Abstract
In order for Staphylococcus aureus to adhere to host extracellular matrix (ECM) substrates, it elicits a wide range of surface proteins. We have characterized a novel ∼1.1-MDa protein in S. aureus, termed Ebh (for ECM-binding protein homologue), which has homology to other ECM-binding proteins. Ebh consists of several domains, including a large central region with 44 imperfect repeats of 126 amino acids. Expression analysis revealed ebh to be growth phase regulated and repressed by agr. A fragment of the central repeat region of Ebh was cloned, overexpressed, and used in ligand-binding studies to determine Ebh function. The recombinant protein was found to specifically bind human fibronectin. Ebh is produced during human infection since serum samples taken from patients with confirmed S. aureus infections were found to contain anti-Ebh antibodies. Localization studies revealed Ebh to be cell envelope associated and is proposed to form a specialized surface structure involved in cellular adhesion.
Staphylococcus aureus is able to cause a wide range of different infections, such as endocarditis, arthritis, and septicemia (55). In order for S. aureus to colonize and disseminate through its host, the bacterium expresses an array of proteins which interact with molecules of the host extracellular matrix (ECM). These bacterial cell surface and extracellular proteins bind to a wide range of host proteins, such as fibronectin (Fn) (15, 21, 25, 42), fibrinogen (Fg) (21, 36, 40, 56), vitronectin (21), collagen (50), thrombospondin (18), bone sialoprotein (51), elastin (8), and von Willebrand factor (16), belying the ability of S. aureus to act as the etiological agent of a variety of pathologies. Fn is an adhesive glycoprotein found on the surface of mammalian cells and in serum (7). Previous studies have implicated staphylococcal Fn-binding proteins with adhesion to different cell types (1, 23, 37, 38, 47). Also, they bind Fn, which acts as an invasin, forming a bridge between S. aureus and an integrin on the surface of nonprofessional phagocytes (1, 9, 23, 33, 46, 48). Most of the ligand-binding proteins that have previously been characterized are found associated with the cell wall and are known as microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (13). The specific interactions that these adhesins undergo with the ECM, coupled with the fact that they are found on the surface of the cell, means that they may be useful targets for prophylaxis or therapy, e.g., as vaccine components, as targets for passive immunotherapy, or for novel antiadhesive strategies.
Recently, the genomes of S. aureus strains N315 and Mu50 have been published (31), and determination of the genomes of five additional strains has either been done or is nearing completion. These studies have revealed the presence of many uncharacterized putative surface proteins. The two largest genes, ebhA and ebhB, encode putative proteins of ca. 722 and 421 kDa, respectively. EbhA shows homology to the major adhesin of Streptococcus defectivus, Emb, a protein that binds to ECM (33). It has been hypothesized that EbhA and EbhB may be surface ligand-binding proteins (31). In the present study, we have examined ebh, a fusion of ebhA and ebhB, and the activity of the Ebh repeat region in vitro. Determination of the role and regulation of Ebh has shown that it is a Fn-binding protein, ionically bound to the cell envelope.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this work are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani medium by using selection with the antibiotics ampicillin (100 μg/ml) and kanamycin (50 μg/ml) where appropriate. S. aureus strains were grown in brain heart infusion (BHI) medium (Oxoid) containing the antibiotics erythromycin (5 μg/ml), tetracycline (5 μg/ml), kanamycin (50 μg/ml), neomycin (50 μg/ml), or lincomycin (25 μg/ml) where appropriate. All bacterial cultures were grown at 37°C. Phage transduction was performed as described by Novick (41), by using φ11 as the transducing phage.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| 8325-4 | Wild-type cured of known prophages | 41 |
| RN4220 | Accepts foreign DNA (r− m+) | 30 |
| COL | Methicillin resistant | 29 |
| PC6911 | agrΔ::tetM Tcr in 8325-4 background | 4 |
| PC1839 | sarA::km Kmr in 8325-4 background | 5 |
| LH9 | ebh::pAZ4353 Emr in 8325-4 background | This work |
| LH10 | ebh::pMUTHEX in 8325-4 background | This work |
| LH11 | ebh::pMUTHEX in PC6911 background | This work |
| LH12 | ebh::pMUTHEX in PC1839 background | This work |
| LH17 | ebh::pAZ4353 Emr in COL background | This work |
| SRC001 | ebh::pMUTHEX in COL background | This work |
| E. coli | ||
| DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Promega |
| BL21(DE3) | F−ompT gal [dcm] [lon] hsdSB (rB− mB−) | Novagen |
| Plasmids | ||
| pAZ106 | Promoterless, transcriptional lacZ fusion vector; Apr (E. coli), Emr (S. aureus) | 27 |
| pAZ4353 | pAZ106-based vector containing a ∼1.6-kb BamHI/EcoRI-cut fragment of ebh; Apr (E. coli), Emr (S. aureus) | This work |
| pMUTIN4 | Promoterless, transcriptional lacZ fusion vector; Apr (E. coli), Emr (S. aureus) | 52 |
| pMUTHEX | pMUTIN4-based vector containing a ∼1.2-kb BamHI/HindIII-cut fragment of ebh, creating an ebh::lacZ fusion; Apr (E. coli), Emr (S. aureus) | This work |
| pET24-d | His6 tag overexpression vector; Kmr | Novagen |
| pETH2 | pET24-d containing ∼1-kb internal ebh fragment encoding H2 polypeptide; Kmr | This work |
Construction of an ebh mutant and ebh::lacZ fusion strain.
All molecular biology and recombinant DNA techniques were carried out by using standard techniques. PCRs were done with Pwo polymerase (Roche). A derivative of pAZ106 (27), an integrating plasmid conferring resistance to erythromycin and containing a promoterless lacZ gene, was constructed to create a mutation in ebh. The forward primer was designed so that it was ∼850 bp downstream of the putative 5′ end of the gene and includes a BamHI site (italicized) (5′-CGGGATCCATGCTAATGATGGTTCCGGG-3′), and the reverse primer contained an EcoRI site (italicized) (5′-GGAATTCGTACCTCTTAATTGCTCAGC-3′). PCR was carried out with S. aureus 8325-4 DNA as a template, creating a ∼1.6-kbp fragment that was cut with BamHI and EcoRI and ligated into the suicide vector pAZ106, which had been cut with the same enzymes. The ligated product, designated pAZ4353, was then transformed into E. coli DH5α. Transformants were selected for ampicillin resistance (Apr) and checked for the correct insert size. This plasmid was then transformed into S. aureus RN4220 by electroporation (46) and selected for by erythromycin resistance (Emr). The fidelity of the insertion of this plasmid into ebh was verified by Southern blot analysis. The mutation was transduced into strains S. aureus 8325-4 and COL, and the resulting strains were designated S. aureus LH9 and LH17, respectively.
