Abstract
The Mi-2/NuRD complex is a multi-subunit protein complex with enzymatic activities involving chromatin remodeling and histone deacetylation. Targeting of Mi-2/NuRD to methylated CpG sequences mediates gene repression. The function of p66α and of p66β within the multiple subunits has not been addressed. Here, we analyzed the in vivo function and binding of both p66-paralogs. Both factors function in synergy, since knocking-down p66α affects the repressive function of p66β and vice versa. Both proteins interact with MBD2 functionally and biochemically. Mutation of a single amino acid of p66α abolishes in vivo binding to MBD2 and interferes with MBD2-mediated repression. This loss of binding results in a diffuse nuclear localization in contrast to wild-type p66α that shows a speckled nuclear distribution. Furthermore, wild-type subnuclear distribution of p66α and p66β depends on the presence of MBD2. Both proteins interact with the tails of all octamer histones in vitro, and acetylation of histone tails interferes with p66 binding. The conserved region 2 of p66α is required for histone tail interaction as well as for wild-type subnuclear distribution. These results suggest a two-interaction forward feedback binding mode, with a stable chromatin association only after deacetylation of the histones has occurred.
INTRODUCTION
DNA methylation at the 5′-position of cytosine within CpG dinucleotides has been shown to mediate long-term transcriptional repression of transposons, imprinted genes and of the inactive X chromosome in female mammals (1). Furthermore, CpG methylation is often found in the context of tissue-specific genes in non-expressing tissues. Similarly, DNA hypomethylation of the genome as well as methylation-dependent silencing of tumor suppressor genes is often found in human cancer (2–5). The molecular mechanism of DNA methylation-mediated gene repression has been shown to involve several aspects. In addition to changing the DNA structure (6), binding of several regulatory factors is influenced or inhibited. This has been shown for the DNA binding factors NF-κB (7), E2F (8), CpG binding protein CGBP (9), GABP (10,11), CREB (12), as well as for the insulator factor CTCF (13,14). Although in these cases an inhibition of DNA binding has been clearly shown to be involved in gene repression, this mechanism seems to be restricted to a small group of genes and regulatory sequences. The most commonly observed mechanism of DNA methylation-mediated transcriptional repression involves modification of the chromatin structure. A family of DNA binding proteins harboring a methyl-CpG-binding domain (MBD) recognizes and binds to methylated CpG sequences (15).
The MBD-containing family members MeCP2, MBD1, MBD2 and MBD3 recruit histone deacetylases (HDAC), which in turn deacetylate histone tails resulting in the generation of a repressive chromatin conformation (16–19). MeCP2 interacts with the Sin3A histone deacetylase complex (20,21). It has recently been shown that MeCP2-mediated repression is associated with the chromatin remodeling complex SWI/SNF, which is possibly involved in changing the chromatin architecture such that histone deacetylases can act on histone tails (22). Similarly, within the MeCP1 complex, MBD2 is associated with the Mi-2/NuRD complex that contains histone deacetylases as well as chromatin remodeling factors (18,23). In contrast to MeCP2, the MBD3 factor of the Mi-2/NuRD complex is unable to bind to methylated CpG sequences in mammals, instead this complex associates with MBD2, which in turn binds to methylated CpGs (18,19). Besides MBD3, the Mi-2/NuRD complex consists of the histone deacetylases HDAC1 and HDAC2 together with the retinoblastoma-associated proteins RbAp46 and RbAp48. These factors are central components of the Mi-2/NuRD complex, the Sin3A complex, as well as of other chromatin associated complexes [reviewed in (24)]. In addition, MTA2 as well as the chromatin remodeling factor Mi-2 together with p66 are components of the Mi-2/NuRD complex (18). There are two members of a small gene family coding for p66α and p66β, both of which interact with MBD2 as well as with MBD3 (25). We wished to analyze the functional interplay between p66α and p66β in the context of MBD2-mediated repression. The results show that p66α and p66β target to DNA bound MBD2 as well as to unmodified, deacetylated histones. Both functions are mediated by separate domains, both of which are required for wild-type distribution within the nucleus.
