Abstract
Staphylococcus epidermidis has been reported to bind to a number of host cell extracellular matrix proteins, including fibronectin. Here we report the identification of a fibronectin-binding protein from S. epidermidis. A phage display library of S. epidermidis genomic DNA was constructed and panned against immobilized fibronectin. A number of phagemid clones containing overlapping inserts were identified, and one of these clones, pSE109FN, contained a 1.4-kb insert. Phage pSE109FN was found to bind to fibronectin but not to collagen, fibrinogen, laminin, or vitronectin. However, pSE109FN also bound to heparin, hyaluronate, and plasminogen, although to a lesser extent than it bound to fibronectin. Analysis of The Institute for Genomic Research S. epidermidis genome sequence database revealed a 1.85-kb region within a putative 30.5-kb open reading frame, to which the overlapping DNA inserts contained within the fibronectin-binding phagemids mapped. We have designated the gene encoding the fibronectin-binding domain embp. A recombinant protein, Embp32, which encompassed the fibronectin-binding domain of Embp, blocked the binding of S. epidermidis, but not the binding of Staphylococcus aureus, to fibronectin. In contrast, a recombinant protein, FnBPB[D1-D4], spanning the fibronectin-binding domain of the S. aureus fibronectin-binding protein FnBPB, blocked binding of S. aureus to fibronectin but had a negligible effect on the binding of S. epidermidis.
Staphylococcus epidermidis, a normal skin commensal, is an important opportunistic pathogen responsible for infections of implanted medical devices and infections at sites of surgery (24). Medical devices infected by S. epidermidis include cardiac pacemakers (1), cerebrospinal fluid shunts (23), continuous ambulatory peritoneal dialysis catheters (14), intravascular catheters (2), orthopedic devices (11), and prosthetic heart valves (13). The accepted paradigm for S. epidermidis adhesion to biomaterial surfaces is a two-step process involving a primary attachment step mediated by hydrophobic interactions, followed by biofilm formation and intercellular interactions (6). However, it is acknowledged that the primary attachment step is mediated not only by hydrophobic interactions but also by bacterial proteins.
Since foreign bodies become rapidly coated with plasma proteins when they are implanted in the body, S. epidermidis may bind to implants through interactions with immobilized extracellular matrix proteins. In vitro studies have shown that S. epidermidis can bind to biomaterials coated with fibronectin and fibrinogen (3, 5,9, 17). The ability of Staphylococcus aureus to bind to extracellular matrix proteins through a family of proteins termed the microbial surface components recognizing adhesive matrix molecules is well documented (for a review see reference 4). However, comparatively little is known about how S. epidermidis interacts with matrix proteins. It has been established that S. epidermidis binds to fibrinogen through SdrG, a microbial surface component recognizing adhesive matrix molecules, which is also known as Fbe (7, 16, 18, 19). The autolysin AtlE of S. epidermidis, which has been implicated as the mediator of primary attachment of this organism to polystyrene surfaces, has also been shown to bind vitronectin (8). Although it has been demonstrated that S. epidermidis binds to fibronectin in vitro (3, 5, 9), no fibronectin-binding protein has been identified yet. In fact, it has been suggested that teichoic acid may mediate binding of S. epidermidis to immobilized fibronectin (10). The aim of the work described in this paper was to use shotgun phage display cloning to identify S. epidermidis genes that code for fibronectin-binding proteins.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The type strain of S. epidermidis, strain NCTC11047, and S. aureus strain 8325-4 were routinely grown in brain heart infusion broth (Oxoid Ltd., Basingstoke, United Kingdom) aerobically at 37°C. The phagemid pG8SAET (a kind gift from Lars Frykberg, Department of Microbiology, Swedish University of Agricultural Sciences, Uppsala, Sweden) and the Escherichia coli host TG1 [supE hsdΔ5 thi Δ(lac-proAB) F′(traD36 proAB+ lacIq lacZΔM15)] were used in the construction of the phage display library. E. coli was grown in nutrient broth no. 2 (Oxoid Ltd.). The medium was supplemented when appropriate with 100 μg of ampicillin per ml to maintain the phagemid. All cultures were grown at 37°C under aerobic conditions.
Construction of the S. epidermidis phage display library.
To construct the phage display library, S. epidermidis chromosomal DNA was sheared by sonication to obtain fragments between 0.5 and 2 kb long. Without further size fractionation, the chromosomal fragments were blunt ended by using the Klenow fragment and T4 DNA polymerase and then ligated to SnaBI-digested and dephosphorylated phagemid vector pG8SAET by using a Ready-to-Go T4 DNA ligase kit (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). The ligated DNA was purified by using a QIAquick PCR purification kit (Qiagen Ltd., Crawley, United Kingdom) and was introduced by electroporation into E. coli TG1. The electroporated cells were allowed to recover for 2 h at 37°C in 10 ml of nutrient broth no. 2 prior to infection with helper phage R408 (Promega, Southampton, United Kingdom) at a multiplicity of infection of 20. The infected cells were grown overnight in 190 ml of nutrient broth no. 2 containing antibiotic. The phage were recovered from the culture supernatant by precipitation with a solution containing 20% (wt/vol) polyethylene glycol 8000 (Sigma-Aldrich Co. Ltd., Poole, United Kingdom) and 1 M NaCl, resuspended in phosphate-buffered saline (PBS) containing calcium and magnesium (PBSCM) (Sigma-Aldrich Co. Ltd.), and sterilized by passage through a 0.45-μm-pore-size filter.
