Abstract
Most illnesses caused by Shiga toxin-producing Escherichia coli (STEC) have been attributed to E. coli serotype O157:H7, but non-O157 STEC infections are now increasingly recognized as public health problems worldwide. The O121:H19 serotype is being isolated more frequently from clinical specimens and has been implicated in one waterborne outbreak. We used multilocus virulence gene profiling, a PCR-based assay, to characterize the virulence gene content of 24 isolates of serotype O121:H19 and nonmotile variants. We also performed multilocus enzyme electrophoresis and multilocus sequencing to establish the clonal relatedness of O121 isolates and to elucidate the relationship of O121 to common STEC clones. The 24 isolates were found to represent a single bacterial clone, as there was no allelic variation across 18 enzyme loci among the isolates. The complete nucleotide sequence of the intimin gene differed by four substitutions from that of the epsilon (Int-ɛ) allele of O103:H2 strain PMK5. The typical O121 virulence gene profile was similar to the profiles of enterohemorrhagic E. coli (EHEC) clones of E. coli: it included a Shiga toxin 2 gene (stx2), two genes on the EHEC plasmid (toxB and ehxA), and the gene encoding intimin (eae). Despite the similarities, putative virulence genes distributed on O islands—large chromosomal DNA segments present in the O157:H7 genome—were useful for discriminating among STEC serotypes and the O121:H19 clone had a composite profile that was distinct from the profiles of the other major EHEC clones of pathogenic E. coli. On the basis of sequencing analysis with 13 housekeeping genes, the O121:H19 clone did not fall into any of the four classical EHEC and enteropathogenic E. coli groups but instead was closely related to two eae-negative STEC strains.
Shiga toxin-producing Escherichia coli (STEC) bacteria are infectious pathogens that have been implicated in outbreaks and sporadic cases of food- and waterborne diarrheal diseases in humans. The clinical symptoms caused by STEC infection include nonbloody diarrhea, hemorrhagic colitis (HC), and the hemolytic-uremic syndrome (HUS) (25, 33). The first recorded STEC outbreak (in 1982) was attributed to E. coli O157:H7 (28), a serotype that still accounts for the greatest proportion of STEC disease in North America, Europe, and Japan. However, more than 200 serotypes have been isolated from humans and non-O157 STEC strains may account for 20 to 70 percent of STEC infections overall in different countries (42). Pathogenic non-O157 STEC bacteria are unlike those of the O157:H7 serotype in that they usually ferment sorbitol, so they are more difficult to differentiate from commensal E. coli; thus, the incidence of disease caused by non-O157 STEC strains is more difficult to determine (35).
The defining characteristic of STEC is the production of Shiga toxins (Stx), a family of bacteriophage-encoded cytotoxins with diverse effects on eukaryotic cells (18, 20). There are two main antigenic types of Stx, Stx1 and Stx2, whose genes are highly divergent at the nucleotide level (40), and STEC strains may produce either or both toxin variants. Members of a subset of STEC strains called enterohemorrhagic E. coli (EHEC) effect a particular pathology called attaching and effacing (A/E) lesions, which are mediated in part by the adhesin intimin (16). Intimin is encoded by the eae gene on a pathogenicity island (the locus of enterocyte effacement [LEE]) whose presence is widespread among enteric bacterial pathogens (14). Other virulence elements are found in STEC strains, but their role in pathogenesis is unclear. For example, an enterohemolysin (ehxA) resides on a large plasmid (pO157) found in E. coli O157:H7 strains, variants of which are distributed among STEC strains of a variety of serotypes (3). The pO157 plasmid also encodes other factors that may contribute to virulence, including a type II secretion system (29) and accessory adhesins (24, 34, 36). Other virulence gene candidates were uncovered when the genomes of two O157:H7 strains (7, 26) were compared to the genome for the nonpathogenic E. coli strain K-12. The comparison revealed large chromosomal segments called O islands that were not present in the K-12 genome (26), and many of these islands contain DNA homologous to virulence factors from other bacterial pathogens. For example, O island 48 encodes an operon for urease activity, a mechanism for acid resistance in Yersinia and Helicobacter. The extent of occurrence of various O islands and their associated genes in non-O157 STEC strains is presently unknown, and their role in virulence remains to be defined.