In order to create an ebh::lacZ fusion for expression analysis, a construct based on plasmid pMUTIN4 (52) which carries a copy of lacZ, was made so as to leave an intact copy of ebh under the control of Pspac. A forward oligonucleotide with a HindIII site (italicized) (5′-CGCAAGCTTGCTAAGGAGTGAACAATAGCTGTG-3′) and a reverse primer, c. 1.2 kb downstream with a BamHI site (italicized) (5′-CGCGGATCCCTGCTCCTGCTCCATGACTC-3′), were used to amplify 8325-4 DNA. The PCR product was cut with BamHI and HindIII and ligated into pMUTIN4 which had been similarly cut. The construct, designated pMUTHEX, was transformed into E. coli DH5α, and transformants were isolated by selection for Apr. Plasmids were checked for correct insert size and transformed into S. aureus RN4220 by electroporation (46), where they integrated into the bacterial chromosome. Transformants were isolated by selection for Emr. The construct was checked by Southern blot analysis and transduced into S. aureus 8325-4, PC6911 (agr), PC1839 (sarA), and COL. The constructs were designated S. aureus LH10, LH11, LH12, and SRC001, respectively.
Murine skin abscess model of infection.
An established murine abscess model of infection was used to assess the virulence of the wild-type and mutant strains (6). Briefly, S. aureus cells were grown to stationary phase in BHI medium (time, 15 h) and washed twice in phosphate-buffered saline (PBS). The cells were then adjusted to 5 × 108 CFU ml−1, and 200 μl of the cell suspension was injected subcutaneously in 6- to 8-week-old female BALB/c mice. The precise inoculum was determined by plating out onto BHI agar. At 7 days postinjection, the mice were euthanized with CO2 and the lesions were removed aseptically and frozen in liquid nitrogen. The lesions were homogenized in a miniblender in 2.5 ml of ice-cold PBS. After 1 h of incubation on ice, the lesions were homogenized again before serial dilution of the resultant suspension. The total number of bacteria was determined by growth on BHI agar. Statistical significance was evaluated based on the percent recovery of strains by using the Student t test with a 5% confidence limit.
Reporter gene (lacZ) fusion assays.
β-Galactosidase assays used MUG (4-methylumbelliferyl-β-d-galactopyranoside) as a substrate as previously described (4). Cultures for LacZ assays were grown as described previously (4). Briefly, 100-μl culture samples were stored at −70°C. Thawed samples were centrifuged at 16,100 × g for 5 min, and the pellets resuspended in 500 μl of ABT buffer (60 mM K2HPO4, 40 mM KH2PO4, 100 mM NaCl). The reactions were started by the addition of 50 μl of MUG (10 mg/ml). The reaction mixture was incubated at 25°C for 1 h and stopped by the addition 500 μl of 0.4 M Na2CO3. The assay mixture was then serially diluted in a 1:1 (vol/vol) mixture of ABT and Na2CO3 in a 96-well microtiter plate (Nunc). Fluorescence was measured by using a Victor plate reader (Wallac). The results shown are representative of experiments that were repeated at least twice and that showed <20% overall variation.
Cloning and overexpression of the H2 polypeptide.
The oligonucleotides H2F (5′-CCATGCCATGGATGTTAACACCAGTGAACC-3′), which included an NcoI restriction site (italicized), and H2R (5′-CCGCTCGAGGCACTTGATTCATCGCTTCAG-3′), which included an XhoI (italicized) site, were used to PCR amplify a section of ebh that corresponds to part of the putative central ligand-binding domain. The resultant ∼1.1-kb DNA fragment was digested with NcoI and XhoI and ligated into similarly cut pET24-d. The ligation mixture was transformed into E. coli DH5α, and transformants were selected for kanamycin resistance (Kmr) and checked by restriction digestion and sequencing to confirm the fidelity of the PCR. A representative plasmid, pETH2, was transformed into E. coli BL21(DE3). His6 tag recombinant H2 was expressed by the addition of 100 μM IPTG to growing cells. Purification was achieved by using the Hi-Trap System (Amersham Biosciences). Eluted fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the protein concentration was determined by using the Bio-Rad protein assay kit.
Anti-Ebh antibodies.
Antibodies were raised in young New Zealand White rabbits against recombinant H2. In order to remove antibodies that cross-react with other S. aureus proteins, a lysate of S. aureus LH17 (COL ebh) was used to absorb the sera. Briefly, a 200-ml overnight culture of LH17 was centrifuged at 16,000 × g for 5 min at 4°C, the pellet was resuspended in 10 ml of 50 mM Tris-HCl [pH 8.0]), and lysostaphin (Sigma) was added to a final concentration of 10 μg/ml. The suspension was incubated at 37°C for 15 min, sonicated on ice, and then centrifuged at 12,000 × g for 10 min at 4°C. Then, 50 μl of immune rabbit serum was diluted 1:10 (vol/vol) in 2% (wt/vol) bovine serum albumin (BSA) in TBST (10 M Tris-HCl [pH 7.4], 0.15 M NaCl, 0.05% [vol/vol] Tween 20) and added to 150 μl of lysate. The mixture was incubated at room temperature for 4 h before NaN3 was added to a 0.05% (wt/vol) final concentration.
Human serum.
Sera samples were collected from individuals with proven S. aureus infections diagnosed at the John Radcliffe Hospital, Oxford, United Kingdom, and the Royal Hallamshire Hospital, Sheffield, United Kingdom.
Preparation of cell wall material.