Panning of the S. epidermidis phage library against fibronectin.
A 5-ml Nunc Maxisorb immunotube (Invitrogen Ltd., Paisley, United Kingdom) was coated overnight at 4°C with 1 ml of a solution containing 1 mg of fibronectin from human plasma (Sigma-Aldrich Co. Ltd.). The tube was blocked with 4 ml of 2% bovine serum albumin (BSA) in PBSCM for 2 h at room temperature. After the tube had been extensively washed with PBSCM containing 0.05% Tween 20 and then with PBSCM, 1 ml of the S. epidermidis phage library was added to the tube and incubated for 2 h at room temperature. All unbound phage were removed by 10 washes with 4 ml of PBSCM containing 0.05% Tween 20, followed by 10 washes with 4 ml of PBSCM. The bound phage were eluted in 1 ml of 0.1 M glycine buffer (pH 2.1) for 10 min at room temperature, which was then neutralized with 0.5 ml of 1 M Tris-HCl buffer (pH 8.0).
Preparation of phage stocks.
Stocks of phages pSE104FN and pSE109FN were prepared by infecting 2 ml of mid-exponential-phase E. coli TG1 containing the recombinant phagemids with the helper phage R408 at a multiplicity of infection of 20. After 30 min of incubation at 37°C, the infected cells were added to 190 ml of nutrient broth no. 2 containing ampicillin and grown overnight at 37°C. The phage particles were recovered from the culture supernatant by precipitation with 20% polyethylene glycol 8000-1 M NaCl, resuspended in PBSCM, and filter sterilized as described above.
Binding activity of recombinant phage.
Wells of Nunc Maxisorp microtiter plates (Invitrogen Ltd.) were coated overnight at 4°C with 0.1-ml aliquots of 0.1-mg/ml solutions of the following test ligands dissolved in PBS: fibronectin, the 30-kDa N-terminal and 45-kDa α-chymotryptic fragments of fibronectin, collagen (type I, acid soluble, from human placenta), fibrinogen (fraction I from human plasma), heparin, hyaluronate, mucin (from bovine submaxillary glands), plasminogen, and vitronectin, all of which were purchased from Sigma-Aldrich Co. Ltd.; mouse laminin (Collaborative Biomedical Products, Bedford, Mass.); and the 120-kDa α-chymotryptic fragment of fibronectin (Invitrogen Ltd.). The wells were each blocked with 0.1 ml of either 1% BSA or 1% Tween 20 in PBS for 1 h. After the wells were rinsed three times with PBS containing 0.05% Tween 20 and then with PBS, 0.1 ml of a 1 × 109-CFU/ml phage solution was added to each well and incubated for 1.5 h. The unbound phage was removed by five washes with 0.2 ml of PBS containing 0.05% Tween 20, followed by five washes with 0.2 ml of PBS. The bound phage were eluted with 0.1 ml of 0.1 M glycine buffer (pH 2.1) for 10 min and neutralized with 50 μl of 1 M Tris-HCl buffer (pH 8.0).
For inhibition studies with Embp32, microtiter plates were coated with fibronectin. After blocking and washing, 0.1 ml of one of a range of concentrations of the recombinant protein was added to each well, and the plates were incubated for 1 h before the phage solution was added as described above.
Titration of phage stocks.
The number of phage were determined as CFU infecting E. coli TG1 grown to the mid-exponential phase with the phage solution. After 60 min of incubation at 37°C, the cells were plated onto nutrient broth no. 2 containing ampicillin.
Western blotting.
Extracellular matrix components and plasma proteins were Western blotted to determine if there was any fibronectin contamination. Ligands (10 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 8% polyacrylamide gels and electroblotted onto Immobilon-P nitrocellulose membranes (Sigma-Aldrich Co. Ltd.). The membranes were blocked by incubating them with 5% milk in PBS containing 0.1% Triton X-100 for 1 h, after which they were incubated for 2 h with a 1:1,000 dilution of anti-human fibronectin mouse monoclonal antibody (Sigma-Aldrich Co. Ltd.). The membranes were washed five times with PBS containing 0.1% Triton X-100 and then incubated with goat anti-mouse immunoglobulin G (whole molecule; Sigma-Aldrich Co. Ltd.) at a 1:2,000 dilution. The membranes were washed as described above and developed with Sigma Fast DAB tablets (Sigma-Aldrich Co. Ltd.).
Inhibition of staphylococcal binding to fibronectin by Embp32 and FnBPB[D1-D4].
