Abstract
The pathogenesis of acute rheumatic fever (ARF) is poorly understood. We identified two contiguous bacteriophage genes, designated speL and speM, encoding novel inferred superantigens in the genome sequence of an ARF strain of serotype M18 group A streptococcus (GAS). speL and speM were located at the same genomic site in 33 serotype M18 isolates, and no nucleotide sequence diversity was observed in the 33 strains analyzed. Furthermore, the genes were absent in 13 non-M18 strains tested. These data indicate a recent acquisition event by a distinct clone of serotype M18 GAS. speL and speM were transcribed in vitro and upregulated in the exponential phase of growth. Purified SpeL and SpeM were pyrogenic and mitogenic for rabbit splenocytes and human peripheral blood mononuclear cells in picogram amounts. SpeL preferentially expanded human T cells expressing T-cell receptors Vβ1, Vβ5.1, and Vβ23, and SpeM had specificity for Vβ1 and Vβ23 subsets, indicating that both proteins had superantigen activity. SpeL was lethal in two animal models of streptococcal toxic shock, and SpeM was lethal in one model. Serologic studies indicated that ARF patients were exposed to serotype M18 GAS, SpeL, and SpeM. The data demonstrate that SpeL and SpeM are pyrogenic toxin superantigens and suggest that they may participate in the host-pathogen interactions in some ARF patients.
Group A streptococcus (GAS) is a human pathogen that causes pharyngitis, necrotizing fasciitis, streptococcal toxic shock syndrome (STSS), and other diseases. GAS is also responsible for the postinfection sequela acute rheumatic fever (ARF), the leading cause of preventable childhood heart disease worldwide (7, 26). ARF and severe invasive disease have reemerged in the United States and elsewhere in the last 15 years, but the cause of the resurgence is unknown (26). Hence, continued research into the molecular mechanisms of pathogenesis and characterization of new proteins that mediate human-GAS interactions is warranted.
Streptococcal pyrogenic exotoxins are thought to contribute to the pathogenesis of some GAS diseases, including scarlet fever, STSS, and ARF (7, 24). Streptococcal pyrogenic exotoxins and related exotoxins made by Staphylococcus aureus belong to a family of molecules known as pyrogenic toxin superantigens (PTSAgs). Amino acid sequence comparisons and crystal structure analyses have shown that PTSAgs have several conserved features, including an N-terminal β-barrel domain and some contain a C-terminal zinc-binding motif (23). PTSAgs simultaneously bind major histocompatibility complex (MHC) class II molecules and T-cell receptors (TCRs). This results in proliferation of T cells with specific variable regions of the β-chain (Vβ) on their TCRs and the subsequent release of inflammatory cytokines. This process is thought to contribute to the high fever and shock observed in some patients with GAS infections and autoimmune-mediated sequelae, such as ARF (7). In addition, variation in speA and speC may contribute to temporal and geographic fluctuations in GAS disease frequency and severity (13, 27).
Recently, we sequenced the genome of strain MGAS8232, a serotype M18 strain recovered from a patient with ARF, and identified two genes encoding undescribed PTSAg homologs (33). These genes, designated speL and speM, are located contiguously in a prophage-like element. Inasmuch as the molecular pathogenesis of ARF and rheumatic heart disease are poorly understood and these genes are present in the genome of a strain recovered from an ARF patient, we studied several attributes of the genes and encoded proteins in the context of ARF.
MATERIALS AND METHODS
Identification of speL and speM.
Secretion signal sequences were identified with SignalP version 2.0 (www.cbs.dtu.dk/services/SignalP). A multiple sequence alignment of the predicted mature forms of GAS PTSAgs was constructed with FASTA (28) and CLUSTALW (37), and gaps were adjusted manually based on known toxin structures. A dendrogram of the putative secreted forms of GAS and staphylococcal PTSAgs was constructed with the neighbor-joining algorithm based on the gamma distance model by using MEGA2.1 software (http://www.megasoftware.net). Ribbon diagrams of SpeL and SpeM were generated with Molscript (16) by using SpeC as a reference.
DNA methods.
GAS strains (Table 1) were grown, and genomic DNA was isolated as described previously (25). To examine the genetic diversity of the speL-speM locus, a PCR product encompassing both open reading frames (ORFs) was generated with primers 1721R and 117 (Fig. 1A and Table 2) and the AdvanTaq Plus Kit (Clontech, Palo Alto, Calif.) as described by the manufacturer. The amplicon was sequenced with internal primers (Table 2) and an ABI 3700 instrument (Applied Biosystems, Inc., Foster City, Calif.). Sequence data were assembled and analyzed with Sequencher version 4.1 (Gene Codes Corp., Ann Arbor, Mich.). Strains that were PCR negative for the speL-speM locus with external primers 1721R and 117 were analyzed with internal primers by using Taq polymerase (Table 2). The PCR conditions used were as follows: initial denaturation at 94°C for 10 min; 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 3 min; and final extension at 72°C for 10 min.
TABLE 1.
GAS strains used in this study
| MGAS no.a | emm type | Source/yr of isolationb | Disease or characteristicc | Presence of speL/speMd |
|---|---|---|---|---|
| 300 | 18 | Washington/1986-1990 | Invasive | +/+ |
| 6157 | 18 | Texas/1998 | Invasive | +/+ |
| 8738 | 18 | Texas/2000 | Invasive | +/+ |
| 7629 | 18 | Illinois/1998 | Invasive | +/+ |
| 156 | 18 | Nebraska/1980s | TSLS | +/+ |
| 1286 | 18 | New York/unknown | ARF associated, LC 4RP104 | +/+ |
| 1582 | 18 | Ohio/1987 | ARF-associated | +/+ |
| 1583 | 18 | Colorado/1968 | Nonhemolytic RF | +/+ |
| 1585 | 18 | Illinois/1940s | RF | +/+ |
| 1856 | 18 | United Kingdom/1931 | RF; NCTC 8320 | +/+ |
| 8230 | 18 | Utah/1987 | ARF associated | +/+ |
| 8232 | 18 | Utah/1987 | ARF | +/+ |
| 7551 | 18 | Utah/1985 | Pharyngeal | +/+ |
| 7655 | 18 | Utah/1985 | Pharyngeal | +/+ |
| 7671 | 18 | Utah/1985 | Pharyngeal | +/+ |
| 7687 | 18 | Utah/1985 | Pharyngeal | +/+ |
| 8039 | 18 | Utah/1998-1999 | Pharyngeal | +/+ |
| 8056 | 18 | Utah/1998-1999 | Pharyngeal | +/+ |
| 7705 | 18 | Utah/1985 | Pharyngeal | +/+ |
| 8077 | 18 | Utah/1998-1999 | Pharyngeal | +/+ |
| 8100 | 18 | Utah/1998-1999 | Pharyngeal | +/+ |
| 8118 | 18 | Utah/1998-1999 | Pharyngeal | +/+ |
| 8138 | 18 | Utah/1998-1999 | Pharyngeal | +/+ |
| 666 | 18 | New York/1963 | Nasopharyngeal | +/+ |
| 667 | 18 | New York/1948 | Nasopharyngeal | +/+ |
| 668 | 18 | New York/1949 | Nasopharyngeal | +/+ |
| 669 | 18 | New York/1952 | Nasopharyngeal | +/+ |
| 7574 | 18 | Illinois/1998 | Pharyngeal | +/+ |
| 7612 | 18 | Illinois/1998 | Pharyngeal | +/+ |
| 9060 | 18 | Illinois/1998 | Pharyngeal | +/+ |
| 8253 | 18 | Lackland AFB/1999 | Pharyngeal | +/+ |
| 8314 | 18 | Lackland AFB/1999 | Pharyngeal | +/+ |
| 6505e | 18 | Lackland AFB/1993 | Carrier-Pharyngeal | +/+ |
| 8610 | 1 | Utah/1998-1999 | Pharyngeal | −/− |
| 8574 | 2 | Utah/1998-1999 | Pharyngeal | −/− |
| 8625 | 3 | Utah/1998-1999 | Pharyngeal | −/− |
| 8612 | 4 | Utah/1998-1999 | Pharyngeal | −/− |
| 8488 | 5 | Utah/1998-1999 | Pharyngeal | −/− |
| 8674 | 6 | Utah/1998-1999 | Pharyngeal | −/− |
| 8602 | 11 | Utah/1998-1999 | Pharyngeal | −/− |
| 8697 | 12 | Utah/1998-1999 | Pharyngeal | −/− |
| 8577 | 22 | Utah/1998-1999 | Pharyngeal | −/− |
| 8494 | 28 | Utah/1998-1999 | Pharyngeal | −/− |
| 8557 | 49 | Utah/1998-1999 | Pharyngeal | −/− |
| 8486 | 77 | Utah/1998-1999 | Pharyngeal | −/− |
| 8656 | 89 | Utah/1998-1999 | Pharyngeal | −/− |
Musser laboratory GAS strain number.
