Abstract
Little is known about the genomic-scale transcriptional responses of bacteria during natural infections. We used whole-genome microarray analysis to assess the transcriptional state of the gram-negative pathogen Pasteurella multocida, the causative agent of fowl cholera, during infection in the natural chicken host. We compared the expression profiles of bacteria harvested from the blood of septicemic chickens experiencing late-stage fowl cholera with those from bacteria grown in rich medium. Independent analysis of bacterial expression profiles from the infection of three individual chickens indicated that 40 genes were differentially expressed in all three individuals, 126 were differentially expressed in two of the three individuals, and another 372 were differentially expressed in one individual. Real-time reverse transcription-PCR assays were used to confirm the expression ratios for a number of genes. Of the 40 genes differentially expressed in all three individuals, 17 were up-regulated and 23 were down-regulated in the host compared with those grown in rich medium. The majority (10 of 17) of the up-regulated genes were involved in amino acid transport and metabolism and energy production and conversion, clearly indicating how P. multocida alters its biosynthetic and energy production pathways to cope with the host environment. In contrast, the majority (15 of 23) of down-regulated genes were of unknown or poorly characterized functions. There were clear differences in gene expression between the bacteria isolated from each of the three chickens, a finding consistent with individual host variation being an important factor in determining pathogen gene expression. Interestingly, bacteria from only two of the three infected animals had a gene expression profile highly similar to that observed during growth under iron-limiting conditions, suggesting that severe iron starvation may not always occur during P. multocida infection.
To survive and multiply within a host organism, bacterial pathogens require the coordinated expression of a range of genes. It is likely that pathogens continually alter their gene expression profiles in response to the innate immune system and environment of their host and as they move from one host niche to another. However, at present, little is known about the genomic-scale transcriptional responses of bacteria during natural infection.
Several methods have been devised to identify genes important or essential for growth within a host. Two of the most powerful methods are in vivo expression technology (IVET) (26) and signature-tagged mutagenesis (STM) (14), both of which have been used to identify Pasteurella multocida virulence genes (9, 17). Recently, the development of DNA microarray methods for studying gene expression on a whole-genome scale has provided the opportunity to analyze gene expression changes directly in response to growth within a host. There is a large amount of published data on genes that are differentially regulated in vitro under conditions that mimic those within host organisms. Such conditions include low iron (27, 31) and growth at different temperatures (38, 41), at different pHs (1, 38), and on different media (32, 43). However, at present, the only large-scale transcription-profiling data available on bacterial growth within a natural host organism are those on the growth of Borrelia burgdorferi within dialysis membranes implanted into rat peritoneal cavities (38). Furthermore, there are only a very limited number of data available on how the transcription of even a small number of pathogen genes responds during growth within host organisms (47, 48).
P. multocida is a gram-negative bacterial pathogen that causes a range of diseases in mammals and birds. It is the etiological agent of several economically important diseases, including fowl cholera, atrophic rhinitis in pigs, snuffles in rabbits, and hemorrhagic septicemia in cattle (24). Despite considerable research, the molecular mechanisms by which P. multocida can survive and multiply within a host are poorly understood. Indeed, only a small number of true bacterial virulence or virulence-associated genes (46) have been definitively identified. The identified true virulence genes of P. multocida include those involved in the production of toxins (restricted to a few toxin-producing strains) (22), capsules (2), and hemagglutinins and hemolysins (9), while the virulence-associated genes include those involved in amino acid, nucleotide, and iron transport and metabolism (8, 9, 15). Thus, it is likely that many important P. multocida virulence genes remain unidentified and uncharacterized.
We are interested in identifying P. multocida virulence genes in order to understand pathogenesis at the molecular level and also because of the potential for vaccine development. To this end, we have undertaken a genomic-scale comparison of P. multocida gene expression during growth in rich medium and growth within the chicken host. We suggest that genes expressed at higher levels during growth within a natural host are likely virulence genes. Therefore, we believe that the genes identified by this analysis will form the basis for future directed vaccine approaches as targets both for attenuating mutations and for producing recombinant antigens. Furthermore, the information will substantially increase our understanding of the fundamental interaction between this bacterial pathogen and its host and provide a broad framework for addressing how bacterial transcription is regulated during the course of infection.
MATERIALS AND METHODS
Bacterial growth conditions.
P. multocida strain X-73 (11) was grown in brain heart infusion broth (BHI; Oxoid) at either 37 or 41°C with constant shaking.
RNA isolation.
Bacteria were harvested from duplicate BHI cultures at late log phase (5 × 109 CFU/ml), added to 0.1 volume of ice-cold killing buffer (0.05 M Tris-HCl [pH 7.5], 15 mg of sodium azide/ml, 0.6 mg of chloramphenicol/ml), and pelleted by centrifugation. RNA was isolated from bacteria by using Trizol reagent (Gibco/BRL) as described by the manufacturer. Purified RNA was treated with DNase (15 U for 10 min at 37°C), and the RNA was further purified on RNeasy minicolumns (Qiagen). For the isolation of in vivo-grown bacteria, chickens (outbred Leghorn cross commercial layers) were infected with 5 × 104 CFU of P. multocida strain X-73 by injection into the breast muscle. Blood was recovered from infected chickens during the final stages of disease (between 17 and 22 h after infection), when the level of bacteremia was observed to be between 109 and 1010 CFU/ml. Clinical signs at the time of blood recovery indicated that each infection was in the terminal phase. Blood (30 to 40 ml) was recovered by terminal heart puncture and added to 0.1 volume of ice-cold killing buffer, and the bacteria were separated from the red blood cells by sucrose density centrifugation for 1 h at 3,000 × g in the presence of 12.5% (wt/vol) sucrose. Bacteria were removed from above the sucrose cushion and concentrated by centrifugation for 5 min at 12,000 × g. All manipulations were carried out on ice or at 4°C and took less than 1.5 h. RNA was purified as described above for in vitro-grown bacteria.
Microarray hybridizations.