Wells of Nunc Maxisorp microtiter plates (Invitrogen Ltd.) were each coated overnight at 4°C with 0.2 ml of a 0.1-mg/ml solution of fibronectin. The wells were blocked with 0.2 ml of 1% Tween 20 in PBS for 1 h. After the wells were rinsed four times with PBS, 0.1-ml aliquots of different concentrations of Embp32 or FnBPB[D1-D4] (prepared as described by Nair et al. [S. P. Nair, H. Khalil, R. J. Williams, S. Ahmed, B. Henderson, and S. Meghji, submitted for publication]) were added to the wells and incubated for 1 h. Then 0.1 ml of either S. epidermidis NCTC11047 or S. aureus 8325-4 (corresponding to 5 × 107 cells) was added in triplicate to the appropriate wells and incubated for 1 h at 37°C. The wells were washed four times with 0.1 ml of PBS. The bound bacteria were harvested by addition of 0.1 ml of 0.25% trypsin to each well and incubation for 5 min at 37°C. The bound bacteria were counted by serial dilution and plate counting on blood agar base no. 2 plates containing 5% horse serum (Oxoid Ltd.).
Cloning of embp32.
The oligonucleotides 5′-GGATCCCTACAACAAGCAAGTGCAACA-3′ and 5′-CTGCAGTAGAAGTGCTCTAGCATCATC-3′ were designed to amplify a 870-bp fragment of DNA encoding the putative fibronectin-binding region of Embp (Fig. 1) and contained recognition sequences (underlined) for the restriction enzymes BamHI and PstI, respectively. The PCR fragment was initially cloned into pCR4-TOPO (Invitrogen Ltd.) and transformed into E. coli TOP10. The insert was extracted from pCR4-TOPO on a BamHI-PstI fragment and ligated to BamHI- and PstI-digested pQE30 (Qiagen Ltd.). The ligation mixture was transformed into E. coli M607(pREP4) (Nair et al., submitted), and transformants were selected by growth at 30°C on Luria-Bertani agar containing 100 μg of ampicillin per ml, 25 μg of kanamycin per ml, 20 μg of spectinomycin per ml, and 20 μg of streptomycin per ml.
FIG. 1.
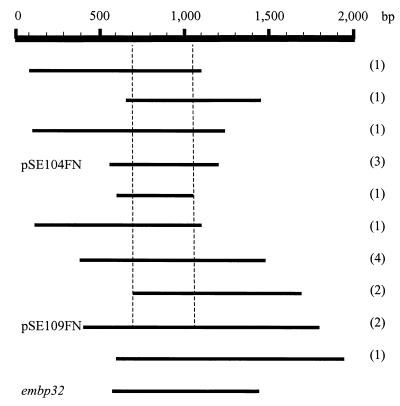
Alignment of the S. epidermidis DNA inserts within the fibronectin-binding phage. The bar at the top indicates the scale of the overlapping sequences. The dashed lines indicate the 364-bp region present in all inserts. The numbers in parentheses are the numbers of clones analyzed that contained the inserts. The inserts in phages pSE104FN and pSE109FN are shown. The region coding for the recombinant protein (Embp32) used in the inhibition studies is also shown.
Expression of embp32 and purification of recombinant Embp32.
For gene expression, positive clones were grown overnight in Luria broth containing antibiotics, after which they were diluted 1:10 in fresh broth and incubated for an additional 2 h at 30°C. Gene expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 30°C. Cells were harvested and then resuspended and lysed for 30 min in B-PER protein extraction reagent (Pierce & Warriner Ltd., Tattenhall, United Kingdom) containing 750 mM ammonium chloride, 20 mM imidazole, 1.5 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin, and 10 μM leupeptin. The lysates were clarified by centrifugation at 23,000 × g for 10 min and then mixed with Ni-nitrilotriacetic acid-agarose (Qiagen Ltd.) and incubated for 1 h at 4°C. After the preparation was loaded onto a column, contaminating proteins were removed by sequential column washes with B-PER containing 500 and 250 mM ammonium chloride, followed by two column washes with PBS containing an additional 0.15 M NaCl. All wash solutions contained 20 mM imidazole. The purified recombinant protein was eluted from the column in PBS containing 250 mM imidazole and 0.15 M NaCl. Finally, the recombinant Empb32 protein was dialyzed against PBS containing an additional 0.15 M NaCl.
Reverse transcription (RT)-PCR analysis of embp expression.
Overnight cultures of S. epidermidis NCTC11047 grown in brain heart infusion broth (Oxoid Ltd.) were diluted to an absorbance at 650 nm of 0.1 in fresh broth and incubated at 37°C with aeration. Aliquots containing 1 × 109 cells were removed at various times, and total cellular RNA was extracted by using an RNeasy mini kit (Qiagen Ltd.) according to the manufacturer's instructions, except that the cells were lysed with 100 μl of a 2.5-mg/ml lysostaphin solution and 20 μl of a 50-mg/ml lysozyme solution for 5 min at 37°C. The purified RNA was treated with RNase-free DNase (Promega), and cDNA was prepared from 2 μg of RNA by using random hexamers and Superscript II (Invitrogen Ltd.) according to the manufacturer's instructions. PCR screening for the embp gene was performed by using 1.5 μl of the cDNA and the oligonucleotides 5′-AGCGGTACAAATGTCAATATC-3′ and 5′-AGAAGTGCTCTAGCATCATCC-3′, which were designed to amplify a 455-bp region of embp.