MATERIALS AND METHODS
Plasmids
pABGal-hp66α and pABGal-hp66β were constructed as described previously (25). pSG5-hp66α was generated by subcloning the EcoRI/XbaI fragment from pOTB7-hp66α (25) into pSG5 (Stratagene) opened with EcoRI/BamHI. pSG5-hp66β was cloned by ligating the EcoRI/BamHI fragment of pABGal-hp66β into pSG5. C-terminal deletions of pSG5-hp66α were generated by digesting the full-length construct with BglII/EheI (amino acids 1–433), BglII/BcuI (amino acids 1–329) or BglII/SacI (amino acids 1–238) and religating the blunted ends. For pSG5-hp66α 134–633 the BspHI/XbaI fragment from pOTB7-hp66α was cloned in-frame into pABGal94 linker (26) to generate pABGal-hp66α 134–633 which was then digested with EcoRI/BamHI and ligated into pSG5. CR2 of hp66α was amplified with primers 5′-GCCGAGGATCCAGCAAGCATGCAGGC-3′ and 5′-GCCGAGATCTGCTGCAGGAGCCGCT-3′, the product was digested with BamHI/BglII and ligated into pSG5 to generate pSG5-hp66α 344–480. For transfections, the 4xUAStk luciferase reporter construct and expression plasmid pCMV-lacZ encoding β-galactosidase were kindly provided by A. Baniahmad. pEGFP-hp66α was constructed by ligating the SalI/BamHI fragment from pAB-Gal94-hp66α into pEGFP-C1 (Clontech). pEGFP-hp66α 1–482 or 1–348 were amplified with the sense primer 5′-TGCAGTCGACCATATGACCGAAGA-3′ and the antisense primers 5′-CGGATCCTGCTGCAGGAGCC-3′ or 5′-GCGGATCCGCTTGCTGGAGACT-3′, respectively. Products were then digested with SalI/BamHI and ligated into pEGFP-C1. Construction of pEGFP-hp66β was described previously (25). C-terminal deletions pEGFP-hp66β 1–489 and 1–354 were generated via PCR with sense primer 5′-TGCAGTCGACATGGATAGAATGACAGA-3′ and antisense primers 5′-GGGATCCCTGCTGCTGTAATCG-3′ and 5′-GCTGGATCCTGTGAGTTGGCAG-3′, respectively. The generated products were treated with SalI/BamHI and cloned into pEGFP-C1. Cloning of pABGal-MBD2b was explained earlier (16). pSil-αX was cloned by annealing oligos 5′-GATCCGGAACAGGAGATTGAGCAGTTCAAGAGACTGCTCAATCTCCTGTTCCTTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAACAGGAACAGGAGATTGAGCAGTCTCTTGAACTGCTCAATCTCCTGTTCCG-3′ and ligating the product into pSilencer 2.1-U6 neo (Ambion) digested with BamHI/HindIII. pSil-βX was created using oligos 5′-GATCCGGAGGATTTGGCAAATCTTTCAAGAGAAGATTTGCCAAATCCTTCCTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAAGGAAGGATTTGGCAAATCTTCTCTTGAAAGATTTGCCAAATCCTTCCG-3′ into pSilencer 2.1-U6 neo opened with BamHI/HindIII. pABGal-p66α with single amino acid substitution K149R was constructed using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions with the sense primers 5′-GGATGATCAAGCAGCTGAGGGAAGAATTGAGGTTAG-3′ and antisense primers 5′-CTAACCTCAATTCTTCCCTCAGCTGCTTGATCATCC-3′. The enhanced green fluorescent protein (EGFP) fusion of p66αK149R was generated by cutting the SalI/BamHI fragment of pABGal-p66αK149R and insertion into pEGFP-p66α cut with SalI/BamHI. pSG5-p66αK149R was created by ligation of the SalI/BamHI fragment from pABGal-p66αK149R into pSG5-p66β digested with SalI/BamHI. Plasmids for the bacterial expression of glutathione S-transferase (GST)-fused histone tails (pGEX1-H2A, pGEX1-H2B, pGEX1-H3, pGEX1-H4) were gratefully received from C. Wu (27); constructs pGEX-2T-p300 and pGEX-5X-PCAF for the expression of the HAT-domains of GST-p300 and GST-PCAF were obtained from S. Berger (28). Generation of pCMV-GST-MBD2b for eukaryotic protein expression was presented previously (25).
