome as a single copy at each site rather than in tandem arrays. Thus, it can be calculated that compared to E. dispar, E. histolytica contains several thousand base pairs of additional DNA, distributed in up to 550-bp portions throughout the genome. ehapt2 elements were regularly found to be located in the vicinity of protein-encoding regions. Gene expression in E. histolytica has been shown to occur in a monocistronic fashion (7), and both 5′ upstream and 3′ downstream regions contribute to transcription regulation (16). It is likely that the location of ehapt2 in close proximity to various gene translation initiation or termination codons influences expression of at least some of the genes. In addition, it remains to be determined whether there are examples in which ehapt2 has disrupted an ORF. In recent years, research on ameba pathogenicity has focused on identification of E. histolytica molecules that are absent or show reduced expression in E. dispar. From the results presented here it is tempting to speculate that the pathogenicity of amebae might be due to loss of function rather than gain of function.
Questions remain about the origin and nature of ehapt2. Database searches did not reveal significant similarities of ehapt2 to DNA sequences of any species other than E. histolytica. Therefore, at present, ehapt2 appears to be specific for E. histolytica, and its origin remains unclear. However, the high copy number and the dispersed localization together with the absence in the closely related species E. dispar support the hypothesis that ehapt2 is a transposable element which was integrated into and quickly distributed throughout the E. histolytica genome after separation of the two ameba species from a common ancestor. Recent acquisition and distribution of ehapt2 are supported by the finding that the various ehapt2 elements exhibit >92% sequence identity. A transposable element that is present in one organism but absent in a closely related species or even in a subgroup of the same species is not without precedence, as shown for various transposable elements like mariner, jockey, or hobo in Drosophila (2). The strongest evidence that ehapt2 is indeed a transposable element was obtained by DNA sequencing of several ehapt2-containing genomic regions and comparison of these regions with orthologous sequences from E. dispar. These analyses indicated that the various elements are flanked by short stretches of identical sequences (direct repeats) that are 8 to 26 bp long and that these repeats were obviously generated by TSD during integration of ehapt2 into the E. histolytica genome. TSDs of variable length are a well-known feature of retrotransposons (11) like the Drosophila I-factor (3) and the human L1 retrotransposon (26). In contrast, DNA transposons usually create smaller and, in particular, inverted duplications during transposition (11, 13). Most likely, ehapt2 belongs to the class of nonautonomous non-LTR retrotansposons, as it is small, lacks any ORF, and does not contain LTRs. Moreover, part of the sequence of the 3′ portion of ehapt2 (positions 476 to 540) is present in the recently identified multicopy retrotransposon-like element EhRLE of E. histolytica. EhRLE is a 4,086-bp DNA element which contains a region coding for reverse transcriptase (RT) of the type found in non-LTR retrotransposons (21). The 3′ sequences of retrotransposons are known to be required for binding of RT (6). Common 3′ sequences of full-length autonomous retrotransposons (long interspersed repetitive elements [LINEs]) and shorter nonautonomous retrotransposons (small interspersed repetitive elements [SINEs]) have been identified in various organisms (17), and it has been suggested that the enzymes encoded by LINEs are required for transposition of SINEs (26). Analogous to this, EhRLE and ehapt2 might represent the LINE-SINE relationship, and the RT encoded by EhRLE might be required for retrotransposition of ehapt2. Interestingly, the RT-encoding region of EhRLE is degenerate and could be reconstructed only by alignment of various copies of the element. So far, no active EhRLE copy has been identified (21). This, however, indicates that ehapt2 is no longer able to retrotranspose, which is consistent with our finding that the locations of the various ehapt2 copies within the ameba genome are well conserved in different E. histolytica isolates.
Another specific property of ehapt2 is its location downstream of longer stretches of pyrimidine-rich DNA (CT content, >78%). Whether this is important for integration of the element remains to be determined. However, in the protozoan parasite Trypanosoma cruzi the short interspersed repetitive element (SIRE) is preferentially found in the vicinity of thymidine-rich sequences. Interestingly, SIRE resembles ehapt2 as it is present at a level of several hundred copies per genome, comprises only 427 bp, contains duplicated target site sequences, and is specific for T. cruzi and absent in other Trypanosoma species (24). In addition, SIRE shares a 3′ sequence with VIPER, an RT-encoding autonomous retrotransposom of T. cruzi.