P. multocida strain PM70 arrays were constructed as described previously (32). These arrays were generated by PCR amplification of each of the 2,015 annotated P. multocida genes and contained triplicate spots for each gene. Fluorescently labeled cDNA was prepared by the incorporation of amino allyl dUTP (Sigma) during reverse transcription (RT) and the subsequent coupling of monofunctional cy3 and cy5 dyes (Amersham) to the amino allyl-labeled cDNA. RT reactions contained 20 μg of total RNA; 30 μg of random hexamers; 10 U of Superscript II (Gibco/BRL) reverse transcriptase; 500 μM concentrations each of dATP, dCTP, and dGTP; 200 μM dTTP; and 300 μM amino allyl dUTP. The amino allyl-labeled cDNA was coupled to either the cy3 or cy5 dyes as described previously (41), and experimental and control probes were hybridized to the same microarray. Typically, 10 μg of each labeled cDNA probe (cy3- and cy5-labeled cDNA) was used in a single hybridization. Microarray hybridizations were carried out at 42°C for 16 h in a solution of 25% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), 0.5 mg of yeast tRNA/ml, and 1 mg of sheared salmon sperm DNA/ml under a coverslip in a humidified chamber. The slides were given one 5-min wash in 2× SSC-0.1% SDS at 42°C, one 10-min wash in 0.1× SSC-0.1% SDS at 20°C, and four 1-min washes in 0.1% SDS at 20°C and then rinsed in distilled H2O and ethanol. Slides were dried by centrifugation at 1,000 × g for 3 min and scanned immediately.
Real-time RT-PCR.
Primers for real-time RT-PCR were designed with Primer Express software (ABI) (Table 1). RT reactions were carried out as described for microarray hybridizations (see above). The synthesized cDNA samples were diluted 40- to 80-fold prior to real-time RT-PCR, which was carried out on an ABI PRISM model 7700 sequence detector with product accumulation quantified by incorporation of the fluorescent dye SYBR Green. Triplicate real-time RT-PCRs were performed on 2.4 μl of cDNA with SYBR Green PCR master mix (ABI) and 50 nM concentrations of each gene-specific primer. The concentration of template in each reaction was determined by comparison with a gene-specific standard curve constructed from known concentrations of P. multocida strain X-73 genomic DNA. gyrB was used as the normalizer for all reactions. All RT-PCRs amplified a single product as determined by melting curve analysis.
TABLE 1.
Sequences of primers used for real-time RT-PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| dcaA | GATATGCCGATTTCAGATTGTCAA | CAGGTGGCTGATTGGCTTGT |
| fecB | CGCCATTCTGCCGTCTATG | TGCGGTTTCAAGGTTTTCTTG |
| fnr | GCGGGTGATCCATTAAATTCTCT | TCGCCTGGTAACTGAAATGAAGT |
| glpQ | ACAAGATCTCGCAATGACGAAAG | CTCGCTCGATTTGGGAATTTT |
| gyrB | GCCCTTTCCGATAAATTGCAA | ATCGCGGCTAATGGTGCTT |
| hexC | GCTCGCGCACAAGATGATACT | AAAACTGGCGGATAGGGATACTTT |
| lpp | AAGCTGATAGCCAAGGAGTTCTTG | CGTTGCAATCATTTGACCAGTAC |
| lppC | GGCGTCTATATTGTTGCCATGA | TACTGCGCGAACTGGCATAA |
| pm0209 | AATTTTGGGTGCTGTTTGAAGTG | ATAAAGACTTTCGCTTGCAATGC |
| pm0453 | CAACATCAAGATAAGCTTCCCGTTA | GCCTGCATTAGCAAACACGAA |
| pm1269 | GAAGAAACGATCCCTTGTCCC | AGGAAAGCGATCACAGCCAT |
| rpoE | TCACAAAACGACATTCCTGATGTT | GCCATGTGTAGAAAGCACTGTCA |
| yfeB | TGCCCTTGCCCAACAAAG | TGATCCAACAATGCCATAATCG |
| yfeC | GGAAAATTAGCGACGATTGCA | TAAACCGCCTGTTGCTCCAT |
Analysis of microarray images.
Microarray hybridizations were scanned with a GMS model 418 microarray scanner (Genetic Microsystems). The cy3 and cy5 images were combined prior to grid location to allow the accurate identification of spot position. The grid location and amount of hybridization of the cy3- or cy5-labeled cDNA to each spot were quantified with Imagene version 4.1 (Biodiscovery). Expression values for each gene were calculated with GeneSight version 3.0.7 (Biodiscovery) by using local background correction and the omission of all negative and very low positive values (<100), and the resultant data were transformed such that the mean expression value for each channel (cy3 and cy5) had a log ratio of zero. Bacterial RNA isolated from each infected chicken (n = 3) was used for two comparative hybridizations with in vitro-derived RNA. Each pair of hybridizations, corresponding to RNA derived from a single infected animal, was analyzed separately. All replicate spots from hybridizations of samples derived from a single animal (up to six values from two hybridizations) were combined, and the average log ratios were used for all further analysis. Identification of differentially regulated genes was carried out in GeneSight with the confidence analyzer (based on the method of Kerr et al. [20]). This statistical analysis uses a comparison of all replicate measurements to determine an experiment-wide noise level. The calculated noise level is then used to determine the statistical measures for the likelihood of false positives above or below a certain expression ratio. Genes were identified as differentially regulated if there was a 95% chance that their average expression value (calculated from the two hybridizations) was above a log2 of 0.6 (up-regulated) or below a log2 of −0.6 (down-regulated). These values correspond to approximately 1.5-fold differences for up- and down-regulated genes, respectively. The statistical significance of similarity between differentially regulated gene sets was assessed by using either the chi-square test or Fisher's exact test (InStat; GraphPad Software Inc.) where appropriate.
RESULTS AND DISCUSSION
Comparison of gene expression profiles of P. multocida during growth in BHI or in the chicken host.
To analyze the expression profiles of P. multocida during growth within a natural host, we used the well-characterized model of P. multocida infection in chickens. P. multocida serotype A strains cause fowl cholera, which usually presents as an overwhelming septicemia in the terminal phase of infection (39). Six chickens were infected with 5 × 104 CFU of P. multocida strain X-73 (serotype A:1). Infected birds were monitored closely for signs of infection, and the level of bacteremia was assessed by regular blood smears. All birds showed clinical signs of fowl cholera within 16 to 18 h, including depression, ruffled feathers, fever, diarrhea, and increased respiratory rate (39). Three chickens, designated c1, c2, and c3, attained high levels of bacteremia at the final stages of infection (approximately 2 × 109 CFU/ml at 22 h, 8 × 109 CFU/ml at 17 h, and 7 × 109 CFU/ml at 21 h, respectively), and bacteria were harvested from each of these infections. At the time of harvest, all chickens showed identical clinical signs (febrile and very depressed) and were all in the terminal phase of disease progression. Despite identical clinical profiles, three chickens failed to attain levels of bacteremia sufficient for RNA purification (>108 CFU/ml). Total RNA was isolated from bacteria from each of the c1, c2, and c3 infections and used to probe a P. multocida strain PM70 microarray that contained all 2,015 of the annotated P. multocida genes (32). Control RNA was isolated from P. multocida strain X-73 grown aerobically at 41°C (the normal body temperature of chickens [30]) in BHI to a cell density similar to that in the in vivo-grown samples (5 × 109 CFU/ml).