Detecting in STEC strains the presence of the accessory virulence genes described above could provide information that is clinically relevant and of epidemiological value (23). For the present investigation, we used multilocus virulence gene profiling (MVGP)—a PCR assay of 15 potential virulence loci—to characterize the diversity and gene content of E. coli O121:H19, an STEC serotype that has been implicated in a waterborne outbreak of HUS (13) and is one of the more common non-O157 serotypes found in sporadic cases of STEC infection (39). We also employed multilocus enzyme electrophoresis (MLEE) to establish the clonal relationships among the O121 isolates. Little is known about the genetic relatedness of E. coli O121:H19 to other serotypes that cause HC and HUS. To address this issue, we used multilocus sequencing of housekeeping genes to determine the evolutionary relationships of the O121:H19 clone to other pathogenic clones of E. coli.
MATERIALS AND METHODS
Bacterial isolates.
A total of 27 of the strains of serogroup O121 that we analyzed were originally isolated from sporadic cases of diarrheal disease. Seventeen strains originated in Montana and were isolated between 1998 and 2000 (11). These included 13 strains of serotype O121:H19 (TW06388, TW08023, TW08032, TW08043, TW08055, TW08062, TW08063, TW08067, TW08068, TW08082, TW08091, TW08096, and TW08105) and 4 for which the flagellar antigen was not determined (TW08036, TW08039, TW08040, and TW08042). An additional five nonmotile strains were included: PT91-4 (TW03508; Washington, 1989), DA-1 (TW07927; District of Columbia, 1998), DA-5 (TW07931; Massachusetts, 1998), DA-37 (TW07972; Ohio, 1998), and DA-69 (TW08004; Ohio, 1998). One O121:H19 isolate, 3056-85, was obtained from the Centers for Disease Control and Prevention (Oklahoma, 1985). For comparisons in the MLEE analysis, we included two strains of serotype O121:H7 that were isolated from cows (90-0364 and 403-3; Canada, 1992) and ECOR 21, a nonmotile strain isolated from a steer (19). The phylogenetic analysis included 20 pathogenic strains previously characterized for multiple gene sequences (27) and the O103:H2 strain TW08101 (Montana, 2000). Additional strains included for comparisons in the MVGP analysis were as follows: O26:H11 strains TW08100 and TW08104 (Montana, 2000); O103:H2 strains TW07877 (Germany, 1998), TW07882 (Washington, 1999), and PMK5 (21); O145:NM strain TW08087 (Montana, 2000); O111:H8 strains 3214-99 and 3215-99 (Texas, 1999); O157:H7 strains Sakai (7) and EDL-933 (26); and the strain K-12 (2), which was used as a negative control. More information about the strains included in this study can be obtained from the STEC Center website (http://www.shigatox.net).
Enzyme electrophoresis.
Protein extracts were obtained by lysing bacterial cells and were subjected to horizontal starch gel electrophoresis and staining, following an established methodology (31). A total of 18 enzymes were analyzed for mobility variants: PGI, IDH, ACO, G3P, PE2, AK, MDH, PGD, M1P, GOT, ADH, MPI, G6P, CAK, NSP, TDH, SKD, and GLU. A multilocus genotype was determined for each strain, and distinct genotypes were classified as electrophoretic types (ETs) as previously described (41).
PCR template preparation.
Luria-Bertani agar plates were inoculated from freezer stocks and incubated overnight at 37°C. Genomic DNA was extracted from single colonies with a Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.). Templates for MVGP were produced by suspending single colonies in 200 ml of 1× TE buffer (10 mM Tris, 1 mM EDTA [pH 7.0]). The suspension was heated to 95°C for 10 min and centrifuged for 2 min. For each strain, three isolated colonies were processed and analyzed by MVGP.
PCR assays for virulence factors and O island genes.
The strains were analyzed by PCR with primers specific for a total of 15 genes that potentially contribute to virulence (Table 1). Nine genes have been found on O islands in the O157:H7 genome; these include the well-characterized virulence factors stx1 (O island 93), stx2 (O island 45), and intimin (O island 148). Three genes occur on O island 48, including a gene involved in tellurite resistance (terC), a locus of the urease gene complex (ureA), and an adherence-conferring gene (iha) encoding a protein homologous to iron-regulated IrgA of Vibrio cholerae (34). Two genes occur on O island 115 and have homology to elements of the Inv-Spa system of Salmonella (spaP and invG), and a homologue of the Shigella enterotoxin ShET2 (Z4326) on O island 122 was also included. Lastly, we screened for two genes (fyuA and irp2) on the high-pathogenicity island (HPI) of Yersinia, the presence of which has been reported for EHEC O26:H11 strains (10). We also screened for four plasmid-borne genes, including ehxA, the bundlin gene (bfpA) of enteropathogenic E. coli (EPEC), and two recently described adhesins, toxB (36, 38) and saa (23). PCR amplification of a conserved housekeeping gene (mdh) was used as a positive locus control. Previously published primer sequences were used for ehxA (8), saa (27), and the two HPI genes (34), except the forward primer for fyuA, which was redesigned (Table 1).
