Abstract
The plasmid-encoded toxin (Pet) of enteroaggregative Escherichia coli (EAEC) belongs to a family of high-molecular-weight serine protease autotransporters of Enterobacteriaceae (SPATEs) which also includes Pic from EAEC and Shigella flexneri, EspC from enteropathogenic E. coli, EspP from enterohemorrhagic E. coli, Sat from uropathogenic E. coli, Tsh from avian pathogenic E. coli, and SepA from S. flexneri. Phylogenetic analysis shows the SPATE proteins to represent a distinct subfamily of autotransporters with amino acid identities ranging from 35 to 55%, providing a powerful resource to direct structure-function studies. In this study, we show that these related proteins are proteases with divergent substrate specificities, suggesting different functions. The cleavage profile of oligopeptides was found to be unique for each SPATE protein. The SPATEs showed proteolytic activity for several substrates, namely mucin, pepsin, human coagulation factor V, and erythroid spectrin. The cleavage of spectrin has been hypothesized as the mechanism through which Pet induces cytopathic effects. However, whereas Pet, Sat, and EspC cleaved spectrin, only Pet and Sat elicited cytopathic effects; the remaining SPATEs did not cause any morphological changes to HEp-2 cell monolayers. EspC and Pet exhibited acid-dissociable binding to HEp-2 cells. However, Pet was more efficient at entering HEp-2 cells, suggesting a basis for the different abilities of these two proteases to damage cells. Our data suggest that, despite the homologies observed among these proteins, the SPATEs have different pathogenetic functions only partly dependent on their substrate specificities.
Pathogenic Escherichia coli and Shigella species are responsible for a variety of illnesses, ranging from urinary tract infections to persistent diarrhea. A common feature of these organisms is their secretion of high-molecular-weight serine protease autotransporters of Enterobacteriaceae (SPATEs) (13). These proteins include EspC from enteropathogenic E. coli (25), EspP from enterohemorrhagic E. coli (EHEC) (5), plasmid-encoded toxin (Pet) from enteroaggregative E. coli (EAEC) (7), Sat from uropathogenic E. coli (10), Tsh from avian pathogenic E. coli (23), Pic from Shigella flexneri and EAEC (12), and SepA also from S. flexneri (2). The SPATEs possess a characteristic GDSGS serine protease motif and are therefore believed to act as serine proteases. Recently, Fink et al. have suggested the presence of specific aspartate and histidine residues, which could comprise the catalytic triad of the Hap protease from Haemophilus influenzae (9); Hap is homologous to the SPATE family, and the proposed catalytic residues are conserved.
Various functions have been proposed for the SPATE proteins. EspC has been shown to act as an enterotoxin on rat jejunal tissue mounted in Ussing chambers (17). EspP cleaves both pepsin and human coagulation factor V (5), the latter effect potentially exacerbating hemorrhagic colitis. In addition, EspP may act as a cytotoxin, causing disruption of the actin network when applied to Vero cells (6). Sat, located on a pathogenicity island in uropathogenic E. coli strain CFT073, elicits cytopathic effects on HEp-2 and Vero monkey kidney cells. These effects include not only elongation of these cells but also vacuolation in other cell lines and animal tissues (10). Pic functions as a mucinase and is thought to inactivate the complement cascade (12). Benjelloun-Touimi and colleagues showed SepA to be an abundant supernatant protein in Shigella spp. and to play a role in causing intestinal inflammation and tissue invasion (2). Examination of the protease activity with oligopeptides conjugated on paranitroanaline (pNA) showed the SepA protease to be highly substrate specific (3). Provence and Curtis described Tsh as a temperature-sensitive hemagglutinin from avian pathogenic E. coli (23). Other researchers have since confirmed the hemagglutination effect of an identical protein, Hbp, and have also demonstrated its ability to cleave hemoglobin and to play a role in abscess formation by Bacteroides fragilis (21, 22).
Pet is believed to contribute to the pathogenesis of EAEC and has been studied extensively by our laboratories (7, 19). Navarro-García et al. showed that concentrated culture supernatants of a minimal clone expressing the Pet protein were able to elicit cytopathic effects on HEp-2 cells in culture in both a dose- and time-dependant manner. Incubation of the Pet protein with these cells caused a loosening of focal contacts, followed by contraction of the cytoplasm and ending with the rounding and detachment of the cell (20). This effect was dependent on the protease activity; however, the exact mode of action has yet to be elucidated. Studies using confocal microscopy and immunological techniques reveal entry and trafficking of the toxin into the cell (18). Moreover, Pet was recently shown to cleave spectrin, a component of the cytoskeleton (26).
The SPATE proteins display high similarity with each other (as high as 58% overall identity), suggesting a common origin (13). Despite the homology, correlations among the different functions of the SPATEs have not yet been established. In this study, we show that each SPATE has unique functions and potentially plays a specialized role in the pathogenesis of the parent organism.
MATERIALS AND METHODS
Bacterial strains.
Minimal clones of the SPATEs are described in Table 1. All DNA constructs were transformed into E. coli strain HB101 (4).
TABLE 1.
SPATE clones used in this study
| Plasmid | SPATE protein | Source | Reference |
|---|---|---|---|
| pJLM174 | espC cloned into pBAD30 (Ampr) | J. B. Kaper | 17 |
| pB9-5 | espP cloned into pK18 (Kanr) | H. Karch | 5 |
| pCEFN1 | pet cloned into pSPORT (Ampr) | J. P. Nataro | 19, 20 |
| pPic | pic cloned into pACYC (Tetr) | J. P. Nataro | 12 |
| pDG7 | sat cloned into pBSKII (Ampr) | H. M. Mobley | 10 |
| pZK15 | sepA cloned into pUC19 (Ampr) | C. Parsot | 2 |
| pAY3108 | tsh cloned into pBSKII (Ampr) | R. Curtis | 23 |
Protein preparation.