Strains from Utah were collected during ARF outbreaks.
Clinical diagnosis is provided when known. LC, Lancefield collection; RF, rheumatic fever; NCTC, National Collection of Type Cultures; TSLS, toxic shock-like syndrome.
PCR was used to detect speL and speM. The genes were subsequently sequenced.
Strain MGAS 6505 was obtained from an asymptomatic recruit enrolled in a study of GAS carriage at Lackland Air Force Base, San Antonio, Tex.
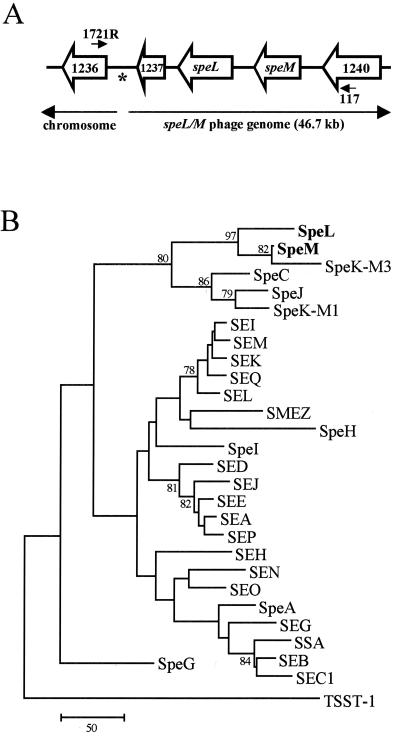
FIG. 1.
Schematic of the speL-speM locus and PTSAg phylogeny. (A) Open arrows denote the direction of transcription of ORFs encoding SpeL, SpeM, and hypothetical proteins in strain MGAS8232 (33). Lines between ORFs represent intergenic spaces. The annealing positions of primers 117 and 1721R are shown. An asterisk marks the position of a 98-bp direct repeat located at the ends of the prophage-like element. (B) The dendrogram was made with amino acid sequences corresponding to predicted mature forms of GAS and staphylococcal PTSAgs. Bootstrap confidence levels (based on 1,000 repetitions) of >70 are shown. The scale bar represents the number of amino acid substitutions per 100 sites, and summation of the entire branch length predicts the number of amino acid substitutions that have occurred along a given lineage.
TABLE 2.
Oligonucleotide primers used in this studya
| Name | Sequence (5′-3′) |
|---|---|
| 1721Rb | TTAAAGACATGTCCGTCAATCC |
| SpeL_Fb | ATGAGAATTTTTTTACACCAG |
| SpeL_Fcutb | ACCATGGGAAGAGACTATTAATATTAAGG |
| SpeL_Rb | CGAATTCATTTTCTTTGTTTGTGAAT |
| 117b | AAATGGCGTCATTTTCTCA |
| SpeM_Fb | ATGTTAGGAGAAAAAATG |
| SpeM_Fcutb | ACCATGGTTTTCAGATGCTGTGTTGG |
| SpeM_Rb | CGGATCCATTTTTAGAAAAATCTTCG |
| SpeL_8169b | ACATATTAACCAAACGACAATTC |
| SpeM_8952b | AGTTATATCTAGCAATGTTTCTCCA |
| Param147b | CCAGCGCAAGAAAGACG |
| Param112b | ACATATTAACCAAACGACAATTC |
| Param134b | AGTTATATCTAGCAATGTTTCTCCA |
| 1220Lb | CAGCACCTTCCTCTTTCTCG |
| 1220Rb | GGAAAAAGAGGGACGCAAG |
| G1295Lb | GGATGAGTGAATAAATCGGTAAAC |
| G5-358D2b | CTTTTGACTGATAGTGACAAGCTG |
| SBBP4004.2b | CTTGTGTGTGTATCGCTTGCTC |
| 1237_Revb | TCATGCGGCAACACATA |
| SpeL_5TQb,c | TCGATGTTTTTGGTCTTGCATT |
| SpeL_3TQb,c | CCGGCTCCTTTTCTCTCATTAA |
| SpeL_probec | 6FAM-AAAGATCAAATACATTCAATCAATGGTGGTCTCGTTAAA-TAMRA |
| SpeM_5TQb,c | AAAGATGTAATTAATAGAACCAATATGAAGATAACA |
| SpeM_3TQb,c | AATTATCATCGTCCCAGACTCTAGTTTT |
| SpeM_probec | 6FAM-AGAAAATTGGCACCCAGTTAATATTTAACACGAATGA-TAMRA |
| SpeB_5TQc | CGCACTAAACCCTTCAGCTCTT |
| SpeB_3TQc | ACAGCACTTTGGTAACCGTTGA |
| SpeB_probec | 6FAM-GCCTGCGCCGCCACCAGTA-TAMRA |
| GyrA_5TQc | CGACTTGTCTGAACGCCAAA |
| GyrA_3TQc | TTATCACGTTCCAAACCAGTCAA |
| GyrA_probec | 6FAM-CGACGCAAACGCATATCCAAAATAGCTTG-TAMRA |
Primers were designed based on the genome sequence of GAS strain MGAS8232 (serotype M18) (33).
Primers used for sequencing the speL-speM locus.
Primers and probes used for real time RT-PCR analysis (TaqMan assays). Probes were labeled at the 5′ end with the reporter dye 6-carboxyfluorescein (6FAM) and at the 3′ end with the quencher 6-carboxy-N,N,N′,N′-tetramethylrhodamine (TAMRA).
Cloning of speL and speM.