Taken together, our results strongly suggest that ehapt2 is a nonautonomous non-LTR retrotransposon of pathogenic E. histolytica, which is absent in the closely related but nonpathogenic species E. dispar. ehapt2 appears to retrotranspose at a very low frequency, if it retrotransposes at all. In this respect it should be emphasized that the majority of E. histolytica infections remain asymptomatic. Only a small proportion of individuals develop amebic disease, and they usually do so months or even years after infection has been established (12, 30). A mobile element with a low transposition frequency would be an ideal candidate to be responsible for the infrequent and late transition of the parasite from a harmless commensal to an invasive pathogen.
Acknowledgments
We thank Britta Weseloh for skillful technical assistance, John Samuelson for providing the genomic E. histolytica library, and Peter Sargeaunt and Rolf Michel for providing various E. histolytica and E. dispar isolates.
This study was supported by Deutsche Forschungsgemeinschaft grant TA 110/4-2.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Bruchhaus, I., M. Leippe, C. Lioutas, and E. Tannich. 1993. Unusual gene organization in the protozoan parasite Entamoeba histolytica. DNA Cell Biol. 12:925-933. [DOI] [PubMed] [Google Scholar]
- 2.Capy, P., D. Anxolabehere, and T. Langin. 1994. The strange phylogenies of transposable elements: are horizontal transfers the only explanations? Trends Genet. 10:7-12. [DOI] [PubMed] [Google Scholar]
- 3.Chaboissier, M.-C., D. Finnegan, and A. Bucheton. 2000. Retrotransposition of the I factor, a non-long terminal repeat retrotransposon of Drosophila, generates tandem repeats at the 3′ end. Nucleic Acids Res. 28:2467-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, C. G., and L. S. Diamond. 1997. Intraspecific variation and phylogenetic relationships in the genus Entamoeba as revealed by riboprinting. J. Eukaryot. Microbiol. 44:142-154. [DOI] [PubMed] [Google Scholar]
- 5.Clark, C. G., and L. S. Diamond. 1991. Ribosomal RNA genes of ‘pathogenic’ and ‘nonpathogenic’ Entamoeba histolytica are distinct. Mol. Biochem. Parasitol. 49:297-302. [DOI] [PubMed] [Google Scholar]
- 6.Craig, N. L. 1997. Target site selection in transposition. Annu. Rev. Biochem. 66:437-474. [DOI] [PubMed] [Google Scholar]
- 7.Cruz-Reyes, J., T. ur-Rehman, W. M. Spice, and J. P. Ackers. 1995. A novel transcribed repeat element from Entamoeba histolytica. Gene 166:183-184. [DOI] [PubMed] [Google Scholar]
- 8.Diamond, L. S., D. R. Hariow, and C. C. Cunnick. 1978. A new medium for axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431-432. [DOI] [PubMed] [Google Scholar]
- 9.Diamond, L. S. 1982. A new liquid medium for xenic cultivation of Entamoeba histolytica and other lumen-dwelling protozoa. J. Protozool. 68:958-959. [PubMed] [Google Scholar]
- 10.Diamond, L. S., and C. G. Clark. 1993. A redescription of Entamoeba histolytica Schaudinn, 1903 (Emended Walker, 1911) separating it from Entamoeba dispar Brumpt, 1925. J. Eukaryot. Microbiol. 40:340-344. [DOI] [PubMed] [Google Scholar]
- 11.Finnegan, J. D. 1989. Eukaryotic transposable elements and genome evolution. Trends Genet. 6:103-107. [DOI] [PubMed] [Google Scholar]
- 12.Knobloch, J., and E. Mannweiler. 1983. Development and persistence of antibodies to Entamoeba histolytica in patients with amebic liver abscess. Analysis of 216 cases. Am. J. Trop. Med. Hyg. 32:727-732. [DOI] [PubMed] [Google Scholar]
- 13.Labrador, M., and V. G. Corces. 1997. Transposable element-host interactions: regulation of insertion and excision. Annu. Rev. Genet. 31:381-404. [DOI] [PubMed] [Google Scholar]