TABLE 1.
Primers and amplicon sizes for loci included in the PCR assay
| Locus | Primer designation | Nucleotide sequence of primer | Amplicon size(s) (bp) |
|---|---|---|---|
| bfpA | bfpA-F11 | 5′-GTCTGCGTCTGATTCCAATA-3′ | 408, 414 |
| bfpA-R1 | 5′-TCAGCAGGAGTAATAGC-3′ | ||
| toxB | toxB.911F | 5′-ATACCTACCTGCTCTGGATTGA-3′ | 602 |
| toxB.1468R | 5′-TTCTTACCTGATCTGATGCAGC-3′ | ||
| stx1 | 1A.251F | 5′-GGGATAGATCCAGAGGAAGG-3′ | 624 |
| 1A.832R | 5′-CCGGACACATAGAAGGAAACTC-3′ | ||
| stx2 | 2A.506F | 5′-CTGGCGTTAATGGAGTTCAG-3′ | 383 |
| 2A.848R | 5′-CCTGTCGCCAGTTATCTGAC-3′ | ||
| eae | eae-F1 | 5′-ACTCCGATTCCTCTGGTGAC-3′ | 389 |
| eae-R3 | 5′-TCTTGTGCGCTTTGGCTT-3′ | ||
| terC | terC.106F | 5′-TATGCACCGTGATGACAAGC-3′ | 210 |
| terC.275R | 5′-GGCGAACCAGGAGAAGATTG-3′ | ||
| ureA | ureA.109F | 5′-TAACTATCCCGAATCCGTGG-3′ | 146 |
| ureA.213R | 5′-GGGATCATTTCTGGTATGCCT-3′ | ||
| iha | iha.138F | 5′-ACGCAGCCGCCAGTGTT-3′ | 186 |
| iha.282R | 5′-CCATCAATCAGTATCAGCGTGTAA-3′ | ||
| invG | invG.481F | 5′-TGACCTGGTCGTTAATGCTG-3′ | 133 |
| invG.572R | 5′-CGCCACGTAACTCATAAGTCC-3′ | ||
| spaP | spaP.143F | 5′-GGACTTCAGCAAGTGCCATC-3′ | 173 |
| spaP.275R | 5′-ACCACTCATGCCTGTCTCAA-3′ | ||
| Z4326 | ShET.1137F | 5′-TCATGCAGACGCAATAAAGG-3′ | 125 |
| ShET.1221R | 5′-GATACGCAAGAAGTAATCCTGG-3′ | ||
| fyuA | fyuA.924F | 5′-GCAGCAGCAGCATTATTCG-3′ | 495 |
| mdh | mdh.269F | 5′-GGTATGGATCGTTCCGACCT-3′ | 304 |
| mdh.530R | 5′-GGCAGAATGGTAACACCAGAGT-3′ |
Synthesized phosphoramidite dye-linked primers (Research Genetics, Huntsville, Ala.) were used at a final concentration of 0.5 μM in a 20-μl reaction mixture with 1 U of AmpliTaq Gold (Applied Biosystems, La Jolla, Calif.) and final concentrations of deoxynucleoside triphosphates of 0.2 mM each and MgCl2 of 2.5 mM. The loci were simultaneously amplified in separate reactions with the following thermal cycle, which was preceded by a 10-min soak at 94°C and run for 35 cycles: 92°C for 30 s, 55°C for 30 s, and 72°C for 45 s. The cycle was followed by a final extension step of 72°C for 7 min. Each run included two negative controls (including DNA from E. coli strain K-12 and no template) and a positive control that was a mixture of strains. Reaction aliquots were pooled for each strain, and the mix was desalted using Multiscreen-HV plates (Millipore, Bedford, Mass.) and a Sephadex G-50 column (Amersham Pharmacia Biotech, Piscataway, N.J.). The amplicon mix was dried under vacuum and suspended in formamide with 0.5 μl of DNA size standard 600 (Beckman Coulter, Fullerton, Calif.). The dye-labeled fragments were electrophoresed for 1 h at 6.0 kV and detected on a Beckman CEQ8000 capillary sequencer. Fragment sizes were calculated automatically, and the presence or absence of fragments was scored for each chromatogram with Fragment Analysis software, version 5.0 (Beckman Coulter).
Intimin allele typing.