Microarrays were hybridized with control and experimental RNA labeled with either cy3 or cy5. Two hybridizations were carried out with each RNA sample; one hybridization had the experimental sample labeled with cy3, and the other hybridization had the experimental sample labeled with cy5. Hybridizations derived from the different chicken infections were analyzed separately. We designated all genes with average expression ratios of 1.5-fold (up or down) differentially expressed. We chose the 1.5-fold value, as other studies have indicated that a 1.5-fold change in gene expression may be biologically relevant (16, 41). Furthermore, we used the Genesight (Biodiscovery) confidence analyzer to identify genes that had a 95% likelihood of being differentially expressed at above or below the 1.5-fold threshold.
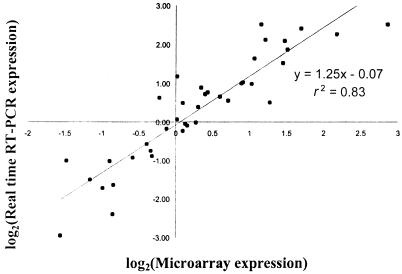
Real-time RT-PCR was used to confirm a number of the expression ratios identified by microarray analysis (Fig. 1). Thirteen genes (including up- and down-regulated and constitutively expressed genes) were analyzed with bacterial RNA from each of the three infected chickens. For RNA derived from all three infections, there was a strong positive correlation between the two methods (r2 = 0.83). The slope of the regression line (m = 1.25) indicated that generally slightly higher expression changes were measured by real-time RT-PCR rather than by microarray hybridization.
FIG. 1.
Correlation between microarray and real-time RT-PCR expression values. Log2-transformed expression values for 13 genes from bacteria isolated from each of the three infected chickens (c1, c2, and c3) were compared. The best-fit linear regression line is shown together with the r2 value and the calculated equation.
Differentially expressed genes.
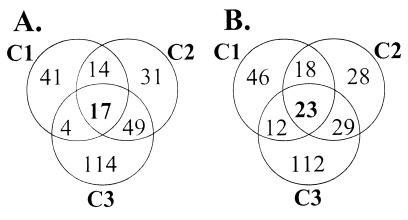
Analysis of the differentially regulated gene sets (Fig. 2) indicated that all three infections were highly similar (chi-square test; P < 0.001). Forty genes (2% of the genome) were differentially expressed in all three chickens. Of these genes, 17 were up-regulated (Table 2) and 23 were down-regulated (Table 3) during growth in the natural chicken host compared with those under in vitro growth conditions. Another 126 genes were differentially regulated in two of the three infections, while 372 genes were identified as differentially regulated in a single chicken.
FIG. 2.
Venn diagrams showing the number of P. multocida genes up-regulated (A) or down-regulated (B) in bacterial populations isolated from three infected chickens (c1, c2, and c3).
TABLE 2.
P. multocida genes up-regulated during growth within all three infected chickens
| Gene | Precise or predicted function (COGs categorya) | Expression valueb
|
||
|---|---|---|---|---|
| c1 | c2 | c3 | ||
| asnA | Ammonia-dependent asparagine synthetase (E) | 3.22 ± 0.77 | 1.24 ± 0.3 | 2.71 ± 0.37 |
| aspC | Aspartate transaminase (E) | 1.09 ± 0.37 | 1.17 ± 0.3 | 1.51 ± 0.32 |
| dcaA | Unknown (R) | 1.27 ± 1.34 | 2.18 ± 0.43 | 2.86 ± 0.9 |
| dppA | Dipeptide transport system permease (E) | 0.88 ± 0.57 | 1.95 ± 0.45 | 3.5 ± 0.33 |
| gdhA | Glutamate dehydrogenase (E) | 3.01 ± 0.27 | 1.51 ± 0.39 | 3.27 ± 0.91 |
| gltA | Citrate synthase (C) | 1.04 ± 0.39 | 1.35 ± 0.43 | 2.05 ± 0.45 |
| ilvH | Acetolactate synthase (E) | 1.21 ± 0.99 | 1.82 ± 0.41 | 1.33 ± 0.23 |
| napA | Periplasmic nitrate reductase (C) | 2.33 ± 0.26 | 2.13 ± 0.51 | 1.69 ± 0.14 |
| napC | Periplasmic nitrate reductase (C) | 1.97 ± 0.53 | 2.28 ± 0.81 | 1.08 ± 0.85 |
| napF | Periplasmic nitrate reductase (C) | 3.7 ± 0.17 | 2.58 ± 0.17 | 2.29 ± 0.43 |
| PM0092 | Sigma-54 modulation protein (J) | 1.4 ± 0.41 | 0.83 ± 0.1 | 1.63 ± 0.14 |
| PM0287 | Oxidoreductase (R) | 0.99 ± 0.32 | 3.07 ± 0.17 | 2.1 ± 0.3 |
| PM0601 | Carbohydrate transport and metabolism (G) | 0.9 ± 1.18 | 0.76 ± 0.72 | 1.36 ± 1.05 |
| PM1460 | Phosphoglycerate transport system activator (T) | 1.26 ± 1.15 | 1.23 ± 0.4 | 1.29 ± 0.61 |
| ppc | PEP carboxylase (C) | 3.07 ± 0.59 | 1.39 ± 0.49 | 3.04 ± 0.61 |
| ptfA | Type IV fimbrial subunit (N) | 0.89 ± 0.79 | 0.83 ± 0.45 | 0.9 ± 0.44 |
| purH | Purine biosynthesis (F) | 1.26 ± 0.12 | 0.87 ± 0.16 | 1.21 ± 0.25 |
COGs functional categories (44). The full functional categories associated with each COGs letter designation are given in the legend to Fig. 3. Where no precise function is apparent from amino acid homology, the general COGs category title is listed under the predicted function.
Expression values are expressed as the log2 of (average experimental intensity/average control intensity) ± 1 standard deviation. Therefore, identical experimental and control expression values would be 0.0.
TABLE 3.