DNA manipulations and sequencing.
All DNA manipulations were carried out by using standard procedures (21). Phagemid DNA was extracted with a QIAprep spin miniprep kit (Qiagen Ltd.). The insert DNA was sequenced from the 5′ and 3′ ends with the oligonucleotides 5′-AGGTACATTACTTATATCTGG-3′ and 5′-CCGCTTTTGCGGGATCGTCAC-3′, respectively, by cycle sequencing performed with a BigDye terminator kit (ABI Perkin-Elmer, Warrington, United Kingdom). The sequencing reactions were performed with an ABI 310 genetic analyzer. Homology searches were carried out against sequence databases by using the BLAST algorithm with either the nucleotide sequence of the S. epidermidis insert DNA or the predicted amino acid sequence in frame with the open reading frame encoded by gene VIII of the phagemid. Preliminary sequence data for S. epidermidis was obtained from The Institute for Genomic Research (TIGR) website (http://www.tigr.org).
Nucleotide sequence accession number.
The nucleotide sequence of embp32 from S. epidermidis strain NCTC11047 has been deposited in the GenBank database under accession number AY101364.
RESULTS
Isolation of phage particles that bound to fibronectin.
To identify fibronectin-binding proteins from S. epidermidis, a phage display library was constructed by using the phagemid vector pG8SAET (16). Electroporation of E. coli TG1 with the S. epidermidis chromosomal DNA-pG8SAET ligation mixture yielded 9 × 107 ampicillin-resistant colonies. Superinfection of these cells with helper phage gave a phage display library containing 5 × 1010 CFU/ml. A total of 5 × 104 phage particles bound to immobilized fibronectin when 1 ml of the library was panned (first round) against this ligand. The bound phage were amplified to give a library containing 5 × 1010 CFU/ml, and 1 ml was used in a second round of panning against fibronectin. The second round of panning resulted in 3 × 105 phage particles binding to fibronectin. The phage from the second round of panning were again amplified (giving 2 × 109 phage particles/ml) and used to pan against fibronectin a third time, which resulted in 2 × 107 phage particles binding to fibronectin. DNA sequencing of 20 randomly chosen phagemids (10 phagemids from the second round of panning and 10 phagemids from the third round of panning) revealed that 17 clones contained inserts with overlapping sequences, while 3 of the clones contained unrelated sequences which were isolated due to artifactual binding. Figure 1 shows an alignment of the overlapping inserts from the clones isolated from the second and third rounds of panning against fibronectin. These inserts encompassed approximately 1.85 kb of DNA, and a 364-bp region was present in all of them. BLAST searches of the TIGR sequence database from the on-going S. epidermidis genome sequencing project revealed that all of the overlapping inserts mapped to a locus coding for a putative 30.5-kb open reading frame. We have designated the gene encoding this fibronectin-binding protein embp (for extracellular matrix binding protein).
Capacity of recombinant phage to bind to fibronectin.
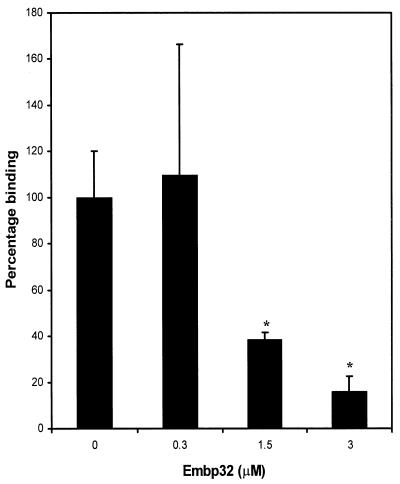
Two of the phagemid clones, pSE104FN and pSE109FN, containing 661- and 1,378-bp inserts, respectively (Fig. 1), were used in studies to confirm that the recombinant phage had the capacity to bind to fibronectin. Phage stocks of pSE104FN and pSE109FN were panned against BSA and fibronectin immobilized on microtiter plate wells. A total of 4.6 × 105 pSE109FN phage particles (range, 4.32 × 105 to 4.86 × 105) were recovered from the fibronectin-coated wells, whereas no phage bound to BSA. Similar results were obtained with phage pSE104FN, suggesting that these phagemids contained S. epidermidis DNA inserts encoding a fibronectin-binding domain. To confirm that the binding capacity was specific, the ability of the recombinant protein Embp32 to inhibit binding of phage pSE109FN to fibronectin was examined. Embp32 contains the predicted fibronectin-binding domain of Embp and has the same N-terminal start point as the peptide displayed by phage pSE104FN (Fig. 1). Preincubation of fibronectin-coated wells with graded concentrations of Embp32 inhibited binding of pSE109FN to fibronectin in a dose-dependent manner (Fig. 2). While no inhibition was evident after preincubation with a 0.3 μM solution of Embp32, 1.5 and 3 μM solutions inhibited binding by 61 and 84%, respectively. These results demonstrated that there was a specific interaction between the recombinant phage and fibronectin and that the phage expressed a fibronectin-binding domain.