Cell culture and transfections
All cells were cultured in DMEM with 10% fetal calf serum at 37°C, 5% CO2. Transfection of HeLa cells was carried out using the CaPO4 method as described elsewhere (29). Cells were cotransfected in 6-well plates (105 cells/well) using 0.75–1.5 µg of 4xUAStk-luciferase reporter plasmid, 0.2 µg of pCMV-lacZ encoding β-galactosidase and 0.75–1.5 µg of expression plasmids. For reporter assays, knock-down of hp66-expression was achieved with 0.5–2.0 µg of pSil-αX or pSil-βX. Cells were harvested 36–96 h after transfection and assayed for luciferase and β-galactosidase activity. All transfection assays shown were performed in duplicates or triplicates and repeated at least twice. Transfection of NIH 3T3 cells and MBD2(−/−) and wild-type tail fibroblasts [kindly provided by A. Bird (30)] was performed using jetPEI (Polyplus transfection) according to the manufacturer's protocol. For fluorescent imaging, cells were grown on cover slips in 6-well dishes (105 cells/well) and transfected with 3 µg of pEGFP-constructs. Images were taken 24–48 h post-transfection using 100-fold magnification after incubation with ‘Hoechst DNA stain’ for 10 min at 37°C. Significance of observed changes in reporter gene activity has been determined by the Dunnett's t-test and Tukey's studentized range (HSD) test, depending on the type of data analyzed.
Immunoblotting
For analysis of reduced protein expression, HeLa cells were transfected in 10 cm dishes (1 × 106 cells/dish) with 15 µg of pSil-αX, pSil-βX and pSil-neg together with 15 µg pBSK (Stratagene). Nuclear extracts were prepared 96 h post-transfection essentially as described previously (31). Cells were resuspended in 3 vol of buffer A (20 mM HEPES, pH 7.9, 10% glycerol, 0.2% NP-40, 10 mM KCl and 1 mM EDTA). After centrifugation at 5000 g for 10 min at 4°C, the supernatant was discarded and the remaining nuclei pellet resuspended in 2 vol buffer B (420 mM NaCl, 20 mM HEPES, pH 7.9, 10 mM KCl and 1 mM EDTA), followed by rotational incubation for 30 min at 4°C. To remove cell debris, the extract was centrifuged at 20 000 g for 15 min at 4°C. The supernatant was separated using a 12% SDS–PAGE and analyzed by western blotting with an anti-p66 antibody (Upstate).
Mammalian pull-down
HEK293 cells were transiently cotransfected with mammalian expression vectors for pCMV-GST, pCMV-GST-MBD2b with Gal, Gal-p66α or Gal-p66αK149R. Cells were collected 48 h after transfection, and nuclear extracts was performed according to the protocol described above. Aliquots containing 400 µg of the nuclear extract were incubated with 40 µl glutathione–Sepharose 4B beads (Amersham Biosciences) for 30 min at room temperature. The beads were washed four times with washing buffer (200 mM NaCl, 20 mM Tris–HCl, pH 8.0, 1 mM EDTA and 0.5% NP-40). Proteins binding to the beads were eluted with SDS sample buffer, separated on SDS–PAGE and subsequently detected by western blotting with anti-Gal4 polyclonal IgG antibody. The ECL™ kit (Amersham Biosciences) was used to visualize the proteins on the membrane following the manufacture's protocol.
GST pull-down of in vitro translated proteins
GST and GST-histone tails were expressed in Escherichia coli BL21. GST pull-downs were carried out essentially as described previously (16). Bacteria were induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 20°C. Recombinant proteins were purified with glutathione–Sepharose beads (Amersham Biosciences) and analyzed on SDS–PAGE to normalize protein amounts. Equivalent amounts of GST fusion proteins were incubated with [35S]methionine-labeled hp66-proteins, produced by the T7/T3 TNT-coupled transcription/translation system (Promega) in 200 µl of binding buffer (100 mM NaCl, 20 mM Tris–HCl, pH 8.0, 1 mM EDTA, 0.5% NP-40, 5 µg of ethidium bromide and 100 µg of BSA). After 30 min of incubation at room temperature, the beads were washed five times with 1 ml of binding buffer without ethidium bromide and BSA. The bound proteins were eluted with SDS sample buffer, fractionated on SDS–PAGE and visualized by fluorography.