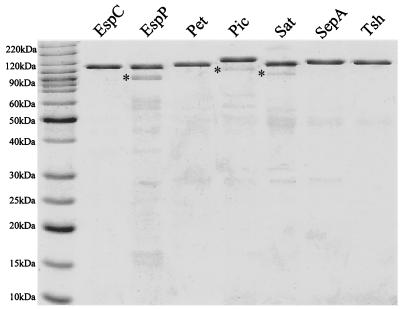
Overnight Luria broth cultures of E. coli HB101 expressing the various SPATE clones were centrifuged at 7,500 × g for 10 min. Culture supernatants were passed through a 0.22-μm-pore size filter to remove residual bacteria and concentrated with 30-kDa-cutoff spin filter devices (Millipore, Bedford, Mass.); the retentates were then separated by anion exchange chromatography using Sepharose Q resin and stepwise elution. Fractions containing SPATEs were again concentrated as described above with spin filters. Proteins were visualized by Coomassie blue staining after separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1) (16). To determine the concentration of each SPATE protein, 5 μl of each preparation was separated by SDS-PAGE and stained with SYPRO Orange stain (Bio-Rad, Hercules, Calif.). The intensity of the SPATE protein bands was measured on a PhosphorImager Storm model 840 apparatus (Amersham Biosciences, Sunnyvale, Calif.) and analyzed with ImageQuant version 5.0 software (Amersham Biosciences). The concentration of protein was estimated by comparing the intensity to that in a standard linear curve generated with known amounts of purified bovine serum albumin. Each preparation was >90% pure.
FIG. 1.
SPATE proteins purified from HB101 harboring high-copy-number clones. One microgram of each protein was separated by SDS-PAGE and stained with Coomassie blue. Asterisks represent the breakdown products of the proteases as confirmed by Western immunoblotting and mass spectroscopy.
Immunologic methods.
Antibodies to Pet were raised as previously described (7). Antibodies to EspC were generated by a similar method, as follows. Two hundred micrograms of EspC protein was excised from a Coomassie blue-stained polyacrylamide gel and injected subcutaneously into an adult New Zealand White rabbit. The rabbit received 100-μg booster injections at 30 and 60 days after primary immunization and was exsanguinated 15 days after the last boost. Antibodies were absorbed with E. coli proteins conjugated to agarose (Sigma Chemical Co., St. Louis, Mo.) according to the manufacturer's instructions. Immunoblotting was performed by following standard methods: proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore), and then detected with rabbit antisera at dilutions of 1:8,000 to 1:15,000. Pet and EspC proteins were visualized with goat anti-rabbit antibodies conjugated with horseradish peroxidase (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) by using the ECL Plus Western blotting detection system (Amersham Biosciences), according to the manufacturer's instructions.
Oligopeptide cleavage profile.
Five micrograms of each SPATE protein was added separately to 1 mM solutions of pNA-conjugated oligopeptides (Sigma Chemical Co.; Bachem, King of Prussia, Pa.; and Calbiochem, La Jolla, Calif.; or custom synthesized oligopeptides from AnaSpec, Inc., San Jose, Calif.) in a buffer containing 0.1 M MOPS (morpholinepropanesulfonic acid), 0.2 M NaCl, and 0.01 mM ZnSO4 (MOPS buffer, pH 7.3). Reactions were performed in a standard volume of 100 μl in microtiter plates. Plates were incubated at 37°C, and absorbance readings at 405 nm were taken at various time points up to 12 h (3). The maximum absorbance reading obtained was set to 100%, and other readings were normalized accordingly with each SPATE. All reactions were performed in triplicate, and the mean ± standard deviation was calculated.
Cleavage of protein substrates.
Spectrin was partially purified from sheep erythrocytes by differential centrifugation as previously described (26). One microgram of spectrin was combined with 1 μg of each SPATE protein in 15 μl of MOPS buffer and incubated overnight at 37°C. Reaction products were separated on 6% PAGE gels (26). Degree of cleavage was calculated using densitometry measured with SYPRO Orange (Bio-Rad) staining as described above. Three micrograms of pepsin (Sigma Chemical Co.) was combined with 1 μg of each SPATE protein in 30 μl of MOPS buffer and incubated for 1 h at 37°C. Reaction products were separated on 12% PAGE gels. A quantity of 2.5 μg of purified human coagulation factor V (Calbiochem) was combined with 2 μg of each SPATE protein in 40 μl of MOPS buffer. In parallel, 2 μg of each SPATE protein was incubated overnight at 37°C with 20 μl of human serum, diluted 1:50. Reaction products were separated by SDS-6% PAGE (5). Ten micrograms of each SPATE protein was incubated overnight at 37°C in wells cut into a medium comprised of 1.5% agarose and 1% bovine submaxillary mucus (Sigma Chemical Co.). Zones of clearing were visualized by staining with 0.1% amido black in 3.5 M acetic acid for 30 min, followed by destaining with 7% acetic acid in 70% methanol (12). All cleavage reactions were controlled using concentrated supernatants of HB101 and HB101 harboring the respective SPATE cloning vectors (Table 1).
Cell studies.