FIG. 2.
Binding of phage pSE109FN to fibronectin preincubated with graded concentrations of Embp32. A binding value of 100% corresponded to the binding measured in wells preincubated with PBS. The results of one representative experiment of at least three experiments are shown. The data are the means and standard deviations for triplicate wells. An asterisk indicates that the P value is <0.05.
In an attempt to identify the region within fibronectin to which the fibronectin-binding domain of pSE109FN bound, the phage stock was panned against immobilized whole fibronectin or one of three proteolytic fragments of this molecule. As expected, pSE109FN bound to whole fibronectin. However, it did not bind to the 45-kDa α-chymotryptic fragment, and there was negligible binding to both the 30- and 120-kDa fragments compared to the binding to whole fibronectin (Table 1).
TABLE 1.
Numbers of recombinant phage binding to fibronectin and proteolytic fragments of fibronectin
| Ligand | No. of bound phagea |
|---|---|
| Whole fibronectin | 2.43 × 105 (1.35 × 105−3.24 × 105) |
| 30-kDa N-terminal fragment | 4.5 × 102 (2.7 × 102−5.4 × 102) |
| 45-kDa α-chymotrypic fragment | 0 |
| 120-kDa α-chymotrypic fragment | 6.3 × 102 (5.4 × 102−8.1 × 102) |
The number of bound phage CFU were determined as CFU infecting E. coli TG1 and plating the infected cells on nutrient broth no. 2 agar plates containing ampicillin. The values are the means for triplicate wells of one representative experiment of at least three experiments. The values in parentheses are the ranges of the numbers of phage recovered from the triplicate wells.
Capacity of recombinant phage to bind to other host matrix components.
To determine if the recombinant phage had the capacity to bind to other host matrix molecules, microtiter plate wells were coated with fibronectin, collagen, fibrinogen, laminin, vitronectin, plasminogen, mucin, heparin, and hyaluronate. As previously determined, pSE109FN bound to fibronectin. Additionally, this recombinant phage bound to plasminogen, heparin, and hyaluronate. However, the numbers of phage recovered from these ligands were more than 100-fold less than the numbers recovered from fibronectin. No binding of pSE109FN to collagen, fibrinogen, laminin, vitronectin, or mucin was detected (Table 2). Western ligand blotting was performed to determine if any of the matrix or plasma components examined were contaminated with fibronectin. The only ligand preparation found to contain any contaminating fibronectin was the fibrinogen preparation, in which low levels were detected (data not shown).
TABLE 2.
Numbers of recombinant phage particles that bound to the immobilized ligands
| Ligand | No. of bound phagea |
|---|---|
| Fibronectin | 5.63 × 105 (5.27 × 105−5.8 × 105) |
| Collagen | 0 |
| Fibrinogen | 0 |
| Laminin | 0 |
| Vitronectin | 0 |
| Plasminogen | 3.38 × 103 (2.84 × 103−3.78 × 103) |
| Mucin | 0 |
| Heparin | 1.58 × 103 (1.08 × 103−2.03 × 103) |
| Hyaluronate | 2.12 × 103 (1.89 × 103−2.3 × 103) |
The number of bound phage CFU were determined as CFU infecting E. coli TG1 and plating the infected cells on nutrient broth no. 2 agar plates containing ampicillin. The values are the means for triplicate wells of one representative experiment of at least three experiments. The values in parentheses are the ranges of the numbers of phage recovered from the triplicate wells.
Inhibition of staphylococcal binding to fibronectin.