Acetylation-dependent interaction
For the acetylation of histone tails GST-p300 and GST-PCAF were expressed in E.coli BL21 cells after induction with IPTG for 5 h at 20°C. The pellet of a 400 ml culture was resuspended in 30 ml STE-buffer (150 mM NaCl, 10 mM Tris–HCl, pH 8.0, 1 mM EDTA) and slowly frozen at −20°C. About 1.2 ml lysis buffer (250 mM MgCl2, 25 mM MnCl2, 250 µg/ml DNase I, 250 µg/ml RNase A and 12 µg/ml lysozyme) were added after thawing and incubated for 30 min at 4°C while rotating. Cell debris was pelleted by centrifugation for (30 min, 10 000 g, 4°C) and the supernatant incubated with 200 µl of 50% glutathione–Sepharose beads (Amersham Biosciences) for 1 h at 4°C. The beads were washed three times with washing buffer (100 mM Tris–HCl, pH 8.0, 100 mM NaCl) and the recombinant proteins eluted with 200 µl of elution buffer (20 mM reduced glutathione, 100 mM Tris–HCl, pH 8.0, 100 mM NaCl) by incubating for 1 h at 4°C and subsequently for 1 h at 20°C. After centrifuging, the glutathione was removed by YM-10 Microcon columns according to the manufacturer's protocol (Millipore). Extracts were stored with 20% glycerol at −80°C.
To determine the acetylation pattern of the purified HAT-extracts, 2–3 µg of recombinant GST-histone tails were incubated for 0.5–2 h at 30°C with 1–2 µg of GST-PCAF or GST-p300 in 30 µl of acetylation buffer (50 mM Tris–HCl, pH 8.0, 5% glycerol, 1 mM DTT and 10 mM Na Butyrate) together with 1 µCi [3H]Acetyl-CoA. Samples were fractionated on SDS–PAGE and visualized by fluorography.
Analysis of acetylation-dependent interaction with histone tails was performed by acetylating glutathione–Sepharose bound GST-histone tails with non-labeled Acetyl-CoA essentially as described above or by treating a second set of histone tails without Acetyl-CoA. Both sets of histone tails were then incubated with in vitro translated [35S]methionine-labeled hp66α or hp66β proteins. Subsequently, all samples were treated according to the GST pull-down protocol described above.
RESULTS
MBD2-mediated repression is increased by p66α and p66β
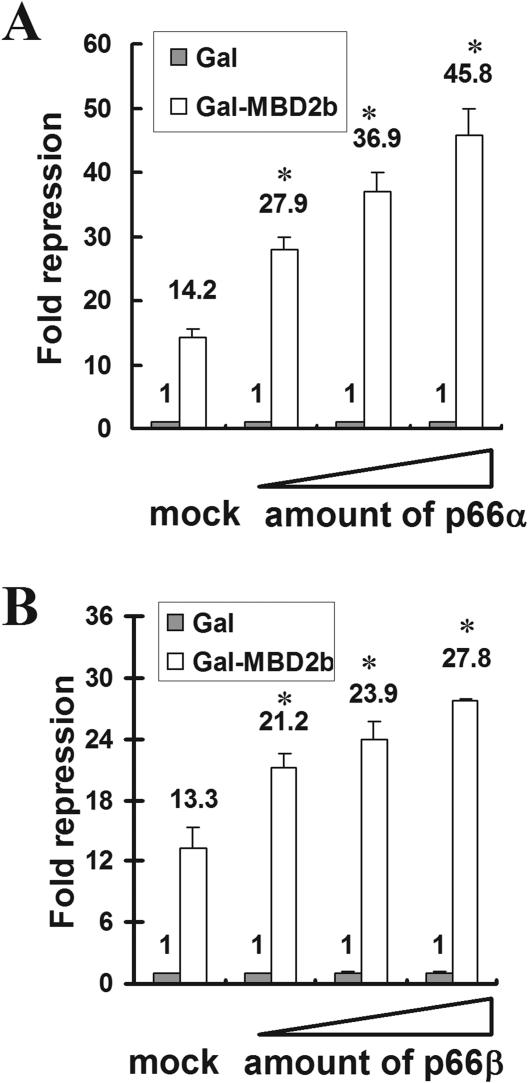
We have previously reported that the paralogous proteins p66α and p66β share a high degree of sequence homology that has been conserved throughout evolution. In addition, both paralogs have been identified as being interaction partners of the methyl-CpG-binding protein MBD2 (25,32). Since Gal-fusions of p66α and p66β were shown to repress transcription of a UAS-reporter construct, we investigated the functional association of these paralogous proteins with MBD2. We first analyzed the influence of p66α and p66β on MBD2-mediated repression. MBD2 was fused to a GAL4 DNA binding domain (Gal-DBD) and transfected into HeLa cells together with increasing amounts of p66α or p66β and an appropriate luciferase reporter containing four GAL4 DNA binding sites (UAS). In agreement with previous reports (16), Gal-MBD2 repressed reporter activity by ∼14-fold in the absence of any cotransfected p66-constructs (Figure 1). MBD2-mediated repression was, however, enhanced in a dose-dependent manner up to 45-fold upon transfection of increasing amounts of p66α. Overexpression of p66β increased the repression of Gal-MBD2 up to ∼28-fold. As a result, both p66-proteins are capable of enhancing MBD2-mediated repression, with the enhancement by p66α being stronger than by p66β.