P. multocida genes down-regulated during growth within all three infected chickens
| Gene | Precise or predicted function (COGs categorya) | Expression valueb
|
||
|---|---|---|---|---|
| c1 | c2 | c3 | ||
| glpB | Glycerol-3-phosphate dehydrogenase (E) | −1.05 ± 0.34 | −1.53 ± 0.21 | −1.23 ± 0.21 |
| glpC | Glycerol-3-phosphate dehydrogenase (C) | −0.94 ± 0.2 | −1.35 ± 0.18 | −0.91 ± 0.15 |
| hbpA | Heme-binding protein A precursor (E) | −3.77 ± 0.37 | −3.02 ± 0.17 | −2.18 ± 0.44 |
| murZ | UDP-GlcNAc enolpyruvyl transferase (F) | −0.95 ± 0.31 | −1.06 ± 0.3 | −1.78 ± 0.23 |
| PM0038 | Amidotransferase (R) | −2.94 ± 0.16 | −2.63 ± 0.32 | −2.51 ± 0.69 |
| PM0159 | Unknown (R) | −2.05 ± 0.23 | −2.55 ± 0.37 | −2.71 ± 0.41 |
| PM0243 | Nucleotide transport and metabolism (F) | −1.24 ± 0.14 | −1.07 ± 0.2 | −1.2 ± 0.14 |
| PM0310 | Unknown (−) | −2.2 ± 0.66 | −1.02 ± 2.3 | −1.81 ± 0.17 |
| PM0335 | Glutamate symport protein (R) | −2.15 ± 0.47 | −2.79 ± 1.12 | −4.44 ± 0.44 |
| PM0402 | Unknown (R) | −1.43 ± 0.31 | −1.44 ± 0.29 | −1.46 ± 0.24 |
| PM0591 | Unknown (S) | −0.94 ± 0.68 | −1.08 ± 0.42 | −1.49 ± 1.59 |
| PM0682 | Unknown (−) | −1.23 ± 0.82 | −1.65 ± 0.91 | −1.7 ± 0.5 |
| PM0761 | Unknown (S) | −1 ± 0.23 | −1.15 ± 0.11 | −2.02 ± 0.26 |
| PM0965 | Unknown (−) | −2.62 ± 0.85 | −2.77 ± 0.45 | −1.45c |
| PM1233 | Amidotransferase (H) | −1.87 ± 1.06 | −2.01 ± 0.63 | −0.92 ± 0.12 |
| PM1566 | Unknown (S) | −1.84 ± 0.41 | −1.66 ± 0.21 | −1.84 ± 0.21 |
| PM1889 | Unknown (S) | −1.22 ± 0.47 | −1.22 ± 0.2 | −1.96 ± 0.48 |
| PM1926 | Nucleotide transport and metabolism (F) | −0.9 ± 0.37 | −1.19 ± 0.16 | −1.69 ± 0.16 |
| PM1932 | Unknown (−) | −0.96 ± 0.38 | −1.27 ± 0.65 | −1.58 ± 0.88 |
| ptsB | Sucrose-specific PTSII (G) | −1.76 ± 0.52 | −2.09 ± 0.69 | −1.84 ± 0.38 |
| rpL34 | Ribosomal protein L34 (J) | −1.07c | −1.67c | −1.15 ± 1.95 |
| rpL35 | Ribosomal protein L35 (J) | −1.95 ± 0.21 | −2.07 ± 0.33 | −1.78 ± 0.35 |
| rseB | Sigma-E regulatory protein (T) | −2.08 ± 0.18 | −1.5 ± 0.16 | −1.21 ± 0.14 |
COGs functional categories (44). The full functional categories associated with each COGs letter designation are given in the legend to Fig. 3. Where no precise function is apparent from amino acid homology, the general COGs category title is listed under the predicted function.
Expression values are expressed as the log2 of (average experimental intensity/average control intensity) ± 1 standard deviation. Therefore, identical experimental and control expression values would be 0.0.
No standard deviation could be calculated as the data were derived from fewer than three values.
Genes up-regulated in all three infections.
Seventeen P. multocida genes were identified as up-regulated during growth within the host in all three infected chickens (Table 2). Of these, most were involved in amino acid transport and metabolism (5 of 17) and energy production and conversion (5 of 17).
The glutamate dehydrogenase gene (gdhA) was very strongly up-regulated in bacteria isolated from all three infected chickens (8-fold, 3-fold, and 10-fold [Table 2]). GdhA converts ammonia from the environment to glutamate (NH3 + α-ketoglutarate + NADPH → glutamate + NADP). In Escherichia coli, this reaction is one of the primary pathways for the assimilation of nitrogen and the glutamate derived from this process is required directly or indirectly for the synthesis of all amino acids; half the nitrogen atoms for the pyrimidine, purine, and imidazole ring; and the amino group of adenine (36). Although glutamate can be synthesized by two different pathways (13), the GdhA pathway is preferred under energy-limited conditions (12, 13). Therefore, in an environment containing ammonia but poor in amino acids and energy limited, this reaction is of critical importance. Glutamate is also a prime cellular defense against osmotic stress and can serve as a carbon source (28). The gene asnA was also strongly up-regulated during growth in all three chickens; this gene encodes an ammonia-dependent asparagine synthetase that produces asparagine from aspartate and ammonia. It has been shown previously in Klebsiella aerogenes that levels of this enzyme are high when cells are grown in an amino acid-limited but ammonia-rich environment (37). The gene aspC, which was also strongly up-regulated in all chickens, is predicted to encode aspartate transaminase, which catalyses the conversion of glutamate to aspartate. In E. coli, this reaction is the prime mechanism of aspartate formation and aspartate is the substrate for the reaction catalyzed by AsnA (see above). It should be noted that glutamate can also function as a major carbon source in E. coli and that the conversion of glutamate to aspartate by AspC is part of the glutamate energy utilization pathway. Furthermore, AspC can function to synthesize phenylalanine and tyrosine (36). Thus, it is likely that the products of gdhA, asnA, and aspC function to convert host-derived ammonia to amino acids. The dppA gene, which was significantly up-regulated in all three chickens, encodes part of a periplasmic dipeptide transport system, while another significantly up-regulated gene, ilvH, encodes an enzyme involved in amino acid biosynthesis. Taken together, these data clearly show that, during growth in the host, P. multocida must modify biosynthetic pathways to cope with an environment that is comparatively amino acid poor but nitrogen (ammonia) rich. To this end, most common up-regulated genes are involved in critical (probably rate-limiting) steps in the transport and biosynthesis of amino acids.
Many of the common up-regulated genes were predicted to be involved in energy production and conversion (Table 2). The nitrate reductase (nap) operon in P. multocida has an arrangement (napFDAGHBC) identical to that found in other gram-negative pathogens (34). The napF, napA, and napC genes were highly up-regulated in the bacteria from all three chickens, while napB was significantly up-regulated in the c2 and c3 chickens and fell just below the level of significance in c1. The napD gene was observed to be significantly up-regulated in the c1 infection but only slightly up-regulated in the other two infections. Taken together, these data indicate that the nitrate reductase operon is significantly up-regulated during growth in the chicken host. The function of the periplasmic nitrate reductase is to reduce nitrate to nitrite, with nitrate being the preferred terminal electron acceptor in the absence of oxygen. Therefore, nitrate reductases are generally assumed to be necessary for anaerobic growth (6). It has also been suggested that an important function of these proteins in bacterial pathogens is to scavenge nitrogen from bodily fluids (34). Given that the nitrite produced by the periplasmic nitrate reductases is rapidly converted to ammonia by nitrite reductases, we hypothesize that this pathway may aid the biosynthesis of glutamate (see above) by increasing the ammonia pool through nitrate scavenging.