Fifty-milliliter batches of BHI broth were inoculated 1:50 with overnight cultures of S. aureus. Growing cultures were harvested and diluted as appropriate to an optical density at 600 nm of 1.0. Then, 50 ml was centrifuged at 16,100 × g for 5 min at 4°C, resuspended, and washed in 1 ml of TBS (50 mM Tris-HCl [pH 7.5], 0.1 M NaCl, 0.5 mM phenylmethylsulfonyl fluoride, 1 mg of iodoacetamide/ml). The samples were centrifuged at 16,100 × g for 5 min at 4°C, and pellets were resuspended in 1 ml of TBS. Next, 0.5 ml of suspension was added to a FastPrep tube (Bio 101) containing glass beads, which was then shaken 10 times in a FastPrep machine (Bio 101) set at speed 6 for 40 s. The tubes were placed on ice and allowed to cool between each cycle. The glass beads were allowed to settle, and the supernatant containing insoluble cell wall material was removed. Insoluble material was recovered by centrifugation at 16,100 × g for 10 min at 4°C and washed twice in 10 volumes of TBS by centrifugation at 16,100 × g for 10 min at 4°C before resuspension in 200 μl of SDS-PAGE buffer. The samples were boiled for 3 min, and insoluble material was removed by centrifugation at 16,100 × g for 10 min at 4°C.
SDS-PAGE and immunoblotting.
Proteins were separated by SDS-PAGE with a 4% (wt/vol) stacking gel and a 6 or 12% (wt/vol) resolving gel for S. aureus-derived proteins and recombinant H2 polypeptide, respectively, in a Mini-Protean II gel apparatus (Bio-Rad). For Western blotting, proteins were transferred to Immun-Blot polyvinylidene difluoride membranes (Bio-Rad) by electroblotting. After overnight blocking at 4°C in TBST containing 6% (wt/vol) skimmed milk powder, probing was carried out with serum (human and rabbit) diluted 1:1,000 for 90 min at room temperature. The blots were then incubated with alkaline phosphatase-conjugated secondary antibodies (anti-human or anti-rabbit; Sigma) diluted 1:30,000 for 30 min at room temperature. Bound antibody was detected by using nitroblue tetrazolium (NBT)-BCIP (5-bromo-4-chloro-3-indolylphosphate) solution (Roche).
Ligand affinity blotting.
Purified H2 polypeptide was electrophoresed on an SDS-PAGE gel as described above, transferred to a supported nitrocellulose membrane (BDH) by electroblotting, and blocked in TBST containing 3% (wt/vol) BSA (Sigma). Either Fn (ICN) or Fg (Calbiochem), labeled by using the Roche biotin labeling kit, was used to probe the membrane at a final concentration of ∼2 μg/ml. Bound ligands were detected by using alkaline phosphatase-conjugated avidin (Bio-Rad) and NBT-BCIP solution (Roche).
ELISA analysis of ligand binding.
An enzyme-linked immunosorbent assay (ELISA) was used to analyze the ability of recombinant H2 to bind ligands. The method used was based on that described by Wann et al. (56). Briefly, 100 μl of appropriate ligand or BSA in PBS (5 μg/ml), was added to wells of a 96-well microtiter plate (Nunc) overnight at 4°C. Plates were washed three times with PBST (PBS containing 0.05% [vol/vol] Tween 20), and the remaining protein-binding sites were blocked with 5% (wt/vol) BSA in PBS for 2 h at room temperature. The plates were again washed three times in PBST, and different concentrations of purified H2 polypeptide diluted in PBS-0.1% (wt/vol) BSA were added. The plates were incubated for a further 1 h before being washed three times, and anti-H2 (diluted 1:1,000) was added in PBS-0.1% (wt/vol) BSA, followed by 1 h of incubation. The plates were washed an additional three times and alkaline phosphatase-conjugated anti-rabbit antibodies diluted 1:30,000 in PBS-0.1% (wt/vol) BSA were added and the plates incubated for 1 h. Finally, bound antibodies were detected by using the Sigma Fast p-nitrophenyl phosphate system (Sigma). Plates were read at 405 nm in a Victor microtiter plate reader (Wallac).
The inhibition ELISA was also carried out as described previously (56). The recombinant H2 was incubated with various concentrations of Fn, Fg, or BSA for 1 h at room temperature in PBS-0.1% (wt/vol) BSA. The reactions were then added to Fn-coated wells, and bound protein was detected as described above.
RESULTS
Sequence analysis of the ebh gene.
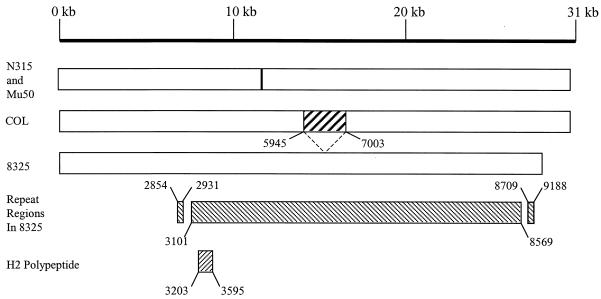
Comparison of ebhA and ebhB with the genome sequences of S. aureus strains 8325 (http://www.genome.ou.edu) and COL (http://www.tigr.org) revealed a single open reading frame, ebh. The ebh gene has a putative GTG start codon and in S. aureus 8325 was found to be 28,605 bp long but 31,494 bp long in COL, as graphically represented in Fig. 1. The gene in 8325 has a 36% GC content, and a predicted pI value of 5.8. Comparison of the peptide sequences from 8325 and COL revealed that they are identical up to amino acid 5944 but, from this point, the COL version of Ebh has an extra 1,058 amino acids not found in the 8325 protein. After these extra residues, the amino acid sequences of Ebh from the two strains are homologous, except that the 8325 version has 94 extra residues, making it 964 amino acids shorter than the COL version. Thus, the 8325 and COL nascent Ebh are 9,535 and 10,498 amino acids, respectively. This makes ebh by far the largest open reading frame in this organism. Similarly, the genome of S. aureus MW2 (http://www.bio.nite.go.jp) possesses a single ebh open reading frame. The sequenced genomes of S. aureus N315 and Mu50 have two adjacent genes, ebhA and ebhB, which are homologous to the C- and N-terminal parts of Ebh, respectively. Sequence analysis of the intergenic DNA between ebhA and ebhB in strains N315 and Mu50 does not reveal any likely ribosome-binding site, and it therefore appears likely that a frameshift mutation has occurred, thus separating ebh into two open reading frames.
FIG. 1.
Organization of ebh in S. aureus strains 8325 and COL and ebhA and ebhB in Mu50 and N315. The crossed box in COL represents the missing sequence in 8325. The positions of the repeat regions and the H2 polypeptide are shown.