Figure 1.
Expression of p66α or p66β increases MBD2-mediated repression. HeLa cells were cotransfected with a 4xUAStk luciferase reporter together with plasmids coding for the Gal-DNA binding domain, Gal-MBD2b and no (mock) or increasing amounts of p66α (A) or p66β (B). Cell extracts were analyzed for reporter gene activity. Fold repression was determined relative to the Gal-DNA binding domain. Error bars represent variations within duplicate transfections, significant changes relative to mock are indicated by asterisk. Luciferase activity was measured 48 h after transfection.
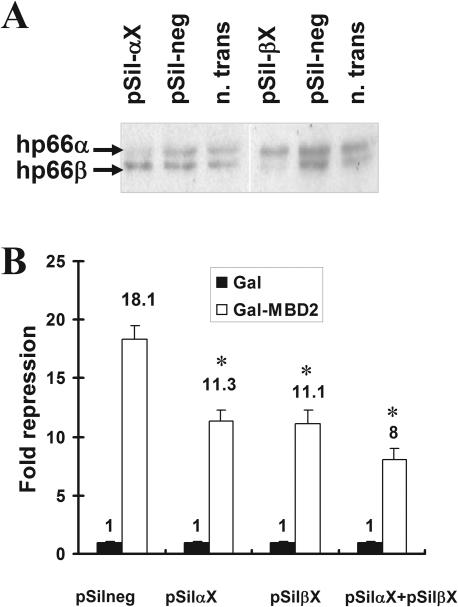
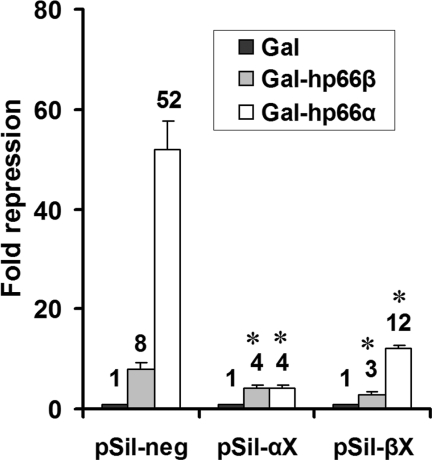
In order to further characterize the influence of the p66-paralogs on MBD2-mediated repression, a second approach that involved knocking down of endogenous p66α and p66β using RNAi and analyzing subsequent alterations of MBD2 function was performed. Using a vector driven shRNAi-procedure, target sequences were identified that resulted in a specific decrease of p66α or p66β expression. HeLa cells were either transfected with constructs targeting p66α (pSil-αX), p66β (pSil-βX) or a non-targeting control sequence (pSil-neg) and analyzed for p66-expression using western blot analyses (Figure 2A). Since p66α and p66β exhibit molecular weights of 68 and 66 kDa, respectively, both proteins could be separated via SDS–PAGE and simultaneously identified using an antibody recognizing both p66α and p66β. Figure 2A demonstrates that the expression of p66α or p66β is clearly reduced 96 h after RNAi treatment. Importantly, knock-down of one p66-protein does not influence the expression of the other homologous partner, demonstrating that the expression levels of both p66-proteins are independent from one another and that each protein can be knocked down specifically using the shRNAi-approach.
Figure 2.
Knock-down of endogenous p66α or p66β decreases MBD2-mediated repression. (A) Endogenous expression of p66α and p66β is reduced 96 h after RNAi treatment. HeLa cells were transfected with either pSilencer constructs targeting p66α (pSil-αX), p66β (pSil-βX), a non-targeting control sequence (pSil-neg) or left untransfected (n.trans). Nuclear extracts were prepared and subjected to western blotting using the p66 antibody. (B) MBD2-mediated repression is reduced by knock-down of endogenous p66α or p66β. HeLa cells were transfected with the indicated pSilencer constructs together with expression vectors for the Gal-DNA binding domain or Gal-MBD2b and the 4xUAStk luciferase reporter. Cell extracts were analyzed for reporter gene activity. Fold repression was determined relative to the Gal-DNA binding domain. Error bars represent variations within duplicate transfections, significant changes relative to pSil-neg are indicated by asterisk.