Two other genes involved in energy production were up-regulated during growth within all of the three infected chickens (Table 2). Citrate synthase (gltA) has been shown to be the rate-limiting step for the tricarboxylic acid (TCA) cycle when E. coli cells are grown on acetate (45). It is likely that up-regulation of this gene in P. multocida leads to a concomitant increase in the rate of energy production (or biosynthetic functions branching from the TCA cycle) under the carbon-limited conditions encountered during growth within the host. The ppc gene is predicted to encode phosphoenolpyruvate (PEP) carboxylase, which functions to convert PEP to oxaloacetate. Oxaloacetate is the precursor of several intermediates of both the TCA cycle and other biosynthetic pathways.
Seven other genes were identified as significantly up-regulated during growth in all three chickens (Table 2). The purH gene is predicted to be involved in purine biosynthesis. Genes involved in purine or pyrimidine biosynthesis have been identified as virulence-associated genes by STM techniques in a number of bacterial species, including P. multocida (9, 33). The ptfA gene is the type IV fimbrial subunit gene (7, 40). Fimbriae are well-characterized virulence factors (10, 19), and there is evidence that some types of fimbriae may be expressed in blood (35). The pm1460 and pm0092 genes are both likely to be involved in gene regulation. The pm0092 gene is predicted to encode a sigma-54 modulation protein, while pm1460 is predicted to encode a phosphoglycerate transport protein that contains a sigma-54 interaction domain. Genes controlled by sigma-54 are generally involved in nitrogen fixation and utilization, although pilin synthesis is also often controlled by sigma-54 (29). The other three up-regulated genes have either been poorly characterized (pm0601) or have no known function (dcaA and pm0287), although dcaA is likely to be transcribed with purH. Taken together, these data indicate that a clear common theme in the genes up-regulated in bacteria during growth in all three chickens is their central importance in the metabolism of nitrogen-containing compounds.
Genes down-regulated in all infections.
In contrast to the genes up-regulated during growth in all three chickens, most (15 of 23) of the genes down-regulated in all three infections (Table 3) were of unknown or poorly characterized function. Two genes, rpL34 and rpL35, encode ribosomal proteins; their lower level of transcription during growth within the host may be an indicator of a reduced bacterial growth rate. The reduced expression of murZ, which encodes UDP-GlcNAc enolpyruvyl transferase, the enzyme which catalyses the first committed step in peptidoglycan synthesis (3), may also indicate reduced growth rate in the chicken host. Two genes, glpB and glpC, encode subunits of the anaerobically expressed glycerol-3-phosphate dehydrogenase, while the gene glpA, which encodes the third subunit, was also significantly down-regulated in two chickens.
Analysis of all differentially expressed genes by functional groupings.
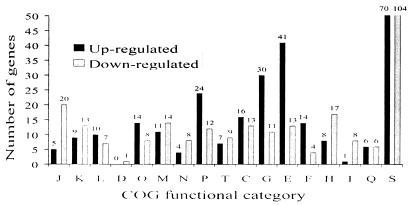
Separate analysis of bacteria derived from each of the three chickens indicated that 522 P. multocida genes (26%) were differentially expressed in bacteria isolated from at least one chicken. The expression ratios of these genes (discussed in the following three sections) in each of the three infections are available online (http://www.med.monash.edu.au/microbiology/staff/adler/PastyArray.html). Of these, 270 were up-regulated and 268 were down-regulated during growth in the chicken compared with those under in vitro growth conditions. Sixteen genes were present in both the up-regulated and down-regulated gene sets. The 522 differentially regulated genes were categorized into their clusters-of-orthologous-groups (COGs) functional categories (44) (Fig. 3). COGs categories highly represented in the up-regulated genes included those involved in amino acid, carbohydrate, nucleotide, and inorganic ion transport and metabolism. COGs categories highly represented in the down-regulated genes included those involved in protein synthesis (translation, ribosomal structure, and biogenesis), coenzyme and lipid metabolism, and cell envelope and outer membrane biogenesis. Notably, there were also 169 genes of unknown function identified as differentially regulated during bacterial growth in at least one chicken; 70 of these were up-regulated.
FIG. 3.
Number of differentially expressed genes by COGs functional categories, including all genes identified as differentially expressed in any of the three animals. The bars for category S have been truncated, and the numbers above each bar indicate the number of genes in each category. The COGs functional categories are as follows: J, translation, ribosomal structure, and biogenesis; K, transcription; L, DNA replication, recombination, and repair; D, cell division and chromosome partitioning; O, posttranslational modification, protein turnover, and chaperone functions; M, cell envelope and outer membrane biogenesis; N, cell motility and secretion; P, inorganic ion transport and metabolism; T, signal transduction mechanisms; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme metabolism; I, lipid metabolism; Q, secondary metabolite biosynthesis, transport, and catabolism; S, unknown function (which includes categories R, general function prediction only, and category −, not in COGs).
Amino acid, carbohydrate, and inorganic ion transport and metabolism genes.
Large percentages of the genes predicted to be involved in amino acid (25%), nucleotide (23%), inorganic ion (22%), and carbohydrate (22%) transport and metabolism were significantly up-regulated in vivo in at least one infection (Fig. 3). By comparison, only 8, 7, 12, and 8% of genes from the respective categories were down-regulated in any of the infections. It is clear that growth in vivo is a more challenging environment for the bacteria to obtain the requisite amino acid, nucleotide, ion, and energy sources than that encountered during growth within BHI. Several genes involved in these processes have been identified previously as necessary for in vivo survival. For example, inactivation of the P. multocida genes aroA (15), purF (9), and purN (our unpublished data) results in the attenuation of virulence because of the reduced ability of the mutants to replicate in vivo. Each of these genes was significantly up-regulated in at least two of the three infections.
Signal transduction and transcription genes.