HEp-2 cells were maintained in humidified 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (HyClone, Logan, Utah), 1% nonessential amino acids, 5 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml) (all from Sigma Chemical Co.). The cells were serially propagated and replated in four-well LabTek chamber slides (VWR, Bridgeport, N.J.) and allowed to grow to 80% confluence (about 2 to 3 days). Twenty micrograms of SPATE protein was added directly to 400 μl of DMEM (0.5 μM final SPATE concentration) containing penicillin and streptomycin. Cells were incubated for 5 h at 37°C and then washed twice with phosphate-buffered saline (PBS), fixed with 70% methanol, and stained with Giemsa stain. Vero cells were grown and treated in a similar manner. Doses of 0.5 and 1 μM were assayed at 5, 10, 18, and 30 h. HT-29 cells were maintained and treated as described above for 5 h.
To assess cell binding, HEp-2 cells were grown as described above, replated in eight-well LabTek chamber slides, and allowed to grow to 90% confluence. The medium was aspirated, and 250 μl of cold DMEM containing penicillin, streptomycin, and 10 μg of EspC or Pet was applied. Cells were incubated for 2 h at 4°C and then washed twice with PBS and a third time in PBS or mild acid containing 50 mM glycine and 100 mM NaCl, pH 2. The third wash was analyzed by enzyme-linked immunosorbent assay with anti-Pet or anti-EspC serum.
To assess internalization, HEp-2 cells treated for various times with either Pet (20 μg/ml) or EspC (40 μg/ml) were fixed with 2% formalin-PBS, washed with PBS, permeabilized by adding 0.1% Triton X-100 in PBS, and stained with 0.05 μg of rhodamine-phalloidin/ml (15). Cells were washed again with PBS and incubated with either anti-Pet or anti-EspC antibodies. Proteins were visualized with fluorescein isothiocyanate-labeled goat anti-rabbit immunoglobulin G. Slides were mounted on Gelvatol (Burkard Scientific, Uxbridge, United Kingdom), covered with a glass slide, and examined under a Bio-Rad MRC600 confocal microscope as previously described (18).
Phylogenetic analyses.
SPATE protein sequences are available in the GenBank database as follows: EspC, accession no. AAC44731; EspP, CAA66144; Pet, AAC26634; Pic, AF097644; Sat, AAG30168; SepA, CAA88252; and Tsh, AA24698. Both full-length passenger domains and the regions comprising the putative proteolytic domains (see below) were aligned by using ClustalX (version 1.8) and adjusted by inspection. The protease domains were defined as lying between the cleavage site of the signal sequence through that residue aligning most closely with amino acid 294 (TYV) of Pet. This stretch was conserved among all SPATEs and Hap from H. influenzae and corresponds to the protease domain predicted by Fink et al. for Hap (9). Dendrograms were constructed from the alignments by using split decomposition with SplitsTree (version 3.1) as described previously (14).
RESULTS
Cleavage of purified protein substrates.
Most of the SPATE proteins have been reported to cleave one or more intact protein substrates with presumed biological relevance (5, 12, 22). To assess the conservation of the ability to cleave these substrates, each SPATE was incubated with each reported substrate under conditions described in the literature.
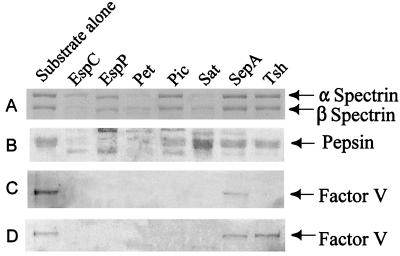
Villaseca and coworkers have shown that the EAEC cytotoxin and enterotoxin Pet cleaves erythroid spectrin in vitro (26). Spectrin cleavage in vivo could explain the exfoliation of colonic enterocytes induced by Pet and its enterotoxic effects on rat tissue in electrophysiological studies (19). We therefore investigated whether other SPATEs also cleave spectrin. Only Pet, Sat, and EspC showed cleavage of the alpha and beta subunits (Fig. 2A). The other SPATEs did not show any detectable cleavage of spectrin despite prolonged incubation, providing evidence for the functional divergence of these proteases.
FIG. 2.
Cleavage of protein substrates by SPATE proteins. (A) Spectrin. One microgram of spectrin purified from sheep red blood cells was incubated with 1 μg of each SPATE protein overnight. The reaction products were separated on SDS-6% PAGE gels. (B) Pepsin. Three micrograms of purified pepsin was incubated with 1 μg of each SPATE for 1 h at 37°C. The reaction products were separated on SDS-12% PAGE gels. (C) Human coagulation factor V. A total of 2.5 μg of purified coagulation factor V was combined with 2 μg of each SPATE protein in a 40-μl total volume and incubated overnight. (D) Human coagulation factor V in serum. Two micrograms of each SPATE protein was added to 20 μl of diluted human plasma (1:50) and incubated overnight. All reaction products were separated on 6% PAGE gels.
Brunder et al. reported that EspP cleaves pepsin in vitro (5). To assay for this effect among the other SPATEs, each protease was incubated individually with purified pepsin. Complete disappearance of the pepsin band in Coomassie blue-stained SDS-PAGE gels was observed after incubation with EspC, EspP, and Pet (Fig. 2B).
EspP was also reported to hydrolyze human coagulation factor V (5). Commercially purified coagulation factor V and, in parallel, diluted human plasma were incubated with each SPATE protein and assayed for the disappearance of the factor V band. With the exception of SepA, all of the SPATEs cleaved the purified factor V. In diluted human plasma, all of the SPATEs except for SepA and Tsh cleaved factor V (Fig. 2C).