Techniques described previously (21) were used to clone speL and speM lacking the regions encoding the putative secretion signal sequence. Primers for speL (5′-GCAGCCATATGGAAGAGACTATTAATATTAAGGATATATACTCTC-3′; 5′-GGGGATCCTTAATTTTCTTTGTTTGTGAATAAATAGAC-3′) and speM (5′-GCAGCCATATGTTTTCAGATGCTGTGTTGG-3′; 5′-GGGGATCCTTAATTTTTAGAAAAATCTTCGTTTAAGTA-3′) contained NdeI and BamHI restriction sites (underlined), respectively. DNA isolated from strain MGAS8232 and the following PCR conditions were used: initial denaturation at 94°C for 2 min; 36 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 2 min; and final extension at 72°C for 5 min. PCR products were purified with the Qiagen PCR Purification Kit (Qiagen, Valencia, Calif.) and cloned in pET28b (Novagen, Madison, Wis.). Recombinant plasmids from Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.) transformants were isolated and used to transform E. coli BL21(DE3) (Novagen) grown in Luria-Bertani broth (Difco Laboratories, Detroit, Mich.) containing kanamycin (50 μg/ml). The cloned DNA was sequenced to rule out the presence of spurious mutations.
Expression and purification of recombinant SpeL (rSpeL) and SpeM (rSpeM).
To assess the production of recombinant proteins, E. coli BL21(DE3) transformants containing speL and speM were grown in Luria-Bertani broth with kanamycin (50 μg/ml) and induced with IPTG (isopropyl-β-d-thiogalactopyranoside) (final concentration, 0.5 mM) at exponential phase. Briefly, cells were pelleted, washed, and suspended in binding buffer (0.5 M NaCl, 20 mM Tris; pH 8.0). rSpeL and rSpeM were purified from E. coli lysates by nickel chelation chromatography (His-Bind Resin, Novagen), and the N-terminal His6 tags were cleaved with the thrombin cleavage kit (Novagen) according to the manufacturer's instructions. Purified proteins (1.0 μg) were analyzed by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-15% PAGE) stained with Coomassie blue and immunoblots to assess apparent molecular weight, homogeneity, and serologic cross-reactivity with SpeA and SpeC (21). All reagents and glassware used for toxin purification and biological assays were pyrogen-free. Briefly, glassware was heated to 160°C for 3 h. Aqueous solutions were made in pyrogen-free glassware and induced 3-h fever responses of <0.5°C in each of three rabbits. The minimum detectable amount of endotoxin by this method is 10 ng/ml.
RNA isolation and in vitro expression analysis.
GAS were harvested in exponential (optical density at 600 nm [OD600] = 0.3) and stationary (OD600 = 0.7) phase and lysed as described previously (35). After incubation for 15 min at 65°C and centrifugation to remove cell debris, lysates were treated with the RNeasy Mini kit (Qiagen). Contaminating DNA was digested with 100 U of DNase I (Roche Molecular Biochemicals, Mannheim, Germany) for 2 h at 37°C. Purification of the RNA and removal of DNase I were achieved by repeating the RNeasy Mini kit protocol. RNA quality was examined spectrophotometrically and with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, Calif.). The absence of contaminating DNA was confirmed by PCR.
TaqMan real-time reverse transcription-PCR (RT-PCR) was used to analyze in vitro expression of speL and speM in both growth phases as described previously (6). TaqMan primer and probe sequences are shown in Table 2. Triplicate assays were performed with RNA isolated from two independent cultures of strain MGAS8232. Standard curves were made with MGAS8232 genomic DNA. Analysis of speB, which encodes a GAS cysteine protease, was used as a control (5). The cDNA quantity of speL, speM, and speB was normalized to gyrA cDNA in each sample, and the average fold difference in transcript quantity in exponential relative to stationary phase was determined.
Assay for superantigen activity.
Mitogenicity of rSpeL and rSpeM was assessed with [3H]thymidine incorporation assays, which were done in the presence of polymyxin B to inactivate potentially contaminating lipopolysaccharide (21, 22). Rabbit splenocytes and human peripheral blood mononuclear cells (PBMCs) (2 × 105 cells/well) were added to 96-well plates, and serial dilutions of rSpeL or rSpeM were added in quadruplicate. Purified rSpeC (22) and phosphate-buffered saline (PBS) were used as controls. The splenocytes were grown at 37°C for 3 days and pulsed with 1 μCi of [3H]thymidine (Amersham, Arlington Heights, Ill.) per well overnight. Cells were harvested onto fiberglass filters, and [3H]thymidine incorporation was measured with a scintillation counter (Beckman Instruments, Fullerton, Calif.).
Analysis of T-cell repertoire.
PBMCs from five healthy human donors were isolated; cultured with anti-CD3 (20 ng/ml), rSpeL (100 ng/ml), or rSpeM (100 ng/ml); and analyzed for TCR Vβ profiles as described previously (19, 21). Briefly, PBMCs (107 cells/ml) were incubated at 37°C for 30 min in the dark in 96-well plates with biotinylated monoclonal antibodies against human Vβ1, 2, 3, 5.1, 5.2, 5.3, 7, 7.2, 8, 11, 12, 13.1, 13.2, 13.6, 14, 16, 17, 18, 20, 21.3, and 22 (Immunotech, Westbrook, Maine), Vβ9, Vβ23 (Pharmingen, San Diego, Calif.) and Vβ6.7 (Endogen, Woburn, Mass.). After two washes, cells were incubated at 4°C for 30 min with anti-CD3 allophycocyanin, anti-CD4-peridinin chlorophyll protein, anti-CD8 fluorescein isothiocyanate (Becton Dickinson, Cockeysville, Md.), and streptavidin phycoerythrin (Southern Biotechnologies, Inc., Birmingham, Ala.). After a final wash, analysis was conducted with four-color flow cytometry (FACSCalibur; Becton Dickinson) as described previously (36).
Pyrogenicity and lethality models of toxic shock syndrome.
American Dutch belted rabbits were used to assess the pyrogenicity and lethality of rSpeL and rSpeM (15, 21). Rabbits were injected in the marginal ear veins with rSpeL, rSpeM, rSpeC (5 μg/kg), or PBS, and temperatures were recorded at 4 h. At 4 h, endotoxin from Salmonella enterica serovar Typhimurium (10 μg/kg, 1/50 lethal dose, 50% endpoint) was injected intravenously to assess the ability of rSpeL and rSpeM to enhance host susceptibility to lethal endotoxin shock (endotoxin enhancement). Animals were monitored for STSS symptoms, and mortality was recorded for 2 days. The miniosmotic pump model of STSS was also used to assess the lethality of rSpeL and rSpeM, since these devices release a constant amount of exotoxin into subcutaneous tissue and hence resemble infection with exotoxin-producing GAS. Miniosmotic pumps (Alza Pharmaceuticals, Palo Alto, Calif.) prefilled with 500 μg of rSpeL, rSpeM, or rSpeC in 200 μl of PBS were implanted subcutaneously into the left flank of rabbits anesthetized with ketamine and xylazine (Phoenix Pharmaceuticals, St. Joseph, Mo.). Rabbits were monitored for signs of STSS, and mortality was recorded for 15 days.
Analysis of immunoreactivity by ELISA.