We determined the nucleotide sequence for the entire eae gene in O121:H19 strain TW08023 by following previously published protocols (32). The program ClustalX (37) was used to align the O121 eae sequence to 10 major eae sequences retrieved from GenBank. The relationship of the O121 sequence to those of other intimin alleles was determined by constructing phylogenetic trees with the program MEGA2 (12) by using the neighbor-joining algorithm. Because the mosaic segments of intimin genes can have different evolutionary histories, eae sequences were partitioned into a conserved central domain (CCD) (residue 187 through 517) and the extracellular (EC) region (amino acid positions 559 to 951) for phylogenetic analysis (15, 32). Two eae sequencing primers (1460F [5′-GGT CTG GAT CGT ATC GTC TG-3′] and 2009R [5′-ATC AGA ACC ATT TGC CTT CG-3′]) were used to amplify a 590-bp fragment, and the sequence was determined in one direction with primer 1460F.
Evolutionary relationships of the O121 clone.
We sequenced ∼500 bp of each of 13 housekeeping genes (arcA, aroE, aspC, clpX, cyaA, dnaG, fadD, grpE, icdA, lysP, mdh, mtlD, and rpoS) for two O121 isolates (TW08023 and TW07927) and an O103:H2 strain (TW08101) and compared the multilocus sequences to those of 20 pathogenic E. coli strains (27). The total length sequenced for each of the gene fragments ranged from 288 bp (aroE) to 561 bp (clpX), with a total of 6,228 bp sequenced across the thirteen loci. Primer sequences and details of the methodology will be published elsewhere. The program ClustalX was used to align the sequences. The 13 gene sequences were concatenated for each strain, and phylogenetic trees were constructed with the program MEGA2 (12). The trees were based on synonymous distances calculated by using the modified Nei-Gojobori method with the Jukes-Cantor correction (17). Bootstrap confidence values for each node of the tree were calculated over 1,000 replicate trees.
RESULTS
MLEE.
A total of three ETs were observed among the 27 strains of the O121 serogroup. All ETs were equally distant from one another, each differing from the other by 8 out of 18 alleles. ECOR 21 was distinct from all other isolates and was designated ET 1. The two O121:H7 strains were identical to each other and were designated ET 2. The remaining O121 strains, including all that express the H19 flagellar antigen, comprised ET 3. We detected no genetic variation within ET 3 at the level of enzyme mobility, which indicates a very close genetic relationship among all O121:H19 strains and related nonmotile variants.
MVGP.
Strains that produce only Stx2 are associated with serious complications, such as HUS, more often than strains that produce either Stx1 or both toxins (22). Of the 24 isolates that we analyzed, all but one (TW03508) tested positive for stx and the 23 positive strains all carried the stx2 gene. One strain (TW08032) also carried stx1.
Proteins encoded by the large EHEC plasmid are thought to contribute to the virulence of STEC strains. We screened for two genes within the EHEC plasmid (ehxA and toxB). The two genes always occurred together, and both genes were present in 22 of the ET 3 O121 isolates. Two ET 3 isolates (TW08032 and TW08042) were negative for both genes, suggesting that they lack the pO157 plasmid. None of the O121 strains was positive for the bundlin gene (bfpA), the presence of which is a characteristic of the EPEC pathotype, and all O121 strains also lacked saa, a gene contained within a large plasmid and usually found in eae-negative STEC strains.
The presence of the LEE island is associated with the ability to cause A/E lesions and is one of the major virulence determinants by which some diarrheagenic E. coli strains cause disease in humans (16). We screened for the LEE island with primers placed in the CCD of the eae gene and detected a copy of eae in each of the 24 isolates. The results of amplification and sequencing of three additional genes (sepZ, tir, and sepL) in one isolate (TW08023) suggested that the O121:H19 clone harbors a complete copy of the LEE pathogenicity island (data not shown).
All ET 3 O121 strains were positive for terC and ureA, two genes found on O island 48 of the E. coli O157:H7 genome (Table 2). This observation suggests that the O121:H19 clone has an O island 48 homologue, although the island differs from that of E. coli O157:H7 because it lacks the iha gene. All ET 3 strains were also positive for Z4326, the gene encoding the ShET2 homologue of O island 122 in E. coli O157:H7. Neither the invG or spaP gene of O island 115 nor either of the two genes (fyuA and irp2) of the HPI (Table 2) was detected in the O121:H19 clone.
TABLE 2.