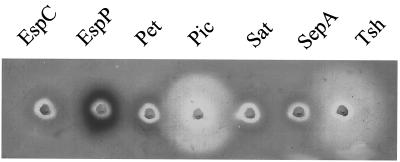
Pic has been shown to cleave bovine submaxillary mucin (12). To assay for mucinolytic effects with the other SPATEs, concentrated supernatants containing each SPATE protein were added to wells in agarose-mucin plates. A zone of clearing around both the Pic and Tsh proteins indicated that these two SPATEs were capable of cleaving mucin (Fig. 3). In a similar assay using hog gastric mucin, none of the SPATEs showed mucinolytic activity (data not shown). These results of the cleavage of biological substrates by the different SPATEs are summarized in Table 2. Culture supernatants of neither HB101 nor HB101 harboring the various cloning vectors used in these studies yielded detectable cleavage of any of the substrates used in these studies.
FIG. 3.
Cleavage of bovine submaxillary mucus. Ten micrograms of each SPATE was incubated overnight at 37°C on medium containing 1.5% agarose and 1% bovine submaxillary mucus. Mucin was stained with 0.1% amido black.
TABLE 2.
Summary of SPATE functions
| SPATE protein | Cleavage of biological substrates
|
Cytopathic effects on HEp-2 cells | |||
|---|---|---|---|---|---|
| Mucin | Pepsin | Factor V | Spectrin | ||
| EspC | − | + | + | + | − |
| EspP | − | + | + | − | − |
| Pet | − | + | + | + | + |
| Sat | − | − | + | + | + |
| SepA | − | − | − | − | − |
| Tsh | + | − | + | − | − |
| Pic | + | − | + | − | − |
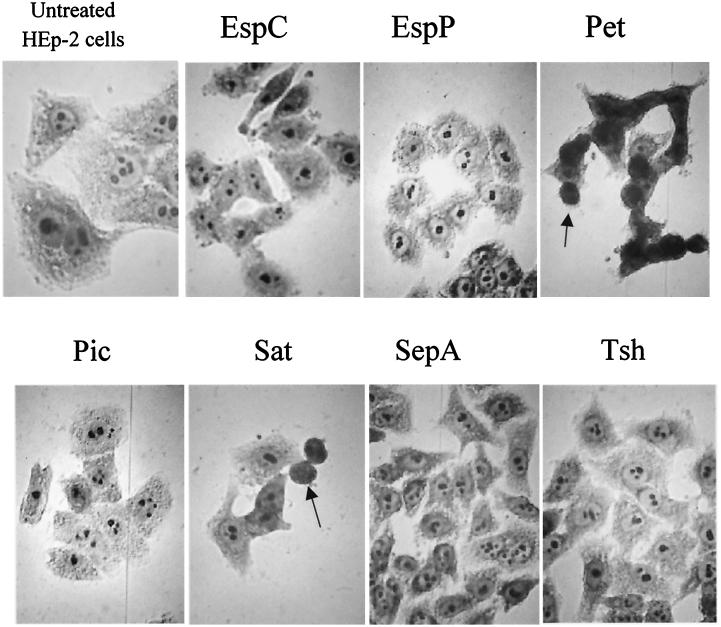
Cytopathic effects induced by SPATEs.
Pet has previously been shown to cause rounding and detachment of HEp-2 cells grown in monolayers. We assayed each of the other SPATEs for their ability to cause cytopathic effects. After 5 h of exposure, HEp-2 cells treated with Pet and Sat exhibited similar morphological changes, characterized by the release of focal contacts from the glass substratum, complete rounding of the cells, and detachment from the glass. The remaining SPATEs or concentrated culture supernatants of HB101 harboring the cloning vectors or pJLM174 grown in the presence of glucose induced no observable effects (Fig. 4). In addition, the SPATE proteins were also incubated with HT-29 and Vero cells. As previously reported, Pet elicited appreciable changes in HT-29 cell morphology that included the condensation and separation of cell clusters (20), but no other SPATE protein induced obvious effects on this cell line. In our hands, only Sat elicited changes upon incubation with Vero cells (data not shown).
FIG. 4.
Effects of SPATEs on HEp-2 cell monolayers are shown by oil immersion light microscopy of Giemsa-stained HEp-2 cells after treatment with SPATE proteins at 500 nM for 5 h. Rounding of cells is shown with Pet and Sat (arrows).
Functional differences between Pet and EspC.
We have shown above that EspC, like Pet, cleaves spectrin in vitro. To investigate the defect in EspC that prevents it from eliciting cytopathic effects in HEp-2 cells, its ability to bind to HEp-2 cells was compared with that of Pet. Cells were treated with either Pet or EspC at 4°C to allow the proteins to bind to the surface without permitting internalization. The cells were then washed twice with PBS to remove any excess protein and washed a third time with PBS or a mild acid wash to remove receptor-bound protein from the cell surface. The third wash was assayed by enzyme-linked immunosorbent assay to quantitate how much Pet or EspC had been removed. Both Pet and EspC showed a significant acid-dissociable binding to the HEp-2 cells (P < 0.05 for both compared with that for the PBS wash). There was no significant difference in the amount of protein specifically bound between Pet and EspC.