To determine whether SpeL and SpeM were produced in vivo during human GAS infections preceding ARF disease, enzyme-linked immunosorbant assays (ELISAs) were used to measure the level of anti-SpeL and anti-SpeM antibody present in convalescent-phase sera from patients with ARF. These sera were drawn from male and female pediatric patients who were diagnosed with ARF at Primary Children's Medical Center in Salt Lake City, Utah, between 1985 and 1990. Assays were conducted as previously described except that all incubations were done at room temperature (11, 17). Briefly, 96-well plates were coated overnight with 250 ng of rSpeL or rSpeM and incubated with human sera diluted 1:500. Plates were washed and incubated with a 1:100,000 dilution of goat anti-human immunoglobulin G (IgG) conjugated to horseradish peroxidase (Bio-Rad Laboratories, Hercules, Calif.). After another wash, the plates were developed with 100 μl of ABTS [2,2′azinobis(3-ethylbenzthiazolinesulfonic acid); Roche Molecular Biochemicals], and absorbances were measured at 405 nm with a SpectraMax 384 Plus microplate spectrophotometer (Molecular Devices, Sunnyvale, Calif.).
Paired acute and convalescent-phase sera from 44 patients with pharyngitis, convalescent-phase sera from 55 patients with ARF, and serum from 20 patients with no documented cases of GAS infection (normal human serum [NHS]) were studied. All sera were obtained from pediatric subjects. Acute-phase sera were obtained upon diagnosis, and convalescent-phase sera were drawn from patients with pharyngitis 10 to 110 days after diagnosis. Patients with pharyngitis were infected with GAS strains of emm gene types emm77 (n = 10); emm75 (n = 9); emm12 and emm28 (n = 5 each); emm4 (n = 4); emm1.0 (n = 3); emm73 and emm89 (n = 2 each); and emm2, emm3, emm6, and emm94 (n = 1 each). GAS isolates are rarely obtained from ARF patients since ARF symptoms occur well after the antecedent infection. Therefore, the M protein serotype of the strains infecting these individuals is rarely known.
Linear B-cell epitope mapping.
Fifty-one overlapping 15-mer synthetic peptides (Chiron Technologies, San Diego, Calif.) spanning the N-terminal variable region of the GAS M18 protein deposited in the GenBank database (accession no. AAB03086) were designed (11). The peptides corresponded to amino acids 29 to 193 except for two amino acid substitutions (Thr79 to Ile79 and Ser107 to Thr107). Peptides were covalently linked to biotin at the N terminus by a serine-glycine-serine-glycine spacer. Analysis of the reactivity of the 55 sera from ARF patients to linear peptides of the M18 protein and determination of positive immunoreactivity for each peptide were done in 96-well streptavidin-coated plates as described previously (11).
RESULTS
Identification and analysis of SpeL and SpeM.
speL and speM were identified in a prophage-like element in the genome of serotype M18 GAS strain MGAS8232 (33) (Fig. 1A). As predicted with SignalP software, SpeL and SpeM have secretion signal sequences. The putative mature forms of SpeL and SpeM, which are 39% identical to each other, are 62 and 77% homologous to SpeK, respectively. This comparison was based on SpeK encoded by an M3 GAS strain and deposited in the GenBank database (accession no. AAL09835) (designated SpeK-M3) rather than the protein fragment described by Ferretti et al. (9) (designated SpeK-M1). The predicted mature forms of SpeL (227 amino acids) and SpeM (211 amino acids) have molecular masses of 26.3 and 25.2 kDa, respectively. Phylogenetic analysis of the amino acid sequences corresponding to mature PTSAgs corroborated that SpeL and SpeM are related to SpeK-M3 (Fig. 1B). Three main evolutionary branches were present: two were comprised of GAS and staphylococcal toxins and one contained only GAS PTSAgs. SpeL and SpeM were located on the latter branch (Fig. 1B).
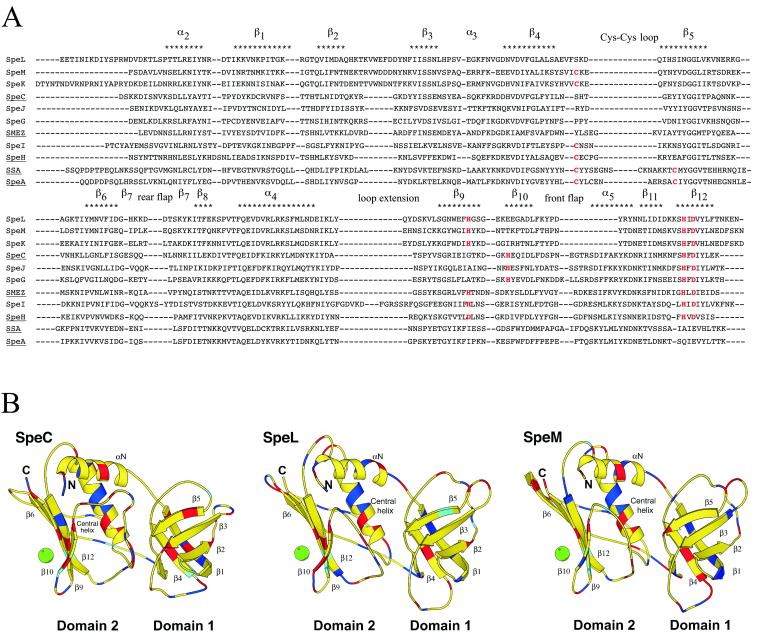
Despite low primary amino acid sequence homology, PTSAgs have similar motifs and a conserved structural architecture (23). The N-terminal domain contains a barrel formed by five β-strands with primarily antiparallel topology. The C-terminal domain consists of a central α-helix packed against a five-stranded β sheet, which forms the β grasp motif. An amino acid sequence alignment of GAS PTSAgs and predicted structural models showed that the putative mature forms of SpeL and SpeM have features in common with previously described PTSAgs (Fig. 2A and B). For example, putative zinc-binding motifs are present in the C-terminal β9 and β12 strands, suggesting that both proteins bind MHC class II molecules in a zinc-dependent manner. The β4-β5 loops in SpeL and SpeM are small and, unlike the β4-β5 loop in the known PTSAgs SpeA and streptococcal superantigen (SSA), lack the cysteines required to form a disulfide bond. In addition, SpeL and SpeM have an acidic and unusually long β2-β3 loop. Finally, SpeL and SpeM are also unique in that they have a short front flap.
FIG. 2.
Sequence alignment and structural analysis of SpeL and SpeM. (A) Amino acid sequences of putative and proven GAS PTSAgs were aligned. Cysteine residues in a Cys-Cys loop (SpeA and SSA) and histidine and aspartate residues in putative or proven zinc-binding motifs are shown in red. Structural elements are shown above the sequences. PTSAgs with known structures are underlined. (B) SpeL and SpeM structural models were predicted based on SpeC. Domains 1 and 2, N and C termini, β-sheets, α-helices, and a zinc ion (green circle) are represented. Red, blue, and cyan represent amino acids aspartate/glutamate, arginine/lysine, and histidine, respectively.
In vitro expression of speL and speM.
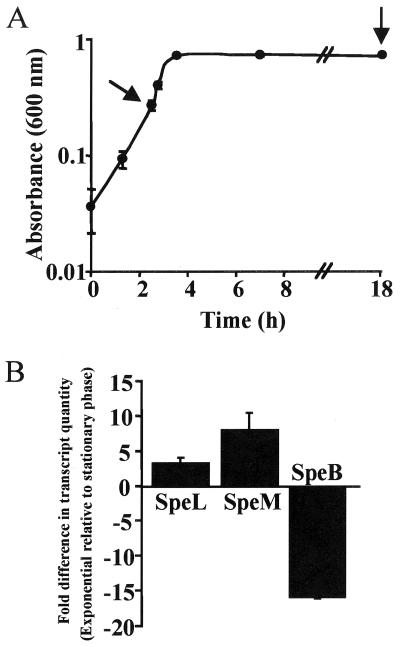
To determine whether the genes were transcribed, TaqMan assays were used to assess in vitro transcript levels of speL and speM in strain MGAS8232. The results indicated that speL and speM were transcribed, and transcript levels were higher in the exponential phase than in the stationary phase (Fig. 3A and B), a finding that is consistent with other GAS superantigen genes (12). Transcript analysis of speB, a GAS gene that encodes a cysteine protease and is known to be upregulated in stationary phase (5), was used as a control. The speB transcript level was greater in the stationary phase than in the exponential phase, as expected (Fig. 3B).