- 14.Leippe, M. 1997. Amoebapores. Parasitol. Today 13:178-183. [DOI] [PubMed] [Google Scholar]
- 15.McCoy, J. J., B. Mann, and W. A. Petri. 1994. Adherence and cytotoxicity of Entamoeba histolytica or how lectins let parasites stick around. Infect. Immun. 62:3045-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nickel, R., and E. Tannich. 1994. Transfection and transient expression of chloramphenicol acetyltransferase gene in the protozoan parasite Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 91:7095-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada, N., M. Hamada, I. Ogiwara, and K. Oshima. 1997. SINEs and LINEs share common 3′ sequences: a review. Gene 205:229-243. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Sargeaunt, P. G., J. E. Williams, and J. D. Greene. 1978. The differentiation of invasive and non-invasive Entamoeba histolytica by isoenzyme electrophoresis. Trans. R. Soc. Trop. Med. Hyg. 72:519-521. [DOI] [PubMed] [Google Scholar]
- 20.Scholze, H., and Tannich, E. 1994. Cysteine endopeptidases of Entamoeba histolytica. Methods Enzymol. 244:512-523. [DOI] [PubMed] [Google Scholar]
- 21.Sharma, R., A. Bagchi, A. Bhattacharya, and S. Bhattacharya. 2001. Characterization of a retrotransposon-like element from Entamoeba histolytica. Mol. Biochem. Parasitol. 116:45-53. [DOI] [PubMed] [Google Scholar]
- 22.Tannich, E. 1998. Entamoeba histolytica and Entamoeba dispar: comparison of molecules considered important for host tissue destruction. Trans. R. Soc. Trop. Med. Hyg. 92:593-596. [DOI] [PubMed] [Google Scholar]
- 23.Tannich, E., and G. D. Burchard. 1991. Differentiation of pathogenic from nonpathogenic Entamoeba histolytica by restriction fragment analysis of a single gene amplified in vitro. J. Clin. Microbiol. 29:250-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vazquez, M., C. Ben-Dov, H. Lorenzi, T. Moore, A. Schijman, and M. J. Levin. 2000. The short interspersed repetitive element of Trypanosoma cruzi, SIRE, is part of VIPER, an unusual retroelement related to long terminal repeat retrotransposons. Proc. Natl. Acad. Sci. USA 97:2128-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh, J. 1986. Problems in recognition and diagnosis of amebiasis: estimation of the global magnitude of morbidity and mortality. Rev. Infect. Dis. 8:228-238. [DOI] [PubMed] [Google Scholar]
- 26.Wei, W., N. Gilbert, S. Oi, J. F. Lawler, E. M. Ostertag, H. H. Kazazian, J. D. Boeke, and J. V. Moran. 2001. Human L1 retrotransposition: cis preferences versus trans complementation. Mol. Cell. Biol. 21:1429-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willhoeft, U., L. Hamann, and E. Tannich. 1999. A DNA sequence corresponding to the gene encoding cysteine proteinase 5 in Entamoeba histolytica is present and positionally conserved but highly degenerated in Entamoeba dispar. Infect. Immun. 67:5925-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willhoeft, U., E. Campos-Góngora, S. Touzni, I. Bruchhaus, and E. Tannich. 2001. Introns in Entamoeba histolytica and Entamoeba dispar. Protist 152:149-156. [DOI] [PubMed] [Google Scholar]
- 29.Willhoeft, U., H. Buß, and E. Tannich. 1999. Analysis of cDNA expressed sequence tags from Entamoeba histolytica: identification of two highly abundant transcripts with no overt open reading frames. Protist 150:61-70. [DOI] [PubMed] [Google Scholar]
- 30.Wynants, H., J. Van den Ende, J. Randaria, A. Van Gompel, E. Van den Enden, C. Brands, P. Coremans, P. Michielsen, L. Verbist, and R. Colebunders. 1995. Diagnosis of amoebic infection of the liver: report of 36 cases. Ann. Soc. Belg. Med. Trop. 75:297-303. [PubMed] [Google Scholar]