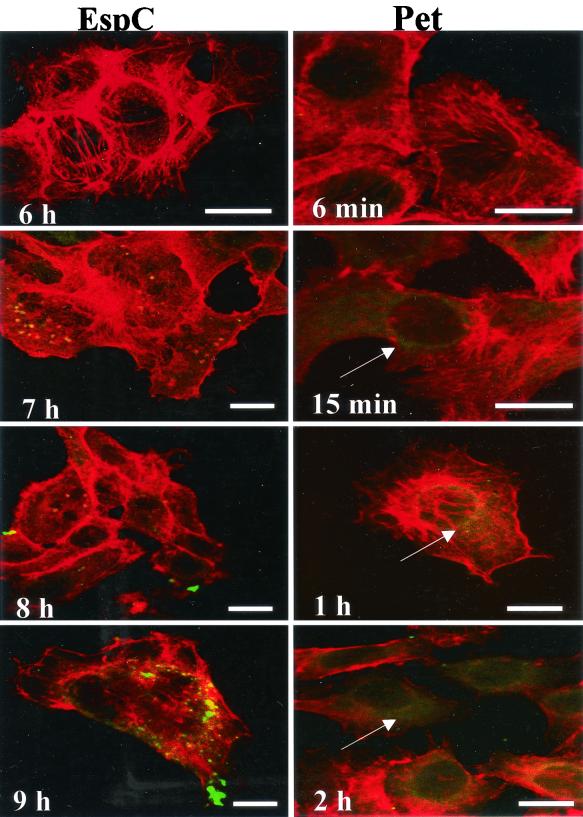
Since both the Pet and EspC proteins appear to bind to HEp-2 cell surfaces, their ability to enter the cell was compared. HEp-2 cells were treated with either Pet or EspC and followed for various amounts of time; both proteins were visualized by immunofluorescence. Pet was observed inside the cells in as little as 15 min, with the cell showing a diffuse green fluorescent staining by 2 h (Fig. 5). In contrast, EspC showed no detectable internalization until 7 h of incubation, with a slight increase in intensity by 9 h (Fig. 5). This variation in uptake efficiency suggests that EspC is impaired in internalization by epithelial cells.
FIG. 5.
Internalization of Pet and EspC by HEp-2 cells. HEp-2 cells were treated with Pet (200 nM) or EspC (400 nM) for various periods of time as indicated. Cells were then fixed, permeabilized, and stained with anti-Pet or anti-EspC rabbit antibodies, followed by fluorescein isothiocyanate-labeled goat anti-rabbit immunoglobulin; cells were simultaneously stained with rhodamine-phalloidin and then subjected to confocal laser scanning microscopy. Actin microfilaments appear red, and Pet and EspC toxins appear green. Pet internalization appears as diffuse straining of the cytoplasm (arrows) as early as 15 min after intoxication. Bars represent 20 μm. EspC shows no detectable internalization until 9 h of incubation.
Cleavage of oligopeptides.
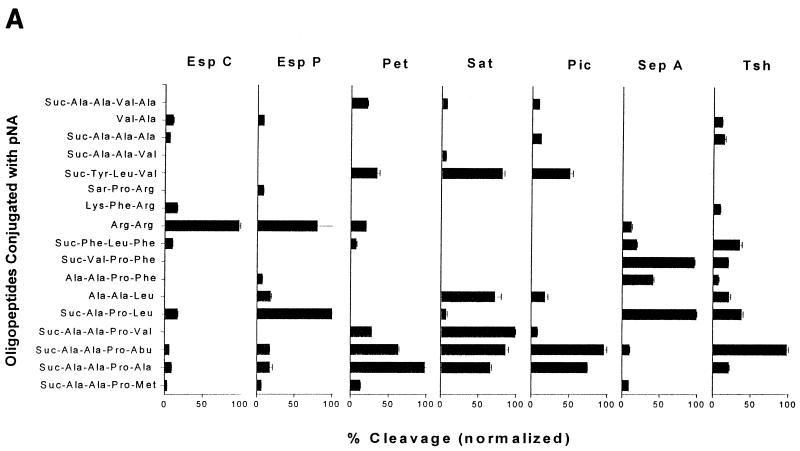
Although the SPATEs have conserved amino acid sequences, comparisons of their abilities to cleave biological substrates have yet to reveal a single shared function. Instead, the data suggest that the protease function appears to be specific for each protein. To gain further insight into their functions, we examined cleavage specificities by using oligopeptides. Each SPATE was reacted with a variety of oligopeptides conjugated at the C terminus with pNA. Oligopeptide cleavage profiles for each SPATE are shown in Fig. 6A. Negative controls included Pet and Pic, with single amino acid substitutions at the catalytic serine, and the supernatants of HB101 carrying the cloning vectors used SPATE expression (Table 1); all negative controls failed to cleave any of the oligopeptides (data not shown).
FIG. 6.
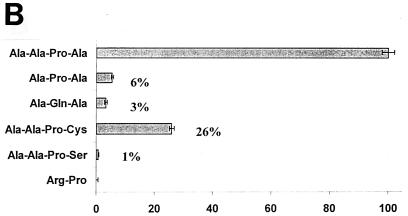
(A) Oligopeptide cleavage profiles of the SPATE proteins. Five micrograms of each SPATE protein was added to 1 mM solutions of oligopeptides conjugated at the C terminus with pNA. Reactions were incubated at 37°C for up to 12 h. The maximum absorbance value obtained was set to 100%, and other readings were normalized accordingly. (B) Cleavage of small, hydrophobic peptides by Pet. pNA-Conjugated oligopeptides were synthesized, and reactions were carried out as described above. Cleavage of Ala-Ala-Pro-Ala was set to 100%, and remaining reactivities were normalized accordingly.