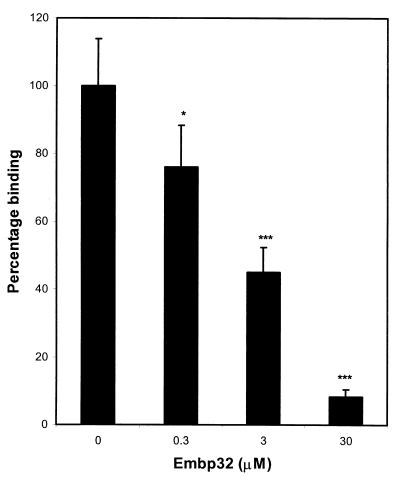
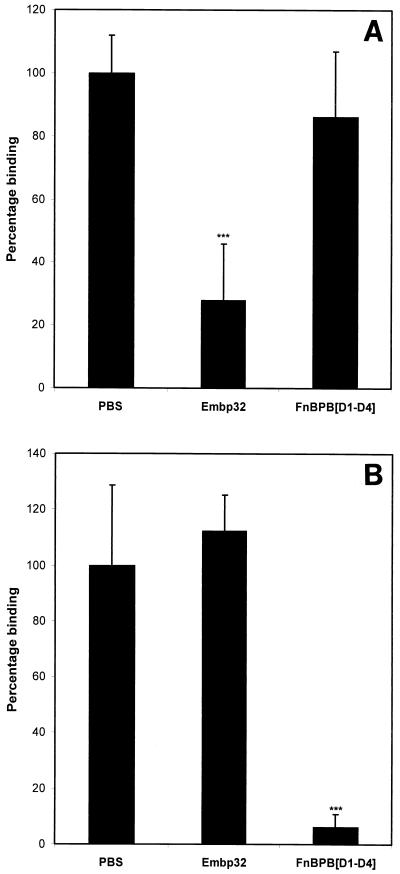
Figure 3 shows the effect of preincubating fibronectin-coated wells with recombinant Embp32 on the binding of S. epidermidis to fibronectin. Recombinant Embp32 inhibited binding of S. epidermidis to fibronectin in a dose-dependent manner, with a 30 μM solution of recombinant protein blocking binding by 91%. To determine the specificity of the Embp inhibition of staphylococcal binding to fibronectin, the effects of preincubating fibronectin-coated wells with 13 μM recombinant Embp32 and recombinant FnBPB[D1-D4] on the binding of S. aureus and S. epidermidis were compared (Fig. 4). Recombinant Embp32 blocked the binding of S. epidermidis to fibronectin by 72%, while recombinant FnBPB[D1-D4] had little effect on binding (Fig. 4A). In contrast, binding of S. aureus to fibronectin-coated wells was blocked by recombinant FnBPB[D1-D4] by 93%, but it was not blocked by recombinant Embp32 (Fig. 4B).
FIG. 3.
Binding of S. epidermidis to fibronectin preincubated with graded concentrations of Embp32. A binding value of 100% corresponded to the binding measured in wells preincubated with PBS. The results of one representative experiment of at least three experiments are shown. The data are the means and standard deviations for triplicate counts from triplicate wells. One asterisk indicates that the P value is <0.05, and three asterisks indicate that the P value is <0.005.
FIG. 4.
Effect of preincubating fibronectin-coated wells with either recombinant Embp32 or FnBPB[D1-D4] on the binding of staphylococci to fibronectin. (A) Binding of S. epidermidis to fibronectin preincubated with 13 μM Embp32 or FnBPB[D1-D4]. (B) Binding of S. aureus to fibronectin preincubated with 13 μM Embp32 or FnBPB[D1-D4]. A binding value of 100% corresponded to the binding measured in wells preincubated with PBS. The results of one representative experiment of at least three experiments are shown. The data are the means and standard deviations for triplicate counts from triplicate wells. Three asterisks indicate that the P value is <0.005.
RT-PCR analysis of the expression of embp.
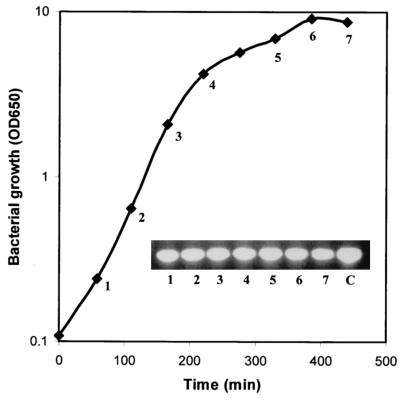
To determine if embp was expressed by S. epidermidis, total cellular RNA extracted from S. epidermidis at various times during growth was subjected to RT-PCR analysis. The RNA was reverse transcribed, and the resultant cDNA was used as the template in PCR screening for a 455-bp fragment covering the fibronectin-binding region of embp. Figure 5 shows that the 455-bp fragment was amplified at each time examined, suggesting that the embp gene was constitutively expressed throughout the growth of S. epidermidis.
FIG. 5.
Expression of the embp gene in S. epidermidis NCTC11047. A 455-bp embp product was PCR amplified from cDNA prepared from total RNA extracted at each of the times shown. OD650, optical density at 650 nm; C, S. epidermidis NCTC11047 genomic DNA used as a template in the PCR.
DISCUSSION
S. epidermidis is a major cause of foreign body infections (20), which has been attributed to its tenacity to adhere to biomaterials. The initial attachment step is generally believed to involve hydrophobic interactions, and these interactions may be mediated by the autolysin (AtlE) of this bacterium (8). It has also been demonstrated that S. epidermidis can bind to implants that have been coated with host extracellular matrix components (9, 22). However, the bacterial proteins that mediate such interactions remain largely unknown. In this paper, we describe the identification, by shotgun phage display cloning, of a fibronectin-binding domain from S. epidermidis. Panning of the S. epidermidis phage display library against fibronectin resulted in affinity selection of phage, 85% of which harbored S. epidermidis inserts coding for overlapping sequences. The overlapping inserts within the affinity-selected phage encompassed approximately 1.85 kb of DNA, and a 364-bp region was present in all of the overlapping sequences (Fig. 1); thus, this region is predicted to be the minimal sequence necessary for fibronectin binding. A comparison of the phagemid insert DNA sequences from overlapping clones with sequences in the TIGR sequence database from the on-going S. epidermidis genome sequencing project resulted in identification of a single 30.5-kb putative open reading frame to which the clones mapped. Whether this putative 30.5-kb open reading frame, which we designated embp, is transcribed as a single transcript remains to be determined, and thus the extent of the embp gene remains to be elucidated. However, RT-PCR analysis demonstrated that the region of embp encoding the fibronectin-binding domain is expressed throughout the growth phases of S. epidermidis.