FIG. 3.
In vitro analysis of speL and speM transcript levels. (A) Arrows denote points when bacteria were removed for RNA isolation and transcript analysis. The mean and standard error of the mean (SEM) of triplicate measurements are shown. (B) TaqMan assays were conducted with two independent RNA preparations to assess the relative quantity of gene-specific transcripts. The cDNA quantity of speL, speM, and speB was normalized to gyrA cDNA in each sample. The average fold difference in transcript quantity in the exponential phase relative to the stationary phase and the SEM are shown.
Cloning, overexpression, and purification of SpeL and SpeM.
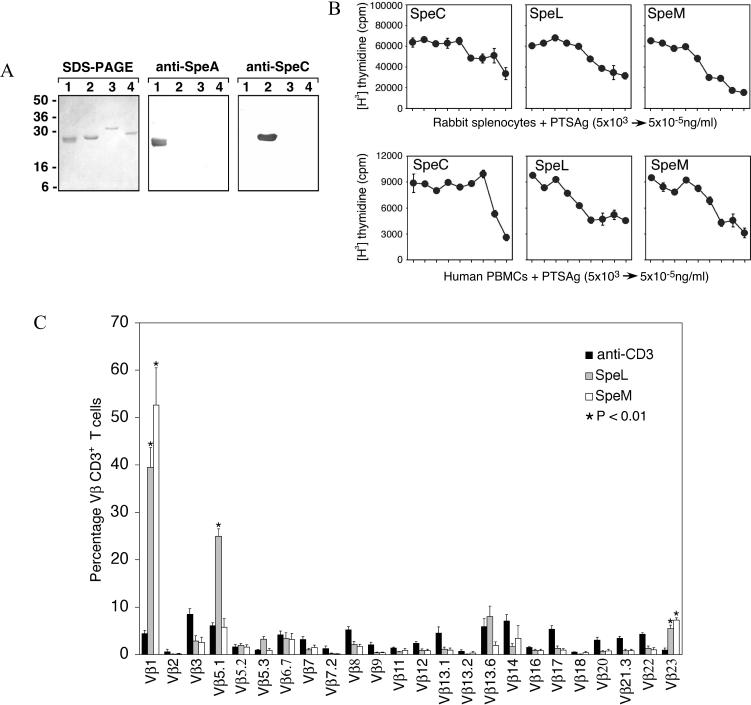
To facilitate functional analysis of SpeL and SpeM, the regions of the genes encoding the predicted mature forms of the proteins were cloned. DNA sequencing revealed that the cloned genes lacked spurious mutations. rSpeL and rSpeM were overexpressed in E. coli and purified to apparent homogeneity. Western immunoblot analysis revealed that neither protein was reactive with antisera raised against known PTSAgs SpeA and SpeC (Fig. 4A).
FIG. 4.
Functional analysis of purified rSpeL and rSpeM. (A) rSpeL and rSpeM were analyzed with SDS-PAGE and immunoblots. Lanes 1 to 4 contain 1.0 μg of purified rSpeA, rSpeC, rSpeL, and rSpeM, respectively. Molecular mass markers are shown. Proteins were analyzed with SDS-PAGE, and immunoblots were probed with specific antisera against rSpeA and rSpeC. (B) Rabbit splenocytes and human PBMCs were incubated with rSpeC, rSpeL, and rSpeM and pulsed with [3H]thymidine to assess mitogenicity. DNA was harvested after 24 h, and the counts per minute (cpm) were determined by scintillation counting. The mean of quadruplicate experiments and the SEM values are shown. (C) PBMCs from five human donors were stimulated with anti-CD3 antibody, rSpeL, and rSpeM to determine the TCR Vβ profile. Cells were stained with monoclonal antibodies against TCR Vβ chains and analyzed by flow cytometry. The percentage of T cells expressing the listed Vβ are shown. P values were determined with a Student t test (P < 0.01). Error bars represent the SEM.
Superantigenicity of SpeL and SpeM.
PTSAgs induce clonal proliferation of T cells with specific TCR Vβ subsets. To determine whether rSpeL and rSpeM had this characteristic, we tested the ability of the proteins to induce proliferation of rabbit splenocytes and human PBMCs. rSpeL and rSpeM caused proliferation of rabbit splenocytes and human PBMCs at concentrations as low as 10−4 ng/ml (Fig. 4B). The results observed for rSpeL and rSpeM were similar to those obtained for rSpeC, a known PTSAg. T-cell proliferation did not occur in response to incubation with PBS alone (data not shown).
Flow cytometric analysis of T-cell repertoire.
PTSAgs preferentially stimulate T cells with specific TCR Vβ subsets. To verify the superantigenicity of rSpeL and rSpeM, the Vβ profile of human PBMCs incubated with rSpeL and rSpeM was determined. rSpeL significantly stimulated CD4+ and CD8+ T cells with Vβ1, Vβ5.1, and Vβ23 regions (P < 0.01), suggesting a PTSAg effect (Fig. 4C). rSpeL induced a very substantial expansion of T cells with Vβ1 and Vβ5.1 (39.5 and 25.0%, respectively). T cells with Vβ5.3 and Vβ13.6 TCR subsets also were stimulated moderately by rSpeL, but the majority of these cells were CD4+. Significant expansion (P < 0.01) of Vβ1 and Vβ23 TCR subsets also occurred in PBMCs treated with rSpeM (Fig. 4C). Consistent with PTSAg-induced activation, these T cells were CD4+ and CD8+. rSpeM also stimulated a massive expansion (52.7%) of T cells with Vβ1 TCR subsets.
Pyrogenicity and lethality.
Previous studies have shown that PTSAgs induce fever and enhance the susceptibility of rabbits to endotoxin shock (23). To determine whether rSpeL and rSpeM have these characteristics, sublethal concentrations of the proteins were injected intravenously into three rabbits, and their temperatures were recorded at 0 and 4 h (Table 3). Average temperature increases of 0.87, 0.67, and 1.50°C were observed in rabbits given rSpeL, rSpeM, and rSpeC, respectively. Temperature increases of ≥0.5°C are considered to be significant in this model (29, 31). No increase in temperature was observed in control rabbits that received PBS. At 4 h, rabbits were injected with sublethal concentrations of endotoxin and monitored for 48 h. All rabbits that received PTSAgs and endotoxin died, indicating that SpeL and SpeM enhanced the susceptibility of rabbits to endotoxic shock (Table 3). In contrast, control rabbits given PBS and endotoxin survived. The lethality of rSpeL and rSpeM was also assessed with the miniosmotic pump model of STSS, which causes death in rabbits in the absence of exogenous endotoxin. Two of the three rabbits infused with 500 μg of rSpeL died on days 5 and 6. However, rabbits that received rSpeM did not die after 15 days (Table 3). The three rabbits given 500 μg of rSpeC (positive control rabbits) died on days 8, 9, and 12.