Pet showed efficient cleavage of Ala-Ala-Pro-Ala, Ala-Ala-Pro-Abu (2-aminobutyric acid), and Ala-Ala-Pro-Val. These experiments suggested that the active site of Pet accommodates small, hydrophobic peptides. When the terminal amino acid of this motif has a larger (i.e., Met, Leu, or Phe) or charged (i.e., Asp) side chain, the efficiency is apparently reduced. To further characterize the requirements for Pet cleavage, we synthesized an additional set of oligopeptides as modifications of Ala-Ala-Pro-Ala (the most efficiently cleaved substrate). When the N-terminal alanine was removed, the resultant tripeptide Ala-Pro-Ala was hydrolyzed at a 20-fold reduction in efficiency. Likewise, when the C-terminal alanine residue was replaced with the more polar cysteine, cleavage efficiency was reduced fourfold. Substitution with a more polar serine reduced cleavage efficiency to 1% of that obtained with a C-terminal alanine (Fig. 6B).
EspC specifically hydrolyzed the dipeptide Arg-Arg and had little reactivity towards the other oligopeptides tested. EspP, however, showed a slightly broader protease activity than EspC by cleaving both the Arg-Arg and Ala-Pro-Leu oligopeptides. Sat and Pet shared some specificity, consistent with their common cleavage of spectrin and common abilities to intoxicate epithelial cells. Like Pet, Sat exhibited preferential cleavage for hydrophobic amino acids but could accommodate slightly larger groups. Maximal cleavage was observed with Ala-Ala-Pro-Val, but Ala-Ala-Pro-Abu was cleaved almost as readily. In contrast to Pet, however, Sat was highly active against Ala-Ala-Leu and Tyr-Leu-Val, which were not well cleaved by Pet.
Surprisingly, Pic showed remarkable similarity to the cleavage specificities of Pet and Sat, despite being distantly related on the phylogenetic tree (Fig. 7) and cleaving different protein substrates. Like Pet, Pic showed maximal activity towards Ala-Ala-Pro-Abu, readily cleaved Ala-Ala-Pro-Ala, and had some activity towards Tyr-Leu-Val. Tsh, which is closely related to Pic, shared Pet's activity towards the small and hydrophobic Ala-Ala-Pro-Abu but differed in most other respects.
FIG. 7.
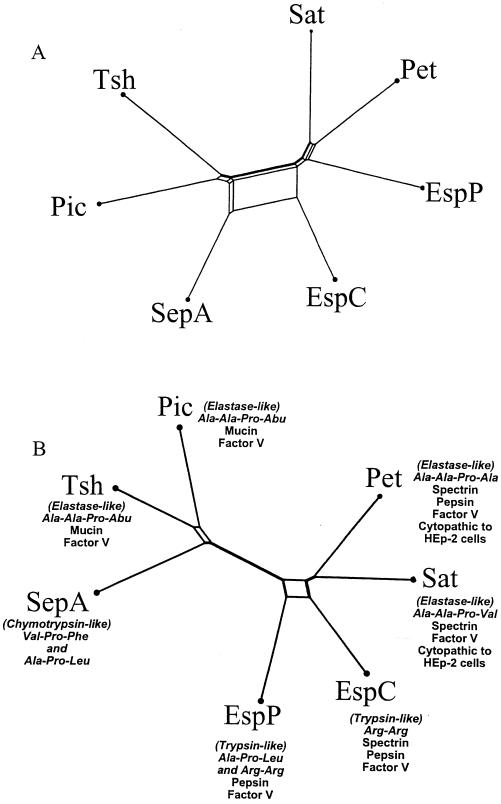
ClustalX phylograms of amino acid sequence alignments of full-length SPATE passenger domains (A) or the N-terminal one-third protease domains (B), corresponding to the first 242 amino acids of Pet. Trees were further tested for reliability by bootstrap analysis, yielding results of 97.3% (A) and 96.8% (B).
In contrast to those of all the other SPATEs assayed, the active site of SepA apparently accommodated larger and more hydrophobic residues, as suggested by the cleavage of Val-Pro-Phe, Ala-Ala-Pro-Phe, and Phe-Leu-Phe. Like EspP, SepA also efficiently cleaved Ala-Pro-Leu.
Phylogenetic analysis of the SPATEs.
We investigated the correlation between the relationships of the SPATE family amino acid sequence and the cleavage profiles observed. By using split decomposition, comparisons of the complete passenger domains and, in parallel, the putative proteolytic domains yielded different unrooted dendrograms. The dendrogram of the complete passenger domains (Fig. 7A) revealed evidence of multiple recombination events among the SPATEs or their progenitors (as evidenced by boxes in the phylogram). Small boxes between Pic and Tsh and between Pet and Sat indicate small recombination events, whereas larger boxes between EspC and SepA suggest more extensive recombination between these two proteins.
When only the putative serine protease regions (essentially the N-terminal third of the passenger domains) were compared, a slightly different dendrogram was generated (Fig. 7B). In this analysis, EspC was seen to be more closely related to Pet and Sat than is EspP, which is consistent with the abilities of only Pet, Sat, and EspC to cleave spectrin.
DISCUSSION
The study of the SPATE proteins and their roles in virulence continues to be a focus of several laboratories. However, the emphasis heretofore has been on each individual SPATE, without the examination of the family as a whole. Here, we have addressed the divergence and conservation of sequence and function within the family. These studies will provide a foundation for comparative structure-function analyses.
Several SPATE proteins, namely Pet, Sat, EspP, and SepA, have previously been suggested to produce cytotoxic effects. Cellular effects have not been reported for other SPATEs. In our hands, only Pet and Sat were able to elicit cytopathic effects on HEp-2 cells, indicating that the SPATE proteins were functionally distinct. We addressed the basis for the inability of some SPATEs to damage cells. Although the mechanism of Pet-induced damage remains to be fully elucidated, previous data suggest that Pet must bind to the HEp-2 cell surface, be internalized, and exert a proteolytic attack on an intracellular host protein (18). Villaseca and coworkers have suggested that this target protein is spectrin and have shown the cleavage of erythroid spectrin in vitro by Pet (18, 26). Spectrin, a heterodimeric cytoskeletal protein, serves to link actin filaments with other cytoskeletal proteins and the plasma membrane (1). Proteolytic attack on spectrin, leading to rearrangements of associated actin microfilaments, is a plausible mechanism behind the effects observed when HEp-2 cells are treated with Pet. Based on this proposed model, it seems likely that the noncytopathic SPATEs may be deficient at one or more of these steps: binding, internalization, or catalysis.