Confirmation that the recombinant phage displayed a fibronectin-binding domain from S. epidermidis was obtained by panning two of the phage, pSE104FN and pSE109FN, against microtiter plates coated with either BSA or fibronectin. Further confirmation of the specificity of binding was obtained by determining that a recombinant protein, Embp32, encompassing the predicted fibronectin-binding domain of Embp, blocked binding of pSE109FN to fibronectin.
The fibronectin-binding proteins of a number of bacteria, including S. aureus and Streptococcus pyogenes, have been shown to bind to a 30-kDa N-terminal region of fibronectin (12, 15). In this study we attempted to map the portion of fibronectin to which the phage encoding the fibronectin-binding domain of S. epidermidis bound. None of the proteolytic fragments of fibronectin tested, including the 30-kDa N-terminal fragment, bound to phage pSE109FN to an appreciable extent. This demonstrates that the S. epidermidis fibronectin-binding domain identified in this study interacts with fibronectin at a site distinct from the site utilized by the S. aureus and S. pyogenes fibronectin-binding proteins. At present it is unclear whether the interaction is with a fragment of fibronectin not examined in this study or whether the interaction site spans two of the fragments examined.
The ability of one of the phages displaying the S. epidermidis fibronectin-binding domain, pSE109FN, to bind to a number of other human matrix and plasma components was examined. No binding to the matrix proteins collagen, laminin, fibrinogen, and vitronectin was observed. Binding to plasminogen was detected, although the level of this binding was more than 100-fold less than that observed for fibronectin. Phage binding to two nonproteinaceous matrix components, heparin and hyaluronate, was also detected, but once again the levels of binding were more than 100-fold less than the level of binding observed with fibronectin. Western ligand blot analysis revealed that most of the matrix and plasma components tested were free of fibronectin contamination; the only exception was fibrinogen, which contained low levels of contaminating fibronectin. Thus, the interactions of pSE109FN with plasminogen, heparin, and hyaluronate were not due to fibronectin contamination. Interestingly it has been reported that heparin can inhibit binding of S. epidermidis to fibronectin-coated implants but not to fibrinogen-coated implants (5). Since the fibronectin-binding domain of S. epidermidis identified in this study binds to other matrix components, we have designated the protein extracellular matrix-binding protein (Embp).
We have also shown that the recombinant protein Embp32 blocks binding of S. epidermidis to fibronectin in a dose-dependent manner. The fact that the binding of S. epidermidis to fibronectin was blocked by 91% at the highest concentration of Embp32 examined suggests that either the Embp protein identified in this study is the major fibronectin-binding protein of S. epidermidis or that any other fibronectin-binding proteins possessed by this bacterium bind to the same region of fibronectin as Embp. Interestingly, the recombinant protein FnBPB[D1-D4], encompassing the D1-D4 repeat region of S. aureus FnBPB, has a negligible effect on the binding of S. epidermidis to fibronectin. Similarly, Embp32 does not inhibit the binding of S. aureus to fibronectin, whereas FnBPB[D1-D4] does. This can be explained by the finding that Embp, as determined by our phage-binding studies, binds to a region of fibronectin distinct from the 30-kDa N-terminal region to which the S. aureus fibronectin-binding proteins bind. These results demonstrate that S. aureus and S. epidermidis have discrete fibronectin-binding proteins that have distinct molecular interactions with fibronectin.
Acknowledgments
We thank the Arthritis Research Campaign for funding this research under program grant H0600.
We thank L. Frykberg for the kind gift of phagemid pG8SAET. Preliminary sequence data were obtained from the TIGR website (http://www.tigr.org). Sequencing of the S. epidermidis genome was accomplished with support from NIH and NIAID.