Many of the genes involved in signal transduction and transcription were differentially regulated in bacteria from at least one infected chicken (Fig. 3). Most of these differentially regulated genes were predicted to be involved in the sensing of the redox state and the presence and/or absence of oxygen and included fnr, oxyR, arcA, and arcB. These findings are consistent with a difference in redox potential between growth in vivo and in aerated broth culture. Surprisingly, the differential regulation of these genes was not consistent across the three infections. Indeed, fnr was significantly up-regulated in vivo in the c2 and c3 infections but significantly down-regulated in the c1 infection. The expression of this gene in the three infections was confirmed by real-time PCR, which gave identical results (data not shown).
Many transcription factors were down-regulated in at least one infection (Fig. 3), including rpoE, rpoH, PM0209, rho, nusA, and nusB. The sigma factors RpoH and RpoE are predicted to be involved in the heat and extracytoplasmic shock responses. Interestingly, rseB, the gene encoding a regulator of RpoE, was also significantly down-regulated during growth in chickens. Paradoxically, the heat shock-induced chaperones HtpG, DnaK, DnaJ, GroES, and PrlC, which are positively regulated by RpoH, were generally up-regulated in vivo. The up-regulation of heat shock proteins during infection has been reported previously (18, 21, 23), and P. multocida has been shown to express heat shock proteins during infection (25). These results may indicate that, in P. multocida, as in E. coli (4), induction of RpoH synthesis is primarily regulated posttranscriptionally. Furthermore, dnaK, dnaJ, and groES transcription is known to be induced by a range of stresses other than heat shock, including oxidative stress, osmotic stress, and amino acid starvation (5, 42).
Differences between infections.
Each of the three infections was initiated with the injection of 5 × 104 CFU of P. multocida strain X-73 into the breast muscle. At the time of bacterial harvest, the attained cell density in the blood varied only slightly (between approximately 2 × 109 and 8 × 109 CFU/ml) in birds from which bacterial RNA was taken for study. Furthermore, each of the three chickens displayed identical clinical signs and was in the terminal phase of disease. It is possible that blood homeostasis may have broken down in some of the animals at this time, but we regard this as a natural consequence of infection. Thus, although it is impossible to know if bacteria were harvested at identical stages during the infection, every effort was taken to isolate the bacteria at the same time with regard to disease progression.
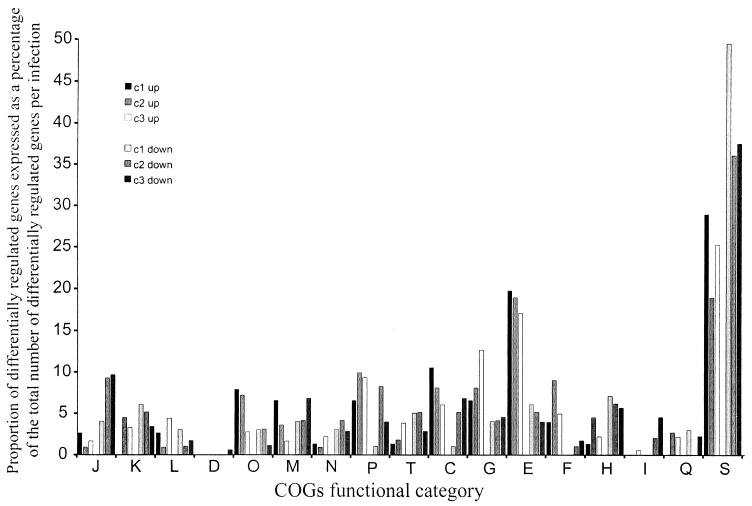
Overall, the differentially expressed gene sets identified for bacteria harvested from each chicken were all highly similar (chi-square test; P < 0.001), although there were also clear differences at the level of functional gene subsets. Cluster analysis indicated clearly that the variation within replicate hybridizations of RNA from the same infection was significantly lower than the variation between hybridizations of RNA samples from different infections (data not shown). The differentially regulated genes from each infection were categorized into their COGs functional categories (44) (Fig. 4).
FIG. 4.
The proportion of differentially regulated genes in each COGs category expressed as a percentage of the total number of differentially expressed genes for each infection. The percentages of genes from each infection that were up-regulated (c1 up, c2 up, and c3 up) or down-regulated (c1 down, c2 down, and c3 down) are shown for each COGs category. The COGs functional categories are as described for Fig. 3.
The majority of genes significantly up-regulated in the c2 and c3 infections, but not in the c1 infection, were involved in nucleotide, carbohydrate, and inorganic ion transport and metabolism (Fig. 4). The htpG, dnaK, dnaJ, groES, and prlC genes, which encode proteins involved in protein turnover and chaperone functions, were all significantly up-regulated in the c1 and c2 infections but not in the c3 infection. Although these genes are generally involved in the heat shock response, rpoH and rpoE were down-regulated in these infections and a number of other known heat shock genes showed no up-regulation.
The response of P. multocida to low iron concentration in vitro has been studied previously (27, 31), and a number of genes have been identified as being differentially regulated in response to limiting free iron. We compared the set of genes identified as up-regulated in vitro under iron limitation (31), with the set of genes up-regulated in bacteria grown within each of the three infected chickens (Table 4). Only 3 of the 71 genes identified as up-regulated in the c1 infection have previously been shown to respond to low iron in vitro, indicating that these gene sets are not significantly similar (Fisher's exact test; P > 0.9). However, 13 of the 93 genes identified as up-regulated in the c2 infection and 25 of the 180 genes identified as up-regulated in the c3 infection have previously been shown to respond to low iron in vitro, indicating that each of these gene sets is significantly similar (Fisher's exact test; P < 0.001). These data indicate that only the c2 and c3 infections displayed a gene expression profile similar to that seen during in vitro iron starvation. It is often assumed that gene expression in vivo would be similar to gene expression under in vitro conditions in the absence of iron. The gene expression levels in the c1 infection indicate that this may not always be the case, although it is possible that some genes involved in growth during iron-starved conditions are expressed only transiently.
TABLE 4.