When spectrin was incubated with each SPATE overnight, only Pet, Sat, and, surprisingly, EspC cleaved this protein. Whereas both EspC and Pet exhibited acid-dissociable binding to HEp-2 cells, EspC was internalized far less efficiently. Previous studies have shown that Brefeldin A inhibits the cytopathic effects of Pet by disrupting its trafficking within HEp-2 cells (18). These data suggest that Pet may employ retrograde transport via the Golgi apparatus, as do several other bacterial toxins; however, no endoplasmic reticulum retrieval signature is apparent in Pet. The details of Pet trafficking are as yet unclear, but further comparisons with EspC may be illuminating.
In addition to spectrin, we assayed the cleavage of other proposed biological substrates that have been reported for the SPATEs. EspP has been shown to cleave pepsin (5). Given that passage through the stomach is required of all enteric pathogens, we hypothesized that all the SPATEs should cleave pepsin. However, only EspC, EspP, and Pet were able to degrade pepsin, indicating that this function is not common to all SPATEs. Notably, it has yet to be proven that these proteins are able to cleave pepsin in vivo. Similarly, mucinases are a growing class of proteins that could be adaptive for all mucosal pathogens. Our lab has shown previously that the Pic protein hydrolyzes bovine submaxillary mucin (12). When the other SPATEs were assayed in a similar manner, only Tsh, in addition to Pic, exhibited mucinolytic activity.
Brunder et al. also reported that EspP cleaved human coagulation factor V, a potentially important function for EHEC. Coupled with the actions of potent cytotoxins that can damage the colonic mucosa, cleavage of factor V could aggravate the hemorrhagic colitis characteristic of EHEC infection (5). We hypothesized that this function could also be adaptive for Shigella spp., which also produce bloody diarrhea. Interestingly, however, the ability to cleave factor V was widespread, as EspC, Pet, Pic, Sat, and Tsh were also able to cleave the purified protein. In whole sera, all but Tsh obliterated the protein. Importantly, cleavage of factor V has not yet been demonstrated in the setting of infection.
In light of these findings, we reexamined the phylogenetic relations for this family of proteins. Our lab has shown here and previously that, when the entire protein sequences are considered, the SPATEs can be divided into two groups (11), exemplified by Pic and Pet. Interestingly, phylogenetic analysis of entire SPATE passenger domains did not reveal a correlation with the biological substrates but did show evidence of significant homologous recombination among family members. Indeed, highly similar proteins such as Pet and Sat (which are 53% identical overall) do not share oligopeptide specificities despite shared abilities to cleave spectrin and cause cytopathic effects in HEp-2 cells. In contrast, proteins that are less similar, such as Pic and Sat (30% identical), do share some specificities for oligopeptides despite their being classified into different groups and cleaving different biological substrates.
In light of these data, we asked whether the SPATE protease domains would correlate better with cleavage profiles. Accordingly, split decomposition analysis was performed on the N-terminal third of each SPATE amino acid sequence. This region was suggested to comprise the proteolytic triad in Hap, a related autotransporter in H. influenzae (9). The resulting phylogram is shown in Fig. 7B (14). The original bifurcating phylogenetic pattern remains, with EspC, EspP, Pet, and Sat comprising one group and Pic, Tsh, and SepA forming the other (11). The major difference in the second tree, however, is that EspC is now grouped closer with Pet and Sat than in the original analysis. This difference is consistent with our hypothesis in that EspC, Pet, and Sat are the SPATEs able to cleave spectrin. These data suggest that significant substrate specificity may be largely determined by protease domains.
However, the phylogenic groupings of the protease regions are not completely consistent with the oligopeptide cleavage profiles. For example, Pet, Sat, Pic, and Tsh show the preferential cleavage of short chains of small, hydrophobic amino acids such as Ala-Ala-Pro-Ala, Ala-Ala-Pro-Abu (2-aminobutyric acid), and Ala-Ala-Pro-Val, thus indicating a strong similarity to the elastase family of proteases. In contrast, both EspC and EspP, which the tree shows to be closely related, have a high affinity for the Arg-Arg oligopeptide. This observation indicates that the active site clefts of these proteases accommodate basic, positively charged amino acids, much like trypsin. SepA, on the other hand, is the most distantly related SPATE and uniquely cleaves large, hydrophobic amino acids (exemplified by cleavage of the oligopeptide Suc-Val-Pro-Phe) (8, 24). Of note, Hap has also been reported to accommodate bulky, hydrophobic residues in its active site (9).
In summary, many pathogenic bacteria secrete proteases to serve multiple functions. These SPATE proteins have been found in a variety of human pathogens, but no common role in virulence has been demonstrated. We show here that these proteins have varying effects on HEp-2 cells and that their specificity extends not only to proposed biological substrates but also to oligopeptides, indicating diversity at the active site. These observations suggest a common ancestral SPATE protein that each pathogenic species has modified to permit adaptation to its specific niche. Further characterization of this diversity will greatly enhance the understanding of structure-function relationships in this family of proteins.