Molecular virulence gene profiles of E. coli isolates
| Serotype (reference isolate) | No. of isolates | Locus
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bfpA | ehxA | toxB | saa | stx1 | stx2 | eae | terC | ureA | iha | invG | spaP | Z4326 | irp2 | fyuA | ||
| O121:H19 (TW08023) | 13 | − | + | + | − | − | + | + | + | + | − | − | − | + | − | − |
| O121:H19 (TW08032) | 1 | − | − | − | − | + | + | + | + | + | − | − | − | + | − | − |
| O121:NM (DA-1) | 5 | − | + | + | − | − | + | + | + | + | − | − | − | + | − | − |
| O121:NM (PT91-4) | 1 | − | + | + | − | − | − | + | + | + | − | − | − | + | − | − |
| O121:ND (TW08039) | 3 | − | + | + | − | − | + | + | + | + | − | − | − | + | − | − |
| O121:ND (TW08042) | 1 | − | − | − | − | − | + | + | + | + | − | − | − | + | − | − |
| O113:H21 (CL-3) | 1 | − | + | − | + | − | + | − | − | − | + | − | + | − | − | − |
| XO3:NM (90-1787) | 1 | − | + | − | + | − | + | − | − | − | + | − | + | − | − | − |
| O103:H2 (TW08101) | 2 | − | + | + | − | + | − | + | − | − | − | − | + | + | − | − |
| O103:H2 (PMK5) | 2 | − | + | − | − | + | − | + | − | − | − | − | + | + | − | − |
| O26:H11 (TW08100) | 1 | − | + | + | − | + | − | + | + | + | + | − | + | + | + | + |
| O111:H8 (3215-99) | 2 | − | + | − | − | + | + | + | + | + | + | − | + | + | − | − |
| O145:NM (TW08087) | 1 | − | + | + | − | + | − | + | + | + | + | + | − | + | − | − |
| K-12 | 1 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| O127:H6 (E2348/69) | 1 | + | − | − | − | − | − | + | − | − | − | − | − | + | − | − |
| O157:H7 (EDL-933) | 3 | − | + | + | − | + | + | + | + | + | + | + | + | + | − | − |
Evolutionary relationship of the O121:H19 clone to other EHEC strains.
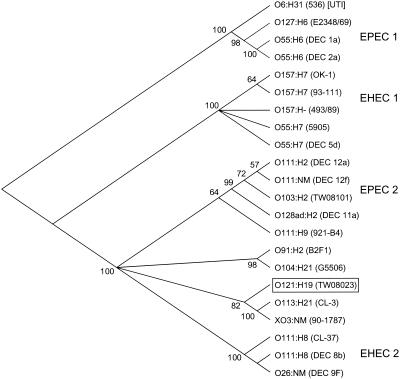
We sequenced segments of 13 housekeeping genes of two ET 3 O121 strains (TW08023 and TW07927) and an STEC O103:H2 strain (TW08101), because O121 and O103 have been reported to have similar intimin sequences (21). The multilocus sequences were aligned and concatenated with homologous sequences from 20 pathogenic strains representing the four groups of major EPEC and EHEC clones (27). There were a total of 281 polymorphic sites among the 23 strains, and the sequences of the two O121 isolates were identical. In the bootstrap analysis of the combined sequences (Fig. 1), the O121:H19 clone did not cluster significantly with either EHEC 1 group (O157:H7) or EHEC 2 group (O26 and O111). The consensus phylogeny indicates that the O121:H19 clonal frame is not significantly allied with STEC O103:H2, clusters at a distant node with the EPEC 2 and EHEC 2 groups, and is most closely related (82% support) to two eae-negative STEC strains (serotypes O113:H21 and XO3:NM).
FIG. 1.
Gene phylogeny inferred from 13 housekeeping gene sequences. Trees were constructed from synonymous distances by using the neighbor-joining algorithm. The number at each branching point indicates the percentage of trees that support a particular node out of 1,000 bootstrapped replicates.
Intimin allele of the O121:H19 clone.
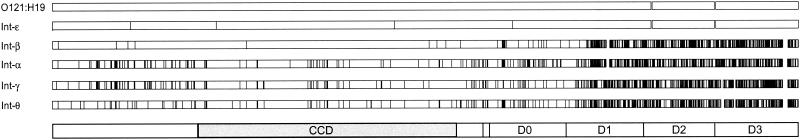
The sequence for the eae gene of O121:H19 strain TW08023 was 2,847 nucleotides in length, which is indicative of a protein of 948 amino acids. A gene phylogeny based on sequences of the CCD indicates that the intimin allele of the O121:H19 clone is virtually the same as the epsilon allele (Int-ɛ) of E. coli O103:H2 (Fig. 2). There are only four nucleotide differences between the two sequences (Fig. 3): two nonsynonymous substitutions (H209R in the CCD and N587D in domain 0 [D0]) and two synonymous substitutions. To confirm this finding, we used primers flanking the replacement location in D0 and found that all 24 isolates of the O121:H19 clone have the Int-ɛ variant allele. Interestingly, Int-ɛ is most similar to Int-β (found in EHEC 2 O26:H11 strains) in the transmembrane region, where the divergence at the amino acid level is less than 1%, but is more than 30% divergent in the EC domains (Fig. 3). Overall, the level of amino acid divergence between Int-ɛ and other intimin alleles ranges from 34% to 40% in the EC domains (Fig. 3).