TABLE 3.
Pyrogenicity and lethality of rSpeL and rSpeM
| Toxin | Pyrogenicitya (avg temp increase in °C) | Models of lethality (no. dead/no. total animals)
|
|
|---|---|---|---|
| Endotoxin enhancementb | Miniosmotic pumpc | ||
| SpeL | 0.87 | 3/3 | 2/3 |
| SpeM | 0.67 | 3/3 | 0/3 |
| SpeC | 1.50 | 3/3 | 3/3 |
| PBS | −0.07 | 0/3 | NDd |
Three rabbits were used for each assay. Animals were injected intravenously with 5 μg of recombinant protein per kg at 0 h, and temperatures were recorded at 4 h. A change of 0.5°C or greater is considered to be significant in this model (29, 31).
Mortality was recorded for a 48-h period after injection of endotoxin.
Mortality was recorded for 15 days after subcutaneous implantation of a miniosmotic pump.
ND, not determined.
Distribution of the speL and speM genes among GAS serotypes and allelic variation.
To gain insight into the molecular population genetics of speL and speM, we studied 46 GAS strains for the presence and sequence divergence of the two genes. We analyzed 33 M18 strains, the predominant serotype associated with ARF outbreaks in Salt Lake City, Utah, and elsewhere in the United States in recent years (26, 34, 39), and 13 strains representing the most common M protein serotypes associated with pharyngitis and invasive disease in the United States, Canada, the United Kingdom, and several other countries (3, 8, 24, 32) (Table 1). The serotype M18 strains were isolated between 1931 and 2000 from either asymptomatic carriers or patients diagnosed with ARF, invasive disease, and pharyngitis. The non-M18 pharyngeal isolates were obtained from patients with pharyngitis during a 1998 to 1999 ARF outbreak in Salt Lake City, Utah.
To determine whether the speL-speM locus was present in these strains, PCR was done with primers that annealed to ORFs SpyM18_1236 (chromosomal DNA) and SpyM18_1240 (phage DNA) (Fig. 1A). A PCR amplicon of the same size was obtained from all 33 serotype M18 strains tested. Moreover, all strains had the identical DNA sequence for this region, which included the putative promoter regions, speL and speM genes, 282-bp intergenic region, and flanking DNA (data not shown). In contrast to the serotype M18 strains, no amplicons were obtained from the non-M18 strains with primers 117 and 1721R or with ORF-specific primers for speL and speM (data not shown). In summary, in this sample of GAS strains studied, speL and speM were uniquely present at the identical chromosomal location and had the same sequence in all serotype M18 strains examined, despite the 69-year time span represented by the organisms studied.
Exposure of ARF patients to serotype M18 GAS, SpeL, and SpeM.
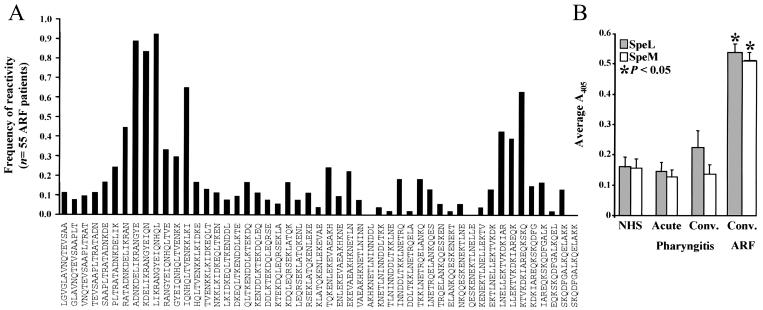
ARF symptoms occur well after an antecedent GAS infection, and the organism rarely is cultured from ARF patients. Hence, it is usually difficult to document the M protein serotype of the strain associated with disease. A recent study demonstrated community-wide coresurgence of two ARF outbreaks and serotype M18 GAS strains in Salt Lake City, Utah (34). To determine whether ARF patients in Salt Lake City had been exposed to serotype M18 GAS, we tested sera obtained from 55 ARF patients for reactivity with 51 overlapping synthetic peptides spanning the N-terminal variable region of the M18 protein. Virtually all patients had antibodies that reacted with M18-specific peptides, indicating that these patients had been exposed to serotype M18 GAS (Fig. 5A). A similar analysis of M3 and M1 overlapping synthetic peptides with the 55 ARF sera revealed that the immunoreactivity profile observed with the M18 protein was unique (data not shown).
FIG. 5.
Human serologic response of patients with ARF. (A) Sera obtained from 55 ARF patients were screened by ELISA against a library of overlapping 15-mer synthetic peptides corresponding to the N-terminal hypervariable region of M18 protein. Frequency of reactivity (the number of serum samples positive for a specific peptide divided by the total sample number) and positive immunoreactivity were calculated as described previously (11). Amino acid sequences of the synthetic peptides are shown. (B) The reactivity of human patient sera to rSpeL and rSpeM was determined by ELISA. Sera were obtained from patients with ARF (n = 55) and pharyngitis (n = 44) and from individuals with no documented case of GAS infection (NHS) (n = 20). The mean ELISA value for triplicate experiments and the SEM values are shown. Statistical differences between ARF and pharyngitis results and ARF and NHS results were determined with a Student t test (P < 0.05).
To determine whether SpeL and SpeM were produced in vivo during human GAS infections preceding ARF disease, ELISAs were used to measure the level of anti-SpeL and anti-SpeM antibodies present in convalescent-phase sera from the 55 ARF patients examined above. Sera obtained from all 55 patients with ARF contained antibodies to SpeL and SpeM. The mean ELISA value obtained with sera from ARF patients was significantly greater (P < 0.05) than the mean ELISA value for sera from patients with uncomplicated pharyngitis (infected with strains representing 12 distinct M protein serotypes), or patients with no recently documented episode of GAS infection (Fig. 5B). Together, the serologic results are consistent with the idea that these 55 humans with ARF were exposed to SpeL and SpeM expressed by serotype M18 GAS.
DISCUSSION
Many extracellular molecules, including PTSAgs, contribute to GAS pathogenesis (7). However, despite extensive research, many aspects of GAS diseases and sequelae such as ARF remain poorly understood. In this report, we characterized two novel PTSAgs (SpeL and SpeM) encoded by recently discovered genes located contiguously in a prophage-like element in the genome of a serotype M18 GAS strain isolated from an ARF patient. SpeL and SpeM were annotated based on their sequence similarity (47 and 48%, respectively) to the known PTSAg SpeC. SpeL and SpeM have predicted secretion signal sequences, suggesting that the mature proteins are present in the extracellular milieu and hence implicating them in host-pathogen interactions.