Having established suitable conditions for p66-knock-down, HeLa cells were transfected with plasmids coding either for Gal-DBD or for a Gal-MBD2 fusion and knock-down plasmids pSil-αX and pSil-βX (Figure 2B). Gal-MBD2 repressed reporter activity by about 18-fold in the absence of targeted knock-down. However, knocking down p66α by cotransfecting pSil-αX relieved MBD2-mediated repression to 11-fold, and similar results were achieved by reducing p66β expression. Notably, combined knock-down of both p66-proteins further abated MBD2-mediated repression down to 8-fold. MBD2-mediated repression is therefore at least partly dependent on p66α as well as on p66β and can be influenced by overexpression or knock-down of both paralogous proteins.
Enhancement of MBD2-mediated repression is dependent on the direct interaction of MBD2 and p66α
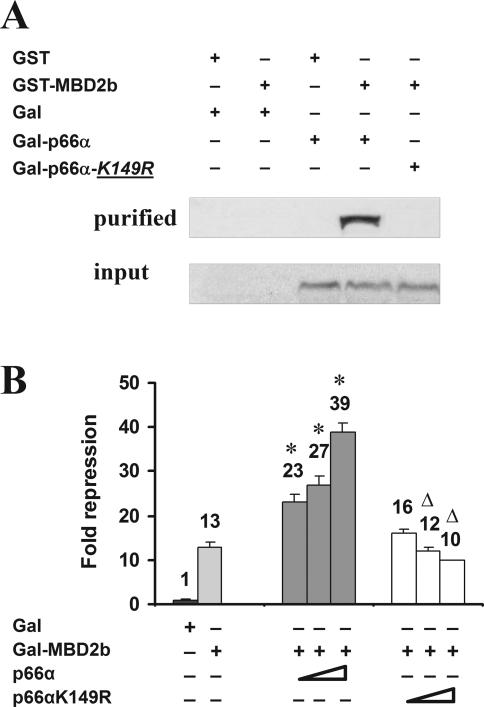
Since both p66α and p66β influence repression by MBD2, we next investigated whether this finding is the result of a direct effect that is mediated by the interaction of the two homologs with MBD2. The conserved region 1 (CR1) located at the N-terminus of both p66-proteins has been shown to be an interaction domain with MBD2, and we had previously identified a minimal overlapping region within the CR1 of p66α that is required for this interaction (25). Database analysis revealed a highly conserved candidate SUMOylation consensus site contained within this sequence that is not present in p66β. In order to disrupt this consensus sequence, we generated the point mutant p66α-K149R that retains the hydrophilic characteristic of the region. Detailed analysis of this mutant did not provide any evidence for SUMOylation of the wild-type site (data not shown). Nevertheless, we analyzed the interaction of p66α-K149R with MBD2 in vivo. GST–MBD2 was coexpressed with Gal-fused p66α-K149R in HEK 293 cells and nuclear extracts were bound to glutathione–Sepharose beads. Coprecipitated proteins were detected on western blots using an antibody directed against Gal-DBD. Figure 3A illustrates that wild-type Gal-p66α precipitates with GST–MBD2. Although the mutant protein is highly expressed in these cells, Gal-p66α-K149R does not coprecipitate with MBD2. Furthermore, this mutant is not able to enhance MBD2-mediated repression as observed for the wild-type protein (Figure 3B). However, Gal-MBD2 repression in part mediated by p66α is slightly reduced, possibly due to a dominant negative effect of the K149R mutant. Taken together, these results suggest that the functional enhancement of MBD2-mediated repression by p66 involves a direct interaction between MBD2 and p66α.
Figure 3.