FIG. 2.
Phylogeny of intimin alleles inferred from the CCD. Trees were constructed from synonymous distances by using the neighbor-joining algorithm. The scale at the lower left shows the horizontal distance that corresponds to two synonymous substitutions per 100 synonymous sites.
FIG. 3.
Plot of the variable nucleotide sites in intimin alleles. Each vertical line represents a site that differs between the O121: H19 epsilon sequence and the allele labeled in the figure. The domain structure of intimin is shown at the bottom of the figure, with the CCD shaded in gray.
DISCUSSION
A key observation from this study is that E. coli O121:H19 represents an independent clone that has acquired particular virulence factors in parallel with the EHEC lineages. The O121:H19 clone has in common with bacteria of the EHEC 1 group (O157:H7 and relatives) and EHEC 2 group (O26, O111, and relatives) at least three virulence characteristics: a Shiga toxin gene, an intimin allele, and genes found on the EHEC plasmid (pO157). However, the phylogenetic tree constructed from the housekeeping gene sequences places the O121:H19 clonal frame on a separate branch distinct from the EHEC 1 and EHEC 2 lineages. Similarly, the EHEC 1 and EHEC 2 clones are not closely related to each other but instead have evolved in parallel through a repeated series of gene acquisitions (27). The results presented here support the hypothesis that E. coli O121:H19 represents a third EHEC clone that has independently accumulated the virulence genes that contribute to the EHEC pathovar.
The comparison of E. coli strain O121:H19 to strains of the other major EHEC serotypes showed that the fragment profiles of the major EHEC serotypes differed in composition when all of the genes were examined. There were few genes that could serve as diagnostic markers for a single clone or serotype (Table 2). The exception was the presence of the genes on the HPI (irp2 and fyuA), which differentiates bacteria of the EHEC O26:H11 clone from those of the other serotypes included here. The HPI is prevalent in E. coli (30) but thus far has not been found in EHEC serotypes other than O26:H11 strains (10). Bacterial clones associated with other serotypes, including O121:H19, were differentiated on the basis of the composite profile rather than that of a single diagnostic gene. It appears that the genomic island markers are more stable than plasmid or phage markers; the latter tended to differ within a serotype and occasionally among the single-colony picks for each strain. The loss of plasmids and phages during subculture is documented (9, 16) and could explain in part the variation in the presence of EHEC plasmid and Stx phage genes within a serotype. The stability of the O island genes makes them useful for differentiating STEC serotypes by PCR.
Intimin and other genes on the LEE pathogenicity island have been shown to be crucial for the full virulence of enteropathogenic E. coli (5), as they confer upon bacterial strains the ability to cause A/E lesions in humans. The intimin gene of the O121:H19 clone is nearly identical in sequence to Int-ɛ of E. coli O103:H2, a strain of the EPEC 2 clonal group (members of which typically carry the Int-β allele) (Fig. 1). Int-ɛ is highly similar to Int-β in the 5′ half of the gene but is highly divergent in the region encoding the EC domains (Fig. 2); it is possible that segments of Int-β and Int-ɛ are ancestral and derive from a similar LEE backbone. To address this hypothesis, we sequenced sepL (a gene downstream of eae on the LEE island) from O121:H19 strain TW08023 and from O103:H2 strain TW08101. The sepL sequences are identical and form a tight cluster (average divergence, 0.4%) with the sepL sequences from strains that carry Int-β (data not shown). These results suggest that the O121:H19 and O103:H2 clones carry a LEE that is closely related by recent descent to the LEE of EPEC2 strains but one lineage of the LEE has undergone recombination with a divergent sequence that converted the 3′ half of the gene.
The O121:H19 clone comprises a single ET, as no polymorphic loci were detected by MLEE. Additional ETs might be present within the clone but were not detected within our limited sample. Even so, E. coli O121:H19 appears to be more homogeneous than other serotypes. In the study by Whittam et al. (41), six serotypes had sample sizes comparable to that of our study (excluding nonmotile strains); among these serotypes, two to five ETs were detected in samples of 10 to 31 isolates and 5 to 70% of the loci were polymorphic. Also, the average genetic diversity (H)—a measure that is not correlated with sample size—was lowest in O121:H19 (where it was zero). The H averaged across 11 serotypes was 0.107, with a range of 0.005 to 0.183. As a comparison, O157:H7 has an H of 0.008 and 95% of the isolates belong to the same ET. The low level of diversity in O121:H19 indicates a relatively recent emergence of this clone from a single ancestral cell.