A conserved tertiary structure and ability to interact with TCRs and MHC class II molecules independent of antigen processing are hallmark features of PTSAgs. The TCR-MHC class II interaction is responsible for subsequent biological activities such as T-cell proliferation and massive cytokine release (23). Importantly, bacterial PTSAgs have been suggested to participate in the pathogenesis of autoimmune manifestations, including ARF (20). We discovered that SpeL and SpeM have structural features that are similar to those of other PTSAgs, including a putative zinc-binding motif, suggesting zinc-dependent binding to MHC class II molecules. We also demonstrated that the proteins have pyrogenic, mitogenic, and lethal properties. SpeL and the known superantigen SpeC were lethal in the endotoxin enhancement and miniosmotic pump assays, but SpeM was lethal in only the former assay. There are several possible explanations for this. First, the amino acid sequence differences between SpeL and SpeM may have rendered SpeM weaker in lethality. The greater sensitivity of the endotoxin enhancement assay is likely why no differences in lethality were observed in the endotoxin enhancement assay. It is also possible that SpeM is less stable in the host. For example, it may be stable enough to cause death in the endotoxin enhancement assays but not stable during the 15 days of the miniosmotic pump assay. In addition, it is also possible that host proteases degrade the toxin over time.
An important biologic function of PTSAgs is their ability to stimulate T cells with specific TCR Vβ subsets. Furthermore, studies have suggested that T cells play a critical role in the pathogenesis of GAS diseases, including ARF (26). Moreover, analysis of the Vβ repertoire in CD4+ and CD8+ T cells isolated from skin biopsy samples from patients with guttate psoriasis, a chronic inflammatory skin disorder frequently associated with antecedent GAS infection, demonstrated a selective accumulation of T cells with Vβ2 TCR subsets (19). Importantly, GAS isolates from these samples contained speC, and SpeC stimulates T cells expressing Vβ2 (38). With respect to ARF, unique Vβ profiles attributable to a PTSAg have not been detected in PBMCs isolated from ARF patients (1), but this issue has not been studied adequately in a strain-specific fashion. It is possible that PTSAgs influence specific Vβ subsets in cardiac tissue rather than peripheral blood and that unique Vβ profiles are present prior to the convalescent onset of ARF (1). In support of this idea, Leung et al. (18) demonstrated expansion of T cells expressing Vβ2 in the myocardium and coronary artery tissues from a patient with Kawasaki syndrome, a disease associated with T-cell activation. Moreover, elevated levels of specific Vβ subsets were present in the acute rather than convalescent phase of the disease (2). SpeL and SpeM stimulated T cells with Vβ1 and Vβ23 regions, and SpeL also had specificity for Vβ5.1 TCR subsets. While Vβ specificity among PTSAgs is overlapping and some Vβs are targeted more frequently than others, no two PTSAgs have identical Vβ profiles (10). Our findings demonstrate an overlap in specificity for Vβ1 and Vβ5.1 with SpeC and SSA, respectively, but PTSAg-induced expansion of T cells with Vβ23 TCR subsets is rare. The biological significance of elevated Vβ23 expression in host-GAS interactions and the possible role of Vβ23 in ARF pathogenesis are not yet known. Of note, Beres et al. (4) recently reported that SpeK, the PTSAg most closely related to SpeM and SpeL, stimulates T cells with Vβ1, Vβ5.1, and Vβ23 TCR subsets. This observation indicates retention of structure-function relationships from a common ancestor PTSAg.
To gain insight into the molecular population genetics of speL and speM, serotype M18 and non-M18 GAS strains were analyzed for the presence of these genes. In the strains analyzed, speL and speM were uniquely present in all serotype M18 strains. Moreover, no allelic variation was found in either gene in the 33 strains, despite the 69-year time span represented by the organisms studied. Further, speL and speM were present at the identical chromosomal location, and the sequence of the flanking chromosomal and phage DNA was identical, suggesting an evolutionarily recent acquisition by a common precursor cell. The presence of perfect 98-bp direct repeats flanking this phage is consistent with the concept of a recent acquisition event. In principle, phage-mediated transduction provides opportunities for PTSAg genes to be transferred horizontally, thus resulting in the generation of GAS strains with a new array of virulence factors. Evidence that horizontal gene transfer has contributed to GAS chromosomal diversity has been summarized previously (14, 30, 33).
Several problems have hindered advancement in our understanding of the molecular mechanism of ARF pathogenesis. First, there is no animal model that fully mimics all aspects of ARF and subsequent rheumatic heart disease. Second, because ARF usually begins several weeks after exposure to GAS, it is relatively uncommon for the inciting strain to be recovered from the patient. Hence, little is known about the virulence determinants that contribute to ARF and their distributions in the GAS strains involved. Our study of SpeL and SpeM was stimulated by the discovery of genes encoding these novel PTSAgs in the genome of a serotype M18 strain recovered from an ARF patient living in Salt Lake City (33). A recent study discovered that two distinct ARF outbreaks in Salt Lake City in the 1980s and 1990s were associated with a striking increase in recovery of serotype M18 strains from patients with pharyngitis (34). Serotype M18 GAS also were implicated in ARF outbreaks in several other localities in the United States in the 1980s (26). These observations led us to test the hypothesis that exposure to serotype M18 strains is a common feature among ARF patients in Salt Lake City. Linear B-cell epitope mapping with M18-specific peptides found that virtually all 55 ARF patients from Salt Lake City had serologic evidence of exposure to the M18 protein. These results are consistent with the observation that sera obtained from some ARF patients in Salt Lake City had opsonic activity against serotype M18 GAS (39). In addition, some ARF patients from Salt Lake City had elevated levels of serum IgG to an M-protein epitope (designated class I) present in serotype M18 strains, also suggesting recent infection with this organism (26). We also demonstrated that the 55 patients with ARF had antibodies to SpeL and SpeM. Taken together, our data are consistent with the idea that these patients were exposed to SpeL and SpeM expressed by serotype M18 GAS.
In summary, the discovery and characterization of two new PTSAgs in ARF-associated organisms provides additional insight into the GAS factors that may contribute to this disease. Analysis of the distribution and allelic variation of speL and speM among serotype M18 GAS strains has provided new information about the molecular population genetics of ARF-associated GAS strains. Our findings may assist in the development of new therapeutics and diagnostics for ARF, a multifactorial disease involving GAS virulence determinants and host genetics.
Acknowledgments
This work was supported in part by Public Health Services grants HL36611 to P. M. Schlievert; AI51607 to D. H. Ohlendorf; and HL36577, AR41256, and HL37260 to D. Y. M. Leung and NIAID contract N01-AO-22749 to H. R. Hill.
We thank G. Hettrick for assistance with preparation of figures and S. Reid for critical review of the manuscript.