A Dotblot analysis (http://www.isrec.isb-sib.ch/java/dotlet.html) was carried out by using the 8325 Ebh sequence to determine whether it contained any repeat regions. The results confirm the presence of three such regions within Ebh. The three putative repeat regions were further analyzed by using CLUSTAL (http://www2.ebi.ac.uk/clustal/). The first region has two 77-amino-acid imperfect repeats (amino acids 2854 to 2931 and 3019 to 3096), the second has 44 imperfect repeats that are 126 amino acids in length (amino acids 3101 to 8569). The third region consists of three 77-amino-acid imperfect repeats (amino acids 8709 to 8863 and 9111 to 9188). Such repeat regions are present in host ligand-binding and peptidoglycan-binding proteins (12, 13).
The peptide sequence of Ebh of S. aureus 8325 was subjected to hydrophobicity analysis by using the TopPred2-Topology prediction of the membrane protein database (http://www.sbc.su.se/∼erikw/toppred2/). The results suggest that it is mainly a hydrophilic protein, except for the first 36 amino acids at the N-terminal end; this is consistent with a signal peptide, a 21-amino-acid stretch just before amino acid 2500, and 61 amino acids at the C-terminal end of the protein, which are indicative of a membrane-spanning domain.
A BLAST-P search using the Ebh sequence from strain 8325 revealed homologies to Mrp (57) (21% identity over 1,357 amino acid residues) and FmtB (28) (20% identity over 1,462 amino acid residues) of S. aureus in the central portion and the C-terminal end of the protein. The second repeat region was also homologous to the Emb protein of S. defectivus (21% identity over 1,412 amino acid residues), which was found to bind to ECM proteins extracted from baby hamster kidneys (33).
Isolation and characterization of an S. aureus ebh mutant.
In order to elucidate the function of ebh in S. aureus, a plasmid carrying an erythromycin resistance marker was introduced into the ebh gene by homologous recombination to disrupt the chromosomal copy. Analysis of the growth rate, proteins present in the whole cell, and wall and supernatant fractions revealed no difference in mutants compared to wild types (results not shown). The pathogenicity of LH9 (8325-4 ebh) in a murine skin abscess model of infection showed no significant alteration compared to 8325-4 (data not shown).
Transcription of ebh is regulated by agr.
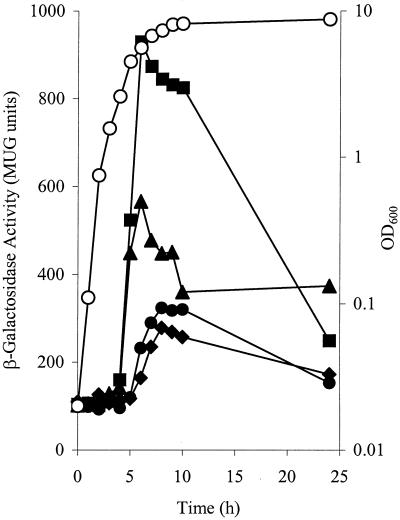
A chromosomal reporter gene (lacZ) fusion was made with ebh so that its expression during growth could be analyzed, along with any regulatory role for the accessory gene regulator (agr) or the staphylococcal accessory regulator (sarA), which act as global regulators of virulence in S. aureus (5, 22). An insertion of the plasmid pMUTHEX, which carries a copy of lacZ, into the chromosome of S. aureus 8325-4 created an ebh::lacZ fusion strain LH10. This mutation was transduced into S. aureus strains COL, PC6911 (agr), and PC1839 (sarA) to create strains SRC001, LH11, and LH12, respectively. Expression of ebh was growth phase dependent in all strains assayed and begins at postexponential phase (Fig. 2). In 8325-4, peak expression is achieved at 8 h with 300 MUG units. Expression of ebh is little affected by sarA mutation but is repressed by agr; as in strain LH11 (agr), expression reaches a maximum of >900 MUG units (Fig. 2). ebh is also expressed in strain COL to a maximum level of ∼550 MUG units.
FIG. 2.
Expression of ebh::lacZ fusion during growth of S. aureus, as described in Materials and Methods. A representative growth curve of LH10 [8325-4] at an optical density at 600 nm is shown (○). β-Galactosidase activity was measured for LH10 (8325-4; •), SRC001 (COL; ▴), LH11 (8325-4; agr; ▪), and LH12 (8325-4, sarA; ⧫).
Human sera contain anti-Ebh immunoglobulin G.
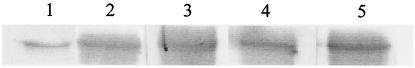
As part of a study to identify antigenic components of S. aureus, antibodies to Ebh were detected (results not shown). In order to show specific Ebh production, the H2 polypeptide was produced. H2 is a polypeptide of ∼44 kDa and spans three repeats (amino acids 3203 to 3595) of the central putative ligand-binding domain (Fig. 1). H2 was produced as a soluble His6-tagged protein in E. coli BL21 and purified by affinity chromatography. The purity (>95%) of the isolated recombinant protein was verified by SDS-PAGE. Purified H2 was shown to have the N-terminal sequence MDVNTVNQKA, which corresponds exactly to the predicted sequence, and ran at ∼46 kDa as determined by SDS-PAGE. Serum samples from patients convalescing from S. aureus infections were used to probe a Western blot of recombinant H2 polypeptide. All of the sera used showed that antibodies to this portion of the protein are present in the sera from S. aureus-infected patients with a range of diseases (Fig. 3).
FIG. 3.
Western immunoblot of H2 polypeptide (each lane containing ∼1 μg), with serum samples collected from four patients recovering from S. aureus infections. Lanes 1, Coomassie blue-stained H2 polypeptide; 2, patient diagnosed with toxic shock syndrome; 3, patient diagnosed with discitis; 4, patient diagnosed with a deep abscess; 5, patient diagnosed with septic arthritis.
Detection of Ebh in the cell envelope fraction.
In order to specifically localize Ebh on S. aureus cells, H2 was used to immunize a rabbit. Anti-Ebh antibodies that are reactive to the H2 polypeptide were absorbed of nonspecific antistaphylococcal antibodies by using a lysate of strain LH17 (COL ebh). By using this preparation, a single reactive band was detected in the cell envelope extract of COL but not 8325-4 (Fig. 4) This band corresponds to Ebh since it is not found in LH17 (COL ebh). The ability to remove the protein from the cell walls by boiling it in SDS-PAGE sample buffer is indicative of the protein being bound ionically to the cell envelope (12). Mechanical breakage of the cells in this manner and subsequent purification of insoluble material provides a pure preparation of cell wall material, free of cellular membranes (12). The reactive band was larger than any size marker available (maximum size, 205 kDa); hence, we were unable to estimate the size of the mature protein. The protein was present in both 3- and 8-h samples. No reactive proteins were detected in samples of extracellular proteins (results not shown).