Recognition of the impact of non-O157 STEC strains on human health is increasing worldwide, but unlike O157:H7, these strains are difficult to differentiate from commensal E. coli. Thus, the prevalence of non-O157 STEC is presently underestimated (8, 35). In 1998, the World Health Organization cited O26, O111, O103, and O145 as the most important non-O157 STEC pathogens in humans. Some of the serotypes appear to have emerged more recently than O157:H7. For example, prior to 1995, O103 was not detected by routine surveillance in a lab in Germany and accounted for less than 2% of the STEC infections in Italy (4). However, in 1996, the frequency of detection of O103 had increased to 18.2% in a German laboratory and accounted for 11% of the HUS cases in Italy in 1996 (4). Similarly, the frequency of infection by O26 STEC increased dramatically in 1996 in Germany and Italy (4).
The O121:H19 clone also may be on a trajectory of emergence and spread. In 1996, Beutin and colleagues reported that among 89 non-O157 STEC strains implicated in human disease in Germany, 40% of the strains were of common STEC serotypes (O26, O103, O111, and O145) but none of the remaining isolates belonged to the O121 serogroup (1). Fey et al. (6) examined 335 specimens from patients in Nebraska in 1998 and isolated four non-O157 serotypes, but there were no cases of disease in which an STEC O121 strain was implicated (6). More recently, however, cases of HC and HUS have been attributed to E. coli O121:H19. In 1999, bacteria of this serotype were implicated as the cause of a waterborne outbreak of HC in Connecticut (13). Also, a characterization of 67 EHEC strains from Swedish patients collected between 1997 and 1999 (39) found that about one-third of the strains were non-O157 and that of these, 17% were O121:H19. In a surveillance study of STEC strains in diarrheal disease, members of the O121:H19 clone group were the third most common STEC serotype (11), accounting for ∼25% of the cases of diarrheal disease caused by non-O157 STEC infections (P. I. Tarr, unpublished data). The molecular data and the recent increase in the incidence of O121:H19 infections suggest that this clone is an emerging pathogen, and surveillance methods should be able to detect this as well as other non-O157 STEC serotypes.
Acknowledgments
This research was supported by the National Institutes of Health (grant AI-47499) (P.I.T.) and the Enteric Pathogen Research Unit (grant N01-AI-65299) at the University of Maryland Medical School.
J. Paton kindly provided primer sequences for saa prior to publication.
Editor: J. T. Barbieri
REFERENCES
- 1.Beutin, L., D. Geier, S. Zimmermann, S. Aleksic, H. A. Gillespie, and T. S. Whittam. 1997. Epidemiological relatedness and clonal types of natural populations of Escherichia coli strains producing Shiga toxins in separate populations of cattle and sheep. Appl. Environ. Microbiol. 63:2175-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glassner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 3.Brunder, W., H. Schmidt, M. Frosch, and H. Karch. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145:1005-1014. [DOI] [PubMed] [Google Scholar]
- 4.Caprioli, A., A. E. Tozzi, G. Rizzoni, and H. Karch. 1997. Non-O157 Shiga toxin-producing Escherichia coli infections in Europe. Emerg. Infect. Dis. 3:578-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnenberg, M. S., and J. B. Kaper. 1992. Enteropathogenic Escherichia coli. Infect. Immun. 60:3953-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fey, P. D., R. S. Wickert, M. E. Rupp, T. J. Safranek, and S. H. Hinrichs. 2000. Prevalence of non-O157:H7 Shiga toxin-producing Escherichia coli in diarrheal stool samples from Nebraska. Emerg. Infect. Dis. 6:530-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 8.Jaeger, J. L., and D. W. Acheson. 2000. Shiga toxin-producing Escherichia coli. Curr. Infect. Dis. Rep. 2:61-67. [DOI] [PubMed] [Google Scholar]
- 9.Karch, H., T. Meyer, H. Rüssmann, and J. Heesemann. 1992. Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect. Immun. 60:3464-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karch, H., S. Schubert, D. Zhang, W. Zhang, H. Schmidt, T. Olschlager, and J. Hacker. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect. Immun. 67:5994-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein, E. J., J. R. Stapp, C. R. Clausen, D. R. Boster, J. G. Wells, X. Qin, D. L. Swerdlow, and P. I. Tarr. 2002. Shiga toxin-producing Escherichia coli in children with diarrhea: a prospective point-of-care study. J. Pediatr. 141:172-177. [DOI] [PubMed] [Google Scholar]
- 12.Kumar, S., K. Tamura, I. Jakobsen, and M. Nei. 2000. MEGA 2: molecular evolutionary genetics analysis program. Version 2.0. Pennsylvania State University, University Park, Pa.