P. multocida genes previously shown to respond to low iron levels in vitro (31) that were up-regulated during growth within infected chickens
| Gene | Predicted function | Expression valuea
|
||
|---|---|---|---|---|
| c1 | c2 | c3 | ||
| aroA | 3-Phosphoshikimate 1-carboxyvinyltransferase | 0.03 | 0.84 | 1.15 |
| cydD | Transport ATP-binding protein | −0.01 | 0.64 | 1.58 |
| fecB | Iron(III) dicitrate-binding periplasmic protein | −0.13 | 0.10 | 1.51 |
| fnr | Fumarate (and nitrate) reduction regulatory protein | −0.99 | 1.45 | 1.21 |
| folB | Dihydroneopterin aldolase | 0.45 | 0.82 | 0.80 |
| hugZ_1 | Heme utilization protein | −0.25 | 0.96 | 2.11 |
| hugZ_2 | Heme utilization protein | −0.25 | 0.96 | 2.11 |
| ilvM | Acetolactate synthase small subunit | 0.14 | 0.80 | 1.71 |
| lldD | l-Lactate dehydrogenase | −0.37 | 2.22 | 2.08 |
| nagB | Glucosamine-6-phosphate isomerase | 0.20 | 0.80 | 0.79 |
| nrdB | Ribonucleoside diphosphate reductase, beta chain | 0.60 | 0.46 | 0.85 |
| PM0298 | Unknown | −0.30 | 0.60 | 0.96 |
| PM0300 | Putative TonB-dependent receptor | −0.22 | 0.68 | 1.14 |
| PM0336 | Putative TonB-dependent receptor | 0.36 | 1.09 | 2.20 |
| PM0424 | Unknown | −0.90 | −0.04 | 0.91 |
| PM0452 | Unknown | −0.21 | 1.04 | 1.67 |
| PM0741 | Hemoglobin receptor precursor | 0.08 | 1.02 | 1.20 |
| PM0803 | Unknown | −0.61 | 0.70 | 1.86 |
| PM1171 | Unknown | −0.49 | 0.04 | 1.03 |
| PM1460 | Phosphoglycerate transport system activator protein | 1.26 | 1.23 | 1.29 |
| PM1604 | Unknown | 0.24 | −0.09 | 1.10 |
| PM1727 | Unknown | 2.29 | 0.21 | −0.46 |
| pqqL | Zinc protease | −0.24 | 0.73 | 2.00 |
| tal | Transaldolase | 0.18 | 0.67 | 1.44 |
| trpG | Anthranilate synthase component II | 1.39 | 0.20 | 0.87 |
| yfeB | Iron(III) dicitrate transport ATP-binding protein | 0.10 | 0.91 | 1.70 |
| yfeC | Iron (chelated) ABC transporter | 0.02 | 0.35 | 1.07 |
| yfeD | Iron (chelated) transporter, permease protein | 0.05 | 0.54 | 1.13 |
Expression values are expressed as the log2 of (average experimental intensity/average control intensity). Therefore, identical experimental and control expression values would be 0.0.
Concluding remarks.
This is the first report of genomic-scale expression profiling of bacteria within a host during infection. The results of our investigations demonstrate that such an analysis identifies a range of genes of fundamental importance to bacterial survival within the host as well as the pathogenesis of infection. Our data explore gene expression at a single time point and in bacteria isolated from a single host tissue (the blood), and the profiles of gene expression may well be different in other tissues or at different times during infection. However, this study has identified a large number of genes whose expression is clearly required substantially more during growth within the blood of septicemic chickens than during growth in vitro. The large number of metabolic genes whose expression was altered in vivo reflects the different nutritional and biochemical environment within the host. A common theme observed in the genes up-regulated during growth in the host is the importance of metabolic pathways involved in nitrogen uptake and metabolism. It is likely that many of the unknown genes up-regulated during growth within the chicken host encode virulence or virulence-associated factors. Therefore, we believe that the genes identified in this study will provide a basis for directed vaccine development, both by the production of attenuated mutants and by the identification of immunogenic in vivo-expressed surface proteins.
A number of large-scale screening methods have been used previously to identify potential virulence genes in P. multocida, including STM (9) and IVET (17). In this study, 7 of the 23 P. multocida genes identified by STM were also identified as differentially regulated while 3 of the 14 genes identified by IVET were identified as up-regulated during growth in chickens. Clearly many genes were identified in our study but not by the STM and IVET methods, indicating that neither of these methods produces a saturation coverage of the genome. One of the clear benefits of whole-genome expression profiling with microarrays is the ability to analyze the gene expression profiles of every gene under the conditions being tested. However, some genes were identified by STM and IVET methods that were not identified in this work. This is not surprising, as STM and IVET analyses were carried out on different P. multocida strains and both used mouse infection models. Thus, we believe our infection model is significantly more relevant to natural fowl cholera. Furthermore, STM identifies genes that are necessary for in vivo survival and it is clear that such genes need not be differentially expressed under in vivo and in vitro conditions. Another clear benefit of the microarray approach, compared to that of IVET, is the ability to apply statistical measures of reproducibility to datasets derived from multiple infections.
A key finding of our work was that the expression patterns of bacteria may vary considerably between infected individuals. The reasons for this are not clear but could include genetic host variability or differences in the breakdown of blood homeostasis in animals. However, we suggest that experiments in which in vivo-derived bacteria are pooled from more than one animal may lead to overly simplified conclusions regarding gene expression in the host. An important finding resulting from this work is the identification of 70 genes of unknown function that were significantly up-regulated in at least one infected individual. Future detailed analyses of these genes and their products will lead to an improved understanding of the pathogenesis of fowl cholera at the molecular level.
Acknowledgments
We thank Vicki Vallance and Ian McPherson for excellent technical assistance, Aidan Sudbury for statistical advice, and Julian Rood and John Davies for critical reading of the manuscript.
This work was funded in part by a project grant from the Australian Research Council. M.P. is supported by an NIH National Institute of General Medical Sciences Training for Future Biotechnology Development grant (T32 GM08347). Funding for the preparation of the microarrays was provided by research grants from the Minnesota Turkey Growers Association, the Minnesota Agricultural Experimentation Station, the University of Minnesota Academic Health Center, and the U.S. Department of Agriculture National Research Initiative (to V.K.).