FIG. 4.
Western immunoblot of S. aureus cell wall extracts with anti-H2 antisera. Samples were taken at time 3 and 8 h postinoculation. Lanes: 1, LH9 (8325-4 ebh) at 3 h; 2, LH9 at 8 h; 3, 8325-4 at 3 h; 4, 8325-4 at 8 h; 5: LH17 (COL ebh) at 3 h; 6, LH17 at 8 h; 7, COL at 3 h; 8, COL at 8 h. The band corresponding to Ebh appears ∼2 mm into the resolving gel (total length, 6 cm).
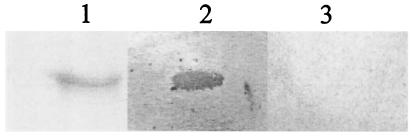
H2 polypeptide binds Fn.
To ascertain any potential ligand-binding activity of the central repeat region of Ebh, human Fg and Fn were biotinylated and used to Western blot H2 polypeptide. The purified H2 bound Fn but not Fg (Fig. 5).
FIG. 5.
Screening for Fn-binding activity by using the Western affinity blotting technique. H2 polypeptide binds human Fn but not Fg. Lanes: 1, Coomassie blue-stained H2 polypeptide (∼1 μg); 2, H2 polypeptide (∼1 μg) blotted with human Fn; 3, H2 polypeptide (∼1 μg) blotted with human Fg.
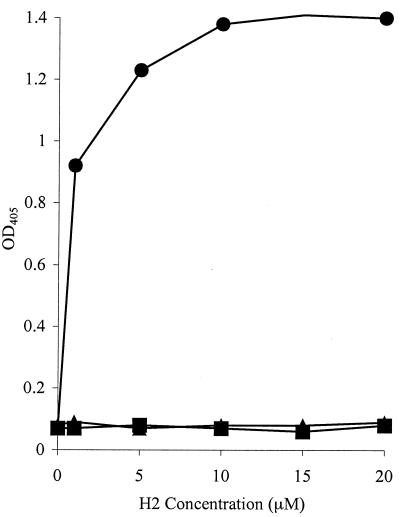
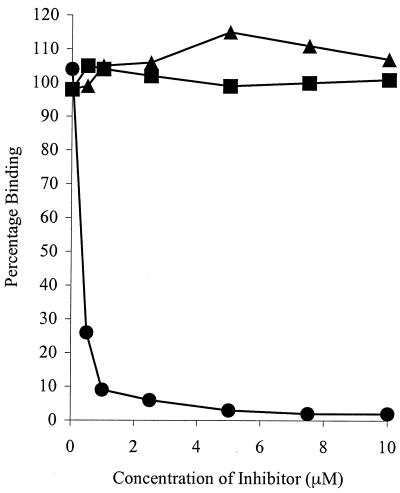
The specific Fn-binding activity of the recombinant H2 polypeptide was assessed by ELISA and compared with its ability to bind Fg and BSA as a control. H2 bound to the immobilized Fn in a dose-dependent and saturable manner. Saturation occurs at ∼10 μM H2 (Fig. 6). An apparent Kd of ∼0.5 μM can be calculated from the concentration of H2 giving half-maximum binding. However, no binding to immobilized Fg or BSA was detected. Similarly, it was possible to inhibit the binding of H2 to immobilized Fn with excess soluble Fn, but not with Fg or BSA, >90% inhibition was observed at 1 μM Fn (Fig. 7). Taken together, these data support the specific nature of the H2-Fn interaction and demonstrate that H2 is capable of binding both immobilized and soluble Fn.
FIG. 6.
Binding of H2 polypeptide to microtiter plate wells coated with Fn (•), Fg (▴), or BSA (▪). Increasing concentrations of H2 were incubated in the wells for 1 h at room temperature. Bound protein was detected with anti-H2 antibodies and anti-rabbit AP-conjugated antibodies. Values represent the means of triplicate wells. H2 polypeptide binds immobilized Fn, but not Fg or BSA.
FIG. 7.
Inhibition of H2 polypeptide binding to immobilized Fn. Wells were coated with Fn. H2 was preincubated for 1 h with increasing concentrations of Fn (•), Fg (▴), and BSA (▪) prior to incubation in Fn-coated wells. Bound proteins was detected with anti-H2 antibodies and anti-rabbit AP-conjugated antibodies. Values represent the means of triplicate wells. H2 polypeptide binds soluble Fn, but not Fg or BSA, inhibiting binding to immobilized Fn.
DISCUSSION
In this study, we describe the Ebh protein of S. aureus, which is found in the cell wall fraction and recognizes human Fn. It is a homologue of Emb, the major adhesin of Streptococcus defectivus (33) and two predicted S. aureus proteins of unknown function, Mrp and FmtB (28, 57). Ebh is encoded by the longest single gene in the S. aureus 8325 and COL genomes. In strains N315 and Mu50, the gene is divided into two open reading frames, ebhA and ebhB, that are the largest and second-largest genes, respectively (31). The large size of Ebh made it impossible to determine the molecular size of the mature protein, but its presence at the top of the 6% (wt/vol) SDS-PAGE gel used for Western blotting indicates that, in the mature form, it is still very large. The predicted size of Ebh is ∼1.1 MDa.
S. aureus possesses a selection of surface proteins that bind Fn, with perhaps the best-characterized being Fn-binding protein A (FnBPA) and FnBPB (13, 15, 25); these two proteins have been shown to facilitate the binding of cells to immobilized Fn. These proteins have similar structural organization to that of the Fn-binding proteins of other bacterial species. Like Ebh, the ligand-binding domains of these two proteins contain repeated amino acid sequences (13). Interestingly, the repeat sequence appears to lack a folded secondary structure but seems to gain such a formation upon binding to Fn (20). The cell surface-located ECM protein, Emp, has been shown to bind Fn, as well as Fg and vitronectin (21). An extracellular ligand-binding protein, Eap, has also been identified; this protein is found in culture supernatant and binds at least seven plasma proteins, including Fn, and also to the surface of S. aureus (42). Furthermore, AtlC and Aas, the autolysins of Staphylococcus caprae and Staphylococcus saprophyticus, have been shown to bind Fn (2, 17).