Editor: D. L. Burns
REFERENCES
- 1.Abbott, W. G., M. A. Skinner, L. Voss, D. Lennon, P. L. Tan, J. D. Fraser, I. J. Simpson, R. Ameratuga, and A. Geursen. 1996. Repertoire of transcribed peripheral blood T-cell receptor beta chain variable-region genes in acute rheumatic fever. Infect. Immun. 64:2842-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abe, J., B. L. Kotzin, K. Jujo, M. E. Melish, M. P. Glode, T. Kohsaka, and D. Y. M. Leung. 1992. Selective expansion of T cells expressing T-cell receptor variable regions V beta 2 and V beta 8 in Kawasaki disease. Proc. Natl. Acad. Sci. USA 89:4066-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall, B., R. Facklam, T. Hoenes, and B. Schwartz. 1997. Survey of emm gene sequences and T-antigen types from systemic Streptococcus pyogenes infection isolates collected in San Francisco, California; Atlanta, Georgia; and Connecticut in 1994 and 1995. J. Clin. Microbiol. 35:1231-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, J. K. McCormick, D. Y. M. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaussee, M. S., E. R. Phillips, and J. J. Ferretti. 1997. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect. Immun. 65:1956-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dicuonzo, G., G. Gherardi, G. Lorino, S. Angeletti, M. De Cesaris, E. Fiscarelli, D. E. Bessen, and B. Beall. 2001. Group A streptococcal genotypes from pediatric throat isolates in Rome, Italy. J. Clin. Microbiol. 39:1687-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser, J., V. Arcus, P. Kong, E. Baker, and T. Proft. 2000. Superantigens: powerful modifiers of the immune system. Mol. Med. Today 6:125-132. [DOI] [PubMed] [Google Scholar]
- 11.Hoe, N. P., P. Kordari, R. Cole, M. Liu, T. Palzkill, W. Huang, D. McLellan, G. J. Adams, M. Hu, J. Vuopio-Varkila, T. R. Cate, M. E. Pichichero, K. M. Edwards, J. Eskola, D. E. Low, and J. M. Musser. 2000. Human immune response to streptococcal inhibitor of complement, a serotype M1 group A Streptococcus extracellular protein involved in epidemics. J. Infect. Dis. 182:1425-1436. [DOI] [PubMed] [Google Scholar]
- 12.Houston, C. W., and J. J. Ferretti. 1981. Enzyme-linked immunosorbent assay for detection of type A streptococcal exotoxin: kinetics and regulation during growth of Streptococcus pyogenes. Infect. Immun. 33:862-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapur, V., K. Nelson, P. M. Schlievert, R. K. Selander, and J. M. Musser. 1992. Molecular population genetic evidence of horizontal spread of two alleles of the pyrogenic exotoxin C gene (speC) among pathogenic clones of Streptococcus pyogenes. Infect. Immun. 60:3513-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kehoe, M. A., V. Kapur, A. M. Whatmore, and J. M. Musser. 1996. Horizontal gene transfer among group A streptococci: implications for pathogenesis and epidemiology. Trends Microbiol. 4:436-443. [DOI] [PubMed] [Google Scholar]
- 15.Kim, Y. B., and D. W. Watson. 1970. A purified group A streptococcal pyrogenic exotoxin: physiochemical and biological properties including the enhancement of susceptibility to endotoxin lethal shock. J. Exp. Med. 131:611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraulis, P. J. 1991. A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 17.Lei, B., L. M. Smoot, H. M. Menning, J. M. Voyich, S. V. Kala, F. R. Deleo, S. D. Reid, and J. M. Musser. 2002. Identification and characterization of a novel heme-associated cell-surface protein made by Streptococcus pyogenes. Infect. Immun. 70:4494-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung, D. Y., R. C. Giorno, L. V. Kazemi, P. A. Flynn, and J. B. Busse. 1995. Evidence for superantigen involvement in cardiovascular injury due to Kawasaki syndrome. J. Immunol. 155:5018-5021. [PubMed] [Google Scholar]
- 19.Leung, D. Y. M., J. B. Travers, R. Giorno, D. A. Norris, R. Skinner, J. Aelion, L. V. Kazemi, M. H. Kim, A. E. Trumble, M. Kotb, and P. M. Schlievert. 1995. Evidence for a streptococcal superantigen-driven process in acute guttate psoriasis. J. Clin. Investig. 96:2106-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marrack, P., and J. Kappler. 1990. The staphylococcal enterotoxins and their relatives. Science 248:705-711. [DOI] [PubMed] [Google Scholar]
- 21.McCormick, J. K., A. A. Pragman, J. C. Stolpa, D. Y. M. Leung, and P. M. Schlievert. 2001. Functional characterization of streptococcal pyrogenic exotoxin J, a novel superantigen. Infect. Immun. 69:1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick, J. K., T. J. Tripp, S. B. Olmsted, Y. V. Matsuka, P. J. Gahr, D. H. Ohlendorf, and P. M. Schlievert. 2000. Development of streptococcal pyrogenic exotoxin C vaccine toxoids that are protective in the rabbit model of toxic shock syndrome. J. Immunol. 165:2306-2312. [DOI] [PubMed] [Google Scholar]
- 23.McCormick, J. K., J. M. Yarwood, and P. M. Schlievert. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 55:77-104. [DOI] [PubMed] [Google Scholar]
- 24.Musser, J. M., A. R. Hauser, M. H. Kim, P. M. Schlievert, K. Nelson, and R. K. Selander. 1991. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. USA 88:2668-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musser, J. M., V. Kapur, J. Szeto, X. Pan, D. S. Swanson, and D. R. Martin. 1995. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect. Immun. 63:994-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musser, J. M., and R. M. Krause. 1998. The revival of group A streptococcal diseases, with a commentary on staphylococcal toxic shock syndrome, p. 185-218. In R. M. Krause (ed.), Emerging infections. Academic Press, New York, N.Y.
- 27.Musser, J. M., K. Nelson, R. K. Selander, D. Gerlach, J. C. Huang, V. Kapur, and S. Kanjilal. 1993. Temporal variation in bacterial disease frequency: molecular population genetic analysis of scarlet fever epidemics in Ottawa and in eastern Germany. J. Infect. Dis. 167:759-762. [DOI] [PubMed] [Google Scholar]
- 28.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pharmacopeia. 2002. Pyrogen test, p. 1914. In The United States pharmacopeia. United States Pharmacopeial Convention, Inc., Rockville, Md.
- 30.Reid, S. D., N. M. Green, J. K. Buss, B. Lei, and J. M. Musser. 2001. Multilocus analysis of extracellular putative virulence proteins made by group A Streptococcus: population genetics, human serologic response, and gene transcription. Proc. Natl. Acad. Sci. USA 98:7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlievert, P. M., and D. W. Watson. 1978. Group A streptococcal pyrogenic exotoxin: pyrogenicity, alteration of blood-brain barrier, and separation of sites for pyrogenicity and enhancement of lethal endotoxin shock. Infect. Immun. 21:753-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharkawy, A., D. E. Low, R. Saginur, D. Gregson, B. Schwartz, P. Jessamine, K. Green, A. McGreer, and the Ontario Group A Streptococcal Study Group. 2002. Severe group A streptococcal soft-tissue infections in Ontario: 1992-1996. Clin. Infect. Dis. 34:454-460. [DOI] [PubMed] [Google Scholar]
- 33.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, L. D. Parkins, S. F. Porcella, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, G. L. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smoot, J. C., E. K. Korgenski, D. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Molecular analysis of group A Streptococcus type emm18 isolates temporally associated with acute rheumatic fever outbreaks in Salt Lake City, Utah. J. Clin. Microbiol. 40:1805-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strickland, I., P. J. Hauk, A. E. Trumble, L. J. Picker, and D. Y. M. Leung. 1999. Evidence for superantigen involvement in skin homing of T cells in atopic dermatitis. J. Investig. Dermatol. 112:249-253. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomai, M. A., P. M. Schlievert, and M. Kotb. 1992. Distinct T-cell receptor V beta gene usage by human T lymphocytes stimulated with the streptococcal pyrogenic exotoxins and pep M5 protein. Infect. Immun. 60:701-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veasy, L. G., S. E. Wiedmeier, G. S. Orsmond, H. D. Ruttenberg, M. M. Boucek, S. J. Roth, V. F. Tait, J. A. Thompson, J. A. Daly, E. L. Kaplan, and H. R. Hill. 1987. Resurgence of acute rheumatic fever in the intermountain area of the United States. N. Engl. J. Med. 316:421-427. [DOI] [PubMed] [Google Scholar]