Editor: D. L. Burns
REFERENCES
- 1.Baddour, L. M., L. P. Barker, C. D. Christensen, J. T. Parisi, and W. A. Simpson. 1990. Phenotypic variation of Staphylococcus epidermidis in infection of transvenous endocardial pacemaker electrodes. J. Clin. Microbiol. 28:676-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheesbrough, J. S., R. G. Finch, and R. P. Burden. 1986. A prospective study of the mechanisms of infection associated with hemodialysis catheters. J. Infect. Dis. 154:579-589. [DOI] [PubMed] [Google Scholar]
- 3.Delmi, M., P. Vaudaux, D. P. Lew, and H. Vasey. 1994. Role of fibronectin in staphylococcal adhesion to metallic surfaces used as models of orthopaedic devices. J. Orthop. Res. 12:432-438. [DOI] [PubMed] [Google Scholar]
- 4.Foster, T. J., and M. Hook. 1998. Surface protein adhesions of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 5.Francois, P., D. Letourneur, D. P. Lew, J. Jozefonwicz, and P. Vaudaux. 1999. Inhibition by heparin and derivatized dextrans of Staphylococcus epidermidis adhesion to in vitro fibronectin-coated or explanted polymer surfaces. J. Biomater. Sci. Polym. 10:1207-1221. [DOI] [PubMed] [Google Scholar]
- 6.Galliani, S., M. Viot, A. Cremieux, and P. Van der Auwera. 1994. Early adhesion of bacteremic strains of Staphylococcus epidermidis to polystrene: influence of hydrophobicity, slime production, plasma, albumin, fibrinogen, and fibronectin. J. Lab. Clin. Med. 123:685-692. [PubMed] [Google Scholar]
- 7.Hartford, O., L. O'Brien, K. Schofield, J. Wells, and T. J. Foster. 2001. The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology 147:2545-2552. [DOI] [PubMed] [Google Scholar]
- 8.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 9.Herrmann, M., P. Vaudaux, D. Pittet, R. Auckenthaler, P. D. Lew, F. Schumacher-Perdreau, G. Peters, and F. A. Waldvogel. 1988. Fibronectin, fibrinogen, and laminin act as mediators of adherence of clinical staphylococcal isolates to foreign material. J. Infect. Dis. 158:693-701. [DOI] [PubMed] [Google Scholar]
- 10.Hussain, M., C. Heilmann, G. Peters, and M. Herrmann. 2001. Teichoic acid enhances adhesion of Staphylococcus epidermidis to immobilized fibronectin. Microb. Pathog. 31:261-270. [DOI] [PubMed] [Google Scholar]
- 11.Inram, R. D., K. V., Gallegos, B. D. Brause, P. B. Redecha, and C. L. Christian. 1984. Clinical and microbial features of prosthetic joint infection. Am. J. Med. 77:47-53. [DOI] [PubMed] [Google Scholar]
- 12.Joh, D., P. Speziale, S. Gurusiddappa, J. Manor, and M. Hook. 1998. Multiple specificities of the staphylococcal and streptococcal fibronectin-binding microbial surface components recognizing adhesive matrix molecules. Eur. J. Biochem. 258:897-905. [DOI] [PubMed] [Google Scholar]
- 13.Karchmer, A. W., G. L. Archer, and W. E. Dismukes. 1983. Staphylococcus epidermidis causing prosthetic valve endocarditis: microbiologic and clinical observations as guides to therapy. Ann. Intern. Med. 98:447-455. [DOI] [PubMed] [Google Scholar]
- 14.Lang, S., M. A. Livesley, P. A. Lambert, J. Elliott, and T. S. Elliott. 1999. The genomic diversity of coagulase-negative staphylococci associated with nosocomial infections. J. Hosp. Infect. 43:187-193. [DOI] [PubMed] [Google Scholar]
- 15.Mosher, D. F., and R. A. Proctor. 1980. Binding and factor XIII-mediated cross-linking of a 27-kilodalton fragment of fibronectin to Staphylococcus aureus. Science 209:927-929. [DOI] [PubMed] [Google Scholar]
- 16.Nilsson, M., L. Frykberg, J.-I. Flock, L. Pei, M. Lindberg, and B. Guss. 1998. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect. Immun. 66:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paulsson, M., M. Kober, C. Freij-Larsson, M. Stollenwerk, B. Wesslen, and Å. Ljungh. 1993. Adhesion of staphylococci to chemically modified and native polymers, and the influence of preadsorbed fibronectin, vitronectin and fibrinogen. Biomaterials 14:845-853. [DOI] [PubMed] [Google Scholar]
- 18.Pei, L., M. Palma, M. Nilsson, B. Guss, and J.-I. Flock. 1999. Functional studies of a fibrinogen binding protein from Staphylococcus epidermidis. Infect. Immun. 67:4525-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pei, L., and J.-I. Flock. 2001. Lack of fbe, the gene for a fibrinogen-binding protein from Staphylococcus epidermidis, reduces its adherence to fibrinogen coated surfaces. Microb. Pathog. 31:185-193. [DOI] [PubMed] [Google Scholar]
- 20.Rupp, M. E., and G. L. Archer. 1994. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect. Dis. 19:231-243. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Vaudaux, P., D. Pittet, A. Haeberli, E. Huggler, U. E. Nydeggar, D. P. Lew, and F. A. Waldvogel. 1989. Host factors selectively increase staphylococcal adherence on inserted catheters: a role for fibronectin and fibrinogen or fibrin. J. Infect. Dis. 160:865-875. [DOI] [PubMed] [Google Scholar]
- 23.Whitehead, W. E., and J. R. Kestle. 2001. The treatment of cerebrospinal fluid shunt infections. Results from a practice survey of the American Society of Pediatric Neurosurgeons. Pediatr. Neurosurg. 4:205-210. [DOI] [PubMed] [Google Scholar]
- 24.Wong, E. S. 1996. Surgical site infections, p. 154. In C. G. Mayhall (ed.), Hospital epidemiology and infection control. Williams & Wilkins, Baltimore, Md.