Transcription of ebh was observed to be under the control of the agr global regulator of virulence. The agr locus encodes a two-component signal transduction system that responds to a quorum-sensing signal (22). Thus, in this system, toxins and extracellular enzymes are in general positively regulated and cell surface proteins are commonly negatively regulated, as seen here with ebh. Studies have shown that agr mutants are able to bind a greater amount of Fn than the corresponding wild-type strains and that FnBPA and FnBPB are regulated at transcription by the agr system (45).
Binding of Fn by gram-positive pathogenic bacteria is common, and these bacteria possess an array of surface proteins with which to bind this ubiquitous eukaryotic ECM protein. Fn is essential for the adhesion of almost all cell types and is abundant in the circulation and at various extracellular sites. The present study showed that Ebh is able to bind Fn, but not Fg, another circulating glycoprotein (7). Not only have Fn-binding proteins been implicated in binding to certain eukaryotic cell types (1, 21, 37, 39, 43), but interactions between Fn and Fn-binding proteins have been implicated in the invasion of nonprofessional phagocytes (1, 9, 23, 34, 47, 48). In such instances, Fn forms a bridge between the bacterial surface protein and an integrin on the surface of the eukaryotic cell (47). It is temping to speculate that Ebh may play a role in such a process, but we were unable to detect any alteration in virulence in an ebh mutant strain. We were also unable to detect Ebh in cell wall preparations of 8325-4 but were able to detect it in COL. Since reporter gene fusion experiments showed transcription of the gene in both strains, the lack of any detectable protein in 8325-4 may be due to the amount of proteases produced by this strain (32). The genotypes of these two strains are known to be similar, although 8325-4 lacks a functional rsbU gene, which encodes the activator of σB (14). It has been shown that several proteases are negatively regulated by σB (58); therefore, since 8325-4 lacks full σB function, it produces more active proteases than COL.
The evaluation of a single adhesin's role in pathogenesis can prove troublesome and needs to be viewed with respect to S. aureus as a complex background, with one or more such ligand-binding proteins being able to substitute for the loss of function in a deletion mutant. Indeed, in previous experimental infections, deletion of one adhesive protein has not completely prevented infection (39) or has had no effect at all (11). The presence of Fn-binding proteins FnBPA, FnBPB, Emp, and Eap would likely substitute for any observable Fn-binding activity by Ebh. However, transposon mutagenesis of emb, the S. defectivus homologue of ebh, abolished the ECM adherence of that organism (33). The functions of single proteins can be elucidated by cloning them into nonpathgenic bacteria such as Lactococcus lactis (44) or Streptococcus gordonii (49). In the case of Ebh it would not be technically feasible to express the whole protein given its very large size, but it may be possible to express a section of it, such as H2, although there would also be a requirement for artificial attachment to the cell surface, which could alter the behavior of the peptide.
Anti-Ebh immunoglobulin G was detected in serum samples obtained from S. aureus-infected individuals, indicating that Ebh is expressed during the infective process. Furthermore, recent studies have identified it in a screen for genes expressed in vivo during human infection (10), and thus it is possible that this protein could have potential as a target in prophylaxis or therapy. Adhesins have been used previously as protective antigens (26), and monoclonal antibodies against the collagen-binding protein have been shown to detach S. aureus from a collagen substrate (54).
Many previously characterized ligand-binding proteins found on the surface of gram-positive bacteria are covalently bound to the peptidoglycan via a Leu-Pro-X-Thr-Gly (LPXTG) pentapeptide consensus sequence at the C terminus (13). Such proteins are attached to the cell wall peptidoglycan by the membrane-associated enzyme sortase, which recognizes the LPXTG motif and joins the Thr residue to the nascent cross-bridge of the peptidoglycan (35). Ebh lacks such a consensus sequence but appears to be tightly associated with the cell surface, since we were unable to detect its presence in the liquid supernatant but were able to extract it from cell envelope wall material.
Gram-positive bacteria employ several different mechanisms to attach proteins to the cell surface. In addition to sortase-mediated systems, InlB of Listeria monocytogenes binds directly to lipoteichoic acid via carboxy-terminal tandem repeats (3, 24). The LytA protein of Streptococcus pneumoniae contains a 20-amino-acid repeat that binds to choline-substituted teichoic acid or lipoteichoic acid (19). The WapA protein of Bacillus subtilis contains a repeat region that binds the protein ionically to cell wall peptidoglycan (12). Ebh does possess a putative C-terminal membrane-spanning domain, which may attach the peptide to the cell membrane, but also a putative peptidoglycan-binding repeat region that may bind it ionically to the peptidoglycan, similarly to what occurs in WapA.
The secretion of such a large protein as Ebh is an interesting biological question. It would be of interest to determine whether secretion is mediated by the Sec pathway, which is common to gram-positive bacteria (53), or if a specialized apparatus exists. Presumably, production and secretion would also be very expensive energetically and, as such, it seems unlikely that it would occur unless the presence of Ebh on the cell surface conferred some benefit to the bacterium.
Why S. aureus should produce such a large protein as Ebh is uncertain. Since it binds to Fn, it can be added to the growing list of proteins potentially involved in host-pathogen interaction. Given the large size of Ebh, it is tempting to speculate that it forms a specialized surface structure involved in cellular adhesion.
Acknowledgments
S.R.C. and L.G.H. contributed equally to this study.
We thank Nick Day and Sharon Peacock, John Radcliffe Hospital, Oxford, United Kingdom, and Robert Read, Royal Hallamshire Hospital, Sheffield, United Kingdom, for providing human serum samples. Oligonucleotide synthesis and DNA sequencing was carried out by Arthur Moir (University of Sheffield). The murine abscess model of infection portion of this study was done in collaboration with Eileen Ingham (University of Leeds).
This work was funded by Biosynexus (S.R.C.), AO Research Foundation (L.G.H. and R.G.R.) grant 99-F55, and the Royal Society (S.J.F.).