- 13.McCarthy, T. A., N. L. Barrett, J. L. Hadler, B. Salsbury, R. T. Howard, D. W. Dingman, C. D. Brinkman, W. F. Bibb, and M. L. Cartter. 2001. Hemolytic-uremic syndrome and Escherichia coli O121 at a lake in Connecticut, 1999. Pediatrics 108:E59. [Online.] http://www.ncbi.nlm.nih.gov. [DOI] [PubMed] [Google Scholar]
- 14.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGraw, E. A., J. Li, R. K. Selander, and T. S. Whittam. 1999. Molecular evolution and mosaic structure of α, β, and γ intimins of pathogenic Escherichia coli. Mol. Biol. Evol. 16:12-22. [DOI] [PubMed] [Google Scholar]
- 16.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, N.Y.
- 18.O'Brien, A. D., V. L. Tesh, A. Donohue-Rolfe, M. P. Jackson, S. Olsnes, K. Sandvig, A. A. Lindberg, and G. T. Keusch. 1992. Shiga toxin: biochemistry, genetics, mode of action, and role in pathogenesis. Curr. Top. Microbiol. Immunol. 180:65-94. [DOI] [PubMed] [Google Scholar]
- 19.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Loughlin, E. V., and R. M. Robins-Browne. 2001. Effect of Shiga toxin and Shiga-like toxins on eukaryotic cells. Microbes Infect. 3:493-507. [DOI] [PubMed] [Google Scholar]
- 21.Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marches, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paton, A. W., and J. C. Paton. 2002. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J. Clin. Microbiol. 40:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paton, A. W., E. Voss, P. A. Manning, and J. C. Paton. 1998. Antibodies to lipopolysaccharide block adherence of Shiga toxin-producing Escherichia coli to human intestinal epithelial (Henle 407) cells. Microb. Pathog. 24:57-63. [DOI] [PubMed] [Google Scholar]
- 25.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 27.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 28.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt, H., B. Henkel, and H. Karch. 1997. A gene cluster closely related to type II secretion pathway operons of gram-negative bacteria is located on the large plasmid of enterohemorrhagic Escherichia coli O157 strains. FEMS Microbiol. Lett. 148:265-272. [DOI] [PubMed] [Google Scholar]
- 30.Schubert, S., A. Rakin, H. Karch, E. Carniel, and J. Heesemann. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66:480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. H. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarr, C. L., and T. S. Whittam. 2002. Molecular evolution of the intimin gene in O111 clones of pathogenic Escherichia coli. J. Bacteriol. 184:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarr, P. I. 1995. Escherichia coli O157:H7: clinical, diagnostic, and epidemiological aspects of human infection. Clin. Infect. Dis. 20:1-10. [DOI] [PubMed] [Google Scholar]
- 34.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarr, P. I., and M. A. Neill. 1996. Perspective: the problem of non-O157:H7 Shiga toxin (Verocytotoxin)-producing Escherichia coli. J. Infect. Dis. 174:1136-1139. [DOI] [PubMed] [Google Scholar]
- 36.Tatsuno, I., M. Horie, H. Abe, T. Miki, K. Makino, H. Shinagawa, H. Taguchi, S. Kamiya, T. Hayashi, and C. Sasakawa. 2001. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tobe, T., T. Hayashi, C. G. Han, G. K. Schoolnik, E. Ohtsubo, and C. Sasakawa. 1999. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect. Immun. 67:5455-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welinder-Olsson, C., M. Badenfors, T. Cheasty, E. Kjellin, and B. Kaijser. 2002. Genetic profiling of enterohemorrhagic Escherichia coli strains in relation to clonality and clinical signs of infection. J. Clin. Microbiol. 40:959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittam, T. S. 1998. Evolution of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains, p. 195-209. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 41.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Ørskov, I. Ørskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.World Health Organization. 1999. Zoonotic non-O157 Shiga toxin-producing Escherichia coli (STEC). Report of a WHO Scientific Working Group meeting, 23-26 June 1998, Berlin, Germany. [Online.] http://www.who.int/emc-documents/zoonoses/whocsraph988c.html.