Editor: J. T. Barbieri
REFERENCES
- 1.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyce, J. D., and B. Adler. 2000. The capsule is a virulence determinant in the pathogenesis of Pasteurella multocida M1404 (B:2). Infect. Immun. 68:3463-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, E. D., E. I. Vivas, C. T. Walsh, and R. Kolter. 1995. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J. Bacteriol. 177:4194-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukau, B. 1993. Immunogold localization of the DnaK heat shock protein in Escherichia coli cells. J. Gen. Microbiol. 139:95-99. [DOI] [PubMed] [Google Scholar]
- 5.Chakrabarti, S., N. Sengupta, and R. Chowdhury. 1999. Role of DnaK in in vitro and in vivo expression of virulence factors of Vibrio cholerae. Infect. Immun. 67:1025-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, J. 1996. Nitrate reduction to ammonia by enteric bacteria: redundancy, or a strategy for survival during oxygen starvation? FEMS Microbiol. Lett. 136:1-11. [DOI] [PubMed] [Google Scholar]
- 7.Doughty, S. W., C. G. Ruffolo, and B. Adler. 2000. The type 4 fimbrial subunit gene of Pasteurella multocida. Vet. Microbiol. 72:79-90. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez de Henestrosa, A. R., I. Badiola, M. Saco, A. M. Perez de Rozas, S. Campoy, and J. Barbe. 1997. Importance of the galE gene on the virulence of Pasteurella multocida. FEMS Microbiol. Lett. 154:311-316. [DOI] [PubMed] [Google Scholar]
- 9.Fuller, T. E., M. J. Kennedy, and D. E. Lowery. 2000. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb. Pathog. 29:25-38. [DOI] [PubMed] [Google Scholar]
- 10.Harel, J., and C. Martin. 1999. Virulence gene regulation in pathogenic Escherichia coli. Vet. Res. 30:131-155. [PubMed] [Google Scholar]
- 11.Heddleston, K. L., and P. A. Rebers. 1972. Fowl cholera: cross-immunity induced in turkeys with formalin-killed in-vivo-propagated Pasteurella. Avian Dis. 16:578-586. [PubMed] [Google Scholar]
- 12.Helling, R. B. 1998. Pathway choice in glutamate synthesis in Escherichia coli. J. Bacteriol. 180:4571-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helling, R. B. 1994. Why does Escherichia coli have two primary pathways for synthesis of glutamate? J. Bacteriol. 176:4664-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 15.Homchampa, P., R. A. Strugnell, and B. Adler. 1997. Cross protective immunity conferred by a marker-free aroA mutant of Pasteurella multocida. Vaccine 15:203-208. [DOI] [PubMed] [Google Scholar]
- 16.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 17.Hunt, M. L., D. J. Boucher, J. D. Boyce, and B. Adler. 2001. In vivo-expressed genes of Pasteurella multocida. Infect. Immun. 69:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kantengwa, S., I. Muller, J. Louis, and B. S. Polla. 1995. Infection of human and murine macrophages with Leishmania major is associated with early parasite heat shock protein synthesis but fails to induce a host cell stress response. Immunol. Cell Biol. 73:73-80. [DOI] [PubMed] [Google Scholar]
- 19.Kennan, R. M., O. P. Dhungyel, R. J. Whittington, J. R. Egerton, and J. I. Rood. 2001. The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosus is essential for virulence, protease secretion, and natural competence. J. Bacteriol. 183:4451-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerr, M. K., M. Martin, and G. A. Churchill. 2000. Analysis of variance for gene expression microarray data. J. Comput. Biol. 7:819-837. [DOI] [PubMed] [Google Scholar]
- 21.Khan, M. A., and R. E. Isaacson. 2002. Identification of Escherichia coli genes that are specifically expressed in a murine model of septicemic infection. Infect. Immun. 70:3404-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lax, A. J., and A. E. Grigoriadis. 2001. Pasteurella multocida toxin: the mitogenic toxin that stimulates signalling cascades to regulate growth and differentiation. Int. J. Med. Microbiol. 291:261-268. [DOI] [PubMed] [Google Scholar]
- 23.Lee, B. Y., and M. A. Horwitz. 1995. Identification of macrophage and stress-induced proteins of Mycobacterium tuberculosis. J. Clin. Investig. 96:245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo, R. Y. C., and P. E. Shewen. 1992. The genus Pasteurella, p. 3331-3338. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, applications, 2nd ed., vol. IV. Springer-Verlag, New York, N.Y.
- 25.Love, B. C., and D. C. Hirsh. 1994. Pasteurella multocida produces heat shock proteins in turkeys. Infect. Immun. 62:1128-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 27.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McFall, E., and E. B. Newman. 1996. Amino acids as carbon sources, p. 358-379. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C.
- 29.Merrick, M. J. 1993. In a class of its own—the RNA polymerase sigma factor sigma 54 (sigma N). Mol. Microbiol. 10:903-909. [DOI] [PubMed] [Google Scholar]
- 30.Mezquita, B., C. Mezquita, and J. Mezquita. 1998. Marked differences between avian and mammalian testicular cells in the heat shock induction and polyadenylation of Hsp70 and ubiquitin transcripts. FEBS Lett. 436:382-386. [DOI] [PubMed] [Google Scholar]
- 31.Paustian, M. L., B. J. May, and V. Kapur. 2001. Pasteurella multocida gene expression in response to iron limitation. Infect. Immun. 69:4109-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paustian, M. L., B. J. May, and V. Kapur. 2002. Transcriptional response of Pasteurella multocida to nutrient limitation. J. Bacteriol. 184:3734-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potter, L., H. Angove, D. Richardson, and J. Cole. 2001. Nitrate reduction in the periplasm of gram-negative bacteria. Adv. Microb. Physiol. 45:51-112. [DOI] [PubMed] [Google Scholar]
- 35.Pourbakhsh, S. A., M. Dhomoulin, A. Bree, C. Desautels, B. Martineaudoize, and J. M. Fairbrother. 1997. Localization of the in vivo expression of P and F1 fimbriae in chickens experimentally inoculated with pathogenic Escherichia coli. Microb. Pathog. 22:331-341. [DOI] [PubMed] [Google Scholar]
- 36.Reitzer, L. J. 1996. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine, p. 391-407. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C.
- 37.Reitzer, L. J., and B. Magasanik. 1982. Asparagine synthetases of Klebsiella aerogenes: properties and regulation of synthesis. J. Bacteriol. 151:1299-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhoades, K. R., and R. B. Rimler. 1989. Fowl cholera. In C. Adlam and J. M. Rutter (ed.), Pasteurella and pasteurellosis. Academic Press Limited, London, England.
- 40.Ruffolo, C. G., J. M. Tennent, W. P. Michalski, and B. Adler. 1997. Identification, purification, and characterization of the type 4 fimbriae of Pasteurella multocida. Infect. Immun. 65:339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. L. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steiner, K. 2001. relA-independent amino acid starvation response network of Streptococcus pyogenes. J. Bacteriol. 183:7354-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 45.Walsh, K., and D. E. Koshland, Jr. 1985. Characterization of rate-controlling steps in vivo by use of an adjustable expression vector. Proc. Natl. Acad. Sci. USA 82:3577-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wassenaar, T. M., and W. Gaastra. 2001. Bacterial virulence: can we draw the line? FEMS Microbiol. Lett. 201:1-7. [DOI] [PubMed] [Google Scholar]
- 47.Yarwood, J. M., J. K. McCormick, M. L. Paustian, V. Kapur, and P. M. Schlievert. 2002. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J. Bacteriol. 184:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yarwood, J. M., J. K. McCormick, M. L. Paustian, P. M. Orwin, and V. Kapur. 2002. Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3. Implications for the evolution of staphylococcal pathogenicity islands. J. Biol. Chem. 277:13138-13147. [DOI] [PubMed] [Google Scholar]