Editor: B. B. Finlay
REFERENCES
- 1.Ahmed, S., S. Meghji, R. J. Williams, B. Henderson, J. H. Brock, and S. P. Nair. 2001. Staphylococcus aureus fibronectin-binding proteins are essential for internalization of osteoblasts but do not account for differences in intracellular levels of bacteria. Infect. Immun. 69:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allignet, J., S. Aubert, K. G. H. Dyke, and N. El Solh. 2001. Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect. Immun. 69:712-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, L., S. Dramsi, P. Dehoux, H. Bierne, G. Lindahl, and P. Cossart. 2001. InlB: and invasion protein of Listeria monocytogenes with a novel type of surface association. Mol. Microbiol. 25:285-294. [DOI] [PubMed] [Google Scholar]
- 4.Chan, P. F., and S. J. Foster. 1998. The role of environmental factors in the regulation of virulence determinant expression in Staphylococcus aureus 8325-4. Microbiology 144:2469-2479. [DOI] [PubMed] [Google Scholar]
- 5.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattwal, G. S., and K. T. Preissner. 2000. Extracellular matrix interactions with gram-positive pathogens, p. 78-86. In V. A. Fischetti (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 8.Downer, R., F. Roche, P. W. Park, R. P. Mecham, and T. J. Foster. 2002. The elastin binding protein of Staphylococcus aureus (EbpS) is expressed at the cell surface as an integral membrane protein and not as a cell wall-associated protein. J. Biol. Chem. 277:243-250. [DOI] [PubMed] [Google Scholar]
- 9.Dziewanowska, K., J. M. Patti, C. F. Deobald, K. W. Bayles, W. R. Trible, and G. A. Bohach. 1999. Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infect. Immun. 67:4673-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etz, H., D. B. Minh, T. Henics, A. Dryla, B. Winkler, C. Triska, A. P. Boyd, J. Söllner, W. Schmidt, U. von Ahsen, M. Buschle, S. R. Gill, J. Kolonay, H. Khalak, C. M. Fraser, A. von Gabain, E. Nagy, and A. Meinke. 2002. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 99:6573-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flock. J., S. A. Hienz, A. Heimdahl, and T. Schennings. 1996. Reconsideration of the role of fibronectin binding in endocarditis caused by Staphylococcus aureus. Infect. Immun. 64:1876-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster, S. J. 1993. Molecular analysis of three major wall-associated proteins of Bacillus subtilis 168: evidence for processing of the product of a gene encoding a 258-kDa precursor two-domain ligand-binding protein. Mol. Microbiol. 8:299-310. [DOI] [PubMed] [Google Scholar]
- 13.Foster, T. J., and M. Höök. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 14.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greene, C., D. McDevitt, P. Francois, P. E. Vaudaux, D. P. Lew, and T. J. Foster. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17:1143-1152. [DOI] [PubMed] [Google Scholar]
- 16.Hartleib, J., N. Köhler, R. B. Dickinson, G. S. Chattwal, J. J. Sixma, O. M. Hartford, T. J. Foster, G. Peters, B. E. Kehrel, and M. Herrmann. 2000. Protein A is the von Willebrand factor binding protein on Staphylococcus aureus. Blood 96:2149-2156. [PubMed] [Google Scholar]
- 17.Hell, W., H. W. Meyer, and S. G. Gatermann. 1998. Cloning of aas, a gene encoding a Staphylococcus saprophyticus surface protein with adhesive and autolytic properties. Mol. Microbiol. 29:871-881. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann, M., S. J. Suchard, L. A. Boxer, F. A. Waldvogel, and P. D. Lew. 1991. Thrombospondin binds Staphylococcus aureus and promotes staphylococcal adherence to surfaces. Infect. Immun. 59:279-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holtje, J. V., and A. Tomasz. 1975. Specific recognition of choline residues in the cell wall teichoic acid by the N-acetylmuramyl-l-alanine amidase of pneumococcus. J. Biol. Chem. 250:6072-6076. [PubMed] [Google Scholar]
- 20.House-Pompeo, K., Y. Xu, D. Joh, P. Speziale, and M. Höök. 1996. Conformational changes in the fibronectin-binding MSCRAMMs are induced by ligand binding. J. Biol. Chem. 271:1379-1384. [DOI] [PubMed] [Google Scholar]
- 21.Hussain, M., K. Becker, C. von Eiff, J. Schrenzel, G. Peters, and M. Herrmann. 2001. Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J. Bacteriol. 183:6778-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joh, D., E. R. Wann, B. Kreikemeyer, P. Speziale, and M. Höök. 1999. Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 18:211-223. [DOI] [PubMed] [Google Scholar]
- 24.Jonquieres, R., H. Bierne, F. Fiedler, P. Gounon, and P. Cossart. 1999. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol. Microbiol. 34:902-914. [DOI] [PubMed] [Google Scholar]
- 25.Jönsson, K., C. Signäs, H. Müller, and M. Lindberg. 1991. Two different genes encode fibronectin-binding proteins in Staphylococcus aureus. Eur. J. Biochem. 202:1041-1048. [DOI] [PubMed] [Google Scholar]
- 26.Josefsson, E., O. Hartford, L. O'Brien, J. M. Patti, and T. Foster. 2001. Protection against experimental Staphylococcus aureus arthritis by vaccination with clumping factor A, a novel virulence determinant. J. Infect. Dis. 184:1572-1580. [DOI] [PubMed] [Google Scholar]
- 27.Kemp, E. H., R. L. Sammons, A. Moir, D. Sun, and P. Setlow. 1991. Analysis of transcriptional control of the gerD spore germination gene of Bacillus subtilis 168. J. Bacteriol. 173:4646-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsuzawa, H., K. Ohta, M. Sugai, T. Fujiwara, P. Glanzmann, B. Berger-Bachi, and H. Suginaka. 2000. Tn551-mediated insertional inactivation of the fmtB gene encoding a cell wall-associated protein abolishes methicillin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 45:421-431. [DOI] [PubMed] [Google Scholar]
- 29.Kornblum, J., B. J. Hartmann, R. P. Novick, and A. Tomasz. 1986. Conversion of a homogeneously methicillin-resistant strain of Staphylococcus aureus to heterogeneous resistance by Tn551-mediated insertional inactivation. Eur. J. Clin. Microbiol. 5:714-718. [DOI] [PubMed] [Google Scholar]