Acknowledgments
We thank James Kaper, Helge Karch, Harry Mobley, Claude Parsot, and Roy Curtis III, who were kind enough to share their SPATE clones with our lab. In addition, we also thank Mardi Reymann, Yenia Azamar, and Adrian Canizalez for their technical assistance and Ian Henderson for his helpful suggestions.
This work was supported by Public Health Service grant AI 43615 to J.P.N. Pinaki R. Dutta was supported in part by National Institutes of Health training grant 5 T32 AI07540-04. Fernando Navarro-García was supported by Consejo Nacional de Ciencia y Tecnología de México (CONACYT, 31028-M).
Editor: J. T. Barbieri
REFERENCES
- 1.Alberts, B., D. Bray, J. Lewis, M. Raff, K. Roberts, and J. Watson. 1994. Molecular biology of the cell, 3rd ed. Garland Publishing, Inc., New York, N.Y.
- 2.Benjelloun-Touimi, Z., P. J. Sansonetti, and C. Parsot. 1995. SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol. Microbiol. 17:123-135. [DOI] [PubMed] [Google Scholar]
- 3.Benjelloun-Touimi, Z., M. S. Tahar, C. Montecucco, P. J. Sansonetti, and C. Parsot. 1998. SepA, the 110 kDa protein secreted by Shigella flexneri: two-domain structure and proteolytic activity. Microbiology 144:1815-1822. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 5.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24:767-778. [DOI] [PubMed] [Google Scholar]
- 6.Djafari, S., F. Ebel, C. Deibel, S. Kramer, M. Hudel, and T. Chakraborty. 1997. Characterization of an exported protease from Shiga toxin-producing Escherichia coli. Mol. Microbiol. 25:771-784. [DOI] [PubMed] [Google Scholar]
- 7.Eslava, C. E., F. Navarro-García, J. R. Czeczulin, I. R. Henderson, A. Cravioto, and J. P. Nataro. 1998. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect. Immun. 66:3155-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fersht, A. 1984. Enzyme structure and mechanism, 2nd ed. W. H. Freeman and Company, New York, N.Y.
- 9.Fink, D., L. D. Cope, E. J. Hansen, and J. W. St. Geme, III. 2001. The Hemophilus [sic] influenzae Hap autotransporter is a chymotrypsin clan serine protease and undergoes autoproteolysis via an intermolecular mechanism. J. Biol. Chem. 276:39492-39500. [DOI] [PubMed] [Google Scholar]
- 10.Guyer, D. M., I. R. Henderson, J. P. Nataro, and H. L. Mobley. 2000. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol. Microbiol. 38:53-66. [DOI] [PubMed] [Google Scholar]
- 11.Henderson, I. R., R. Cappello, and J. P. Nataro. 2000. Autotransporter proteins, evolution and redefining protein secretion. Trends Microbiol. 8:529-532. [DOI] [PubMed] [Google Scholar]
- 12.Henderson, I. R., J. Czeczulin, C. Eslava, F. Noriega, and J. P. Nataro. 1999. Characterization of Pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 67:5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huson, D. H. 1998. SplitsTree: a program for analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 15.Knutton, S., A. D. Phillips, H. R. Smith, R. J. Gross, R. Shaw, P. Watson, and E. Price. 1991. Screening for enteropathogenic Escherichia coli in infants with diarrhea by the fluorescent-actin staining test. Infect. Immun. 59:365-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. 1971. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-683. [DOI] [PubMed] [Google Scholar]
- 17.Mellies, J. L., F. Navarro-Garcia, I. Okeke, J. Frederickson, J. P. Nataro, and J. B. Kaper. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navarro-García, F., A. Canizalez-Roman, J. Luna, C. Sears, and J. P. Nataro. 2001. Plasmid-encoded toxin of enteroaggregative Escherichia coli is internalized by epithelial cells. Infect. Immun. 69:1053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navarro-García, F., C. Eslava, J. M. Villaseca, R. López-Revilla, J. R. Czeczulin, S. Srinivas, J. P. Nataro, and A. Cravioto. 1998. In vitro effects of a high-molecular-weight heat-labile enterotoxin from enteroaggregative Escherichia coli. Infect. Immun. 66:3149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navarro-García, F., C. Sears, C. Eslava, A. Cravioto, and J. P. Nataro. 1999. Cytoskeletal effects induced by Pet, the serine protease enterotoxin of enteroaggregative Escherichia coli. Infect. Immun. 67:2184-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto, B. R., S. J. van Dooren, C. M. Dozois, J. Luirink, and B. Oudega. 2002. Escherichia coli hemoglobin protease autotransporter contributes to synergistic abscess formation and heme-dependent growth of Bacteroides fragilis. Infect. Immun. 70:5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otto, B. R., S. J. van Dooren, J. H. Nuijens, J. Luirink, and B. Oudega. 1998. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J. Exp. Med. 188:1091-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Provence, D. L., and R. I. Curtis. 1994. Isolation and characterization of a gene involved in hemagglutination by an avian pathogenic Escherichia coli strain. Infect. Immun. 62:1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawlings, N. D., and A. J. Barrett. 1994. Families of serine peptidases. Methods Enzymol. 244:19-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stein, M., B. Kenny, M. A. Stein, and B. B. Finlay. 1996. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J. Bacteriol. 178:6546-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villaseca, J. M., F. Navarro-García, G. Mendoza-Hernández, J. P. Nataro, A. Cravioto, and C. Eslava. 2000. Pet toxin from enteroaggregative Escherichia coli produces cellular damage associated with fodrin disruption. Infect. Immun. 68:5920-5927. [DOI] [PMC free article] [PubMed] [Google Scholar]