- 30.Kreiswirth, B., S. Löfdahl, M. J. Betley, M. O'Reilly, P. Schleivert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 31.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 32.Lindsay, J. A., and S. J. Foster. 1999. Interactive regulatory pathways control virulence determinant production and stability in response to environmental conditions in Staphylococcus aureus. Mol. Gen. Genet. 262:323-331. [DOI] [PubMed] [Google Scholar]
- 33.Manganelli, R., and I. van de Rijn. 1999. Characterization of emb, a gene encoding the major adhesin of Streptococcus defectivus. Infect. Immun. 67:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massey, R. C., M. N. Kantzanou, T. Fowler, N. P. J. Day, K. Schofield, E. R. Wann, A. R. Berendt, M. Höök, and S. J. Peacock. 2001. Fibronectin-binding protein A of Staphylococcus aureus has multiple, substituting, binding regions that mediate adherence to fibronectin and invasion of endothelial cells. Cell. Microbiol. 3:839-851. [DOI] [PubMed] [Google Scholar]
- 35.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 36.McDevitt, D., P. François, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 37.Miyamoto, Y. J., E. R. Wann, T. Fowler, E. Duffield, M. Höök, and B. W. McIntyre. 2001. Fibronectin binding protein A of Staphylococcus aureus can mediate human T lymphocyte adhesion and coactivation. J. Immunol. 166:5129-5138. [DOI] [PubMed] [Google Scholar]
- 38.Mongodin. E., O. Bajolet, J. Cutrona, N. Bonnet, F. Dupuit, E. Puchelle, and S. de Bentzmann. 2002. Fibronectin-binding proteins of Staphylococcus aureus are involved in adherence to human airway epithelium. Infect. Immun. 70:620-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreillon. P., J. M. Entenza, P. Francioli, D. McDevitt, T. J. Foster, P. François, and P. Vaudaux. 1995. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect. Immun. 63:4738-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni Eidhin, D., S. Perkins, P. François, P. Vaudaux, M. Höök, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 41.Novick, R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 42.Palma, M., A. Haggar, and J. Flock. 1999. Adherence of Staphylococcus aureus is enhanced by an endogenous secreted protein with broad binding activity. J. Bacteriol. 181:2840-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peacock, S. J., T. J. Foster, B. J. Cameron, and A. R. Berendt. 1999. Bacterial fibronectin-binding proteins and endothelial cell surface fibronectin mediate adherence of Staphylococcus aureus to resting human endothelial cells. Microbiology 145:3477-3486. [DOI] [PubMed] [Google Scholar]
- 44.Que, Y., P. François, J. Haefliger, J. Entenza, P. Vaudaux, and P. Moreillon. 2001. Reassessing the role of Staphylococcus aureus clumping factor and fibronectin-binding protein by expression in Lactococcus lactis. Infect. Immun. 69:6296-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saravia-Otten, P., H. Müller, and S. Arvidson. 1997. Transcription of Staphylococcus aureus fibronectin-binding protein genes is negatively regulated by agr and an agr-independent mechanism. J. Bacteriol. 197:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schenk, S., and R. A. Laddaga. 1992. Improved methods for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 94:133-138. [DOI] [PubMed] [Google Scholar]
- 47.Sinha, B., P. P. François, O. Nüsse, M. Foti, O. M. Hartford, P. Vaudaux, T. J. Foster, D. P. Lew, M. Herrmann, and K. Krause. 1999. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell. Microbiol. 1:101-117. [DOI] [PubMed] [Google Scholar]
- 48.Sinha, B., P. P. François, Y. Que, M. Hussain, C. Heilmann, P. Moreillon, D. Lew, K. Krause, G. Peters, and M. Herrmann. 2000. Heterologously expressed Staphylococcus aureus fibronectin-binding proteins are sufficient for invasion of host cells. Infect. Immun. 68:6871-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stutzmann-Meier, P., J. M. Entenzak, P. Vaudaux, P. Francioli, M. P. Glauser, and P. Moreillon. 2001. Study of Staphylococcus aureus pathogenic genes by transfer and expression in the less virulent organism Streptococcus gordonii. Infect. Immun. 69:657-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Switalski, L. M., J. M. Patti, W. Butcher, A. G. Gristina, P. Speziale, and M. Höök. 1993. A collagen receptor on Staphylococcus aureus strains isolated from patients with septic arthritis mediates adhesion to cartilage. Mol. Microbiol. 7:99-107. [DOI] [PubMed] [Google Scholar]
- 51.Tung, S., B. Guss, U. Hellman, L. Persson, K. Rubin, and C. Ryden. 2000. A bone sialoprotein-binding protein from Staphylococcus aureus: a member of the staphylococcal Sdr family. Biochem. J. 345:611-619. [PMC free article] [PubMed] [Google Scholar]
- 52.Vagner, V., E. Dervyn, and S. D. Erlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 53.van Wely, K. H. M., J. Swaving, R. Freudl, A. J. M. Driessen. 2001. Translocation of proteins across the cell envelope of gram-positive bacteria. FEMS Microbiol. Rev. 25:437-454. [DOI] [PubMed] [Google Scholar]
- 54.Visai, L., Y. Xu, F. Casolini, S. Rindi, M. Höök, and P. Speziale. 2000. Monoclonal antibodies to CNA, a collagen-binding microbial surface component recognizing adhesive matrix molecules, detach Staphylococcus aureus from a collagen substrate. J. Biol. Chem. 275:39837-39845. [DOI] [PubMed] [Google Scholar]
- 55.Waldvogel, F. A. 1995. Staphylococcus aureus (including toxic shock syndrome), p. 1754-1777. In G. L. Mandell, J. E. Bennett, and R. Dolio (ed.), Principles and practice of infectious diseases. Churchill Livingstone, New York, N.Y.
- 56.Wann, E. R., S. Gurusiddappa, and M. Höök. 2000. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds fibrinogen. J. Biol. Chem. 275:13863-13871. [DOI] [PubMed] [Google Scholar]
- 57.Wu, S. W., and H. De Lencastre. 1999. Mrp: a new auxiliary gene essential for optimal expression of methicillin resistance in Staphylococcus aureus. Microb. Drug Resist. 5:9-18. [DOI] [PubMed] [Google Scholar]
- 58.Ziebrandt, A., H. Weber, J. Rudolph, R. Schmid, D. Höper, S. Engelman, and M. Hecker. 2001. Extracellular proteins of Staphylococcus aureus and the role of SarA and σB. Proteomics 1:480-493. [DOI] [PubMed] [Google Scholar]