K149 of p66α is required for the MBD2 interaction as well as for the MBD2-mediated repression. (A) HEK293 cells were harvested 48 h after transfection with various combinations of DNA constructs, as indicated above the figure. Nuclear protein extracts were prepared (input) and purified with glutathione–Sepharose beads. The bound protein together with the input fractions were analyzed by western blotting using the anti-Gal antibody. (B) K149R mutant of p66α decreases MBD2-mediated repression. HeLa cells were cotransfected with a 4xUAStk luciferase reporter together with vectors expressing the Gal-DNA binding domain, or Gal-MBD2b and increasing amount of pSG5-p66α or pSG5-p66αK149R. Fold repression was determined relative to the Gal-DNA binding domain, significant changes relative to Gal-MBD2b (asterisk) and relative to comparable amounts of p66α (open triangle) are indicated.
p66α and p66β repress transcription downstream of MBD2
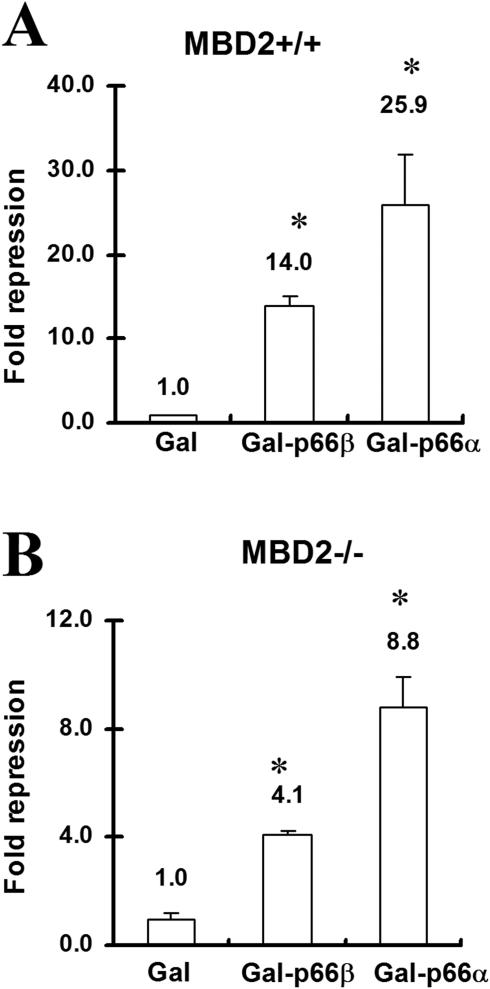
p66α and p66β are interaction partners with MBD2 and have been identified as being components of the Mi-2/NuRD complex. Functioning as the methyl-CpG-binding component, MBD2 is thought to target the Mi-2/NuRD complex to methylated DNA. It should be possible to replace the MBD2 function with the Gal-DNA binding domain fused directly to either p66α or p66β in such a model. In order to test this, GAL-fusion constructs of p66α or p66β were transfected into wild-type mouse fibroblast cells or into cells in which MBD2 has been knocked-out (30). As illustrated inFigure 4A, both p66-paralogs were able to repress transcription of a cotransfected luciferase reporter gene in wild-type fibroblast cells. Repression mediated by p66β was about 2-fold lower than that mediated by p66α. This difference between p66α and p66β is less pronounced when compared with CV-1 cells (25) and may reflect a cell-type specificity. Interestingly, both Gal-p66α and Gal-p66β were able to repress transcription in an MBD2−/− environment (Figure 4B). MBD2−/− cells were treated using identical conditions as used for the wild-type cells. Both Gal-p66-proteins repressed transcription, with p66α-mediated repression also being about 2-fold stronger as compared with p66β-mediated repression. While the overall repressive capacity of both proteins is lower than in wild-type fibroblasts, Gal-p66α and Gal-p66β clearly still maintain their ability to repress transcription. MBD2 is therefore not essential for the repression by either Gal-p66α or Gal-p66β, and thus does not function as a downstream factor for p66-mediated repression. Instead, MBD2 acts as the DNA binding factor targeting p66α and p66β to DNA.
Figure 4.
p66α- or p66β-mediated repression acts downstream of MBD2. Mouse fibroblasts (30) expressing wild-type MBD2 (MBD2+/+) (A) or MBD2 knock-out fibroblasts (MBD2−/−) (B) were transfected with vectors expressing the Gal-DNA binding domain, Gal-p66α or Gal-p66β together with a 4xUAStk luciferase reporter. Significant changes relative to Gal are indicated by asterisk.
p66α or p66β requires the paralogous p66 partner for repression
Different approaches to purify the chromatin remodeling complex Mi-2/NuRD have resulted in the identification of two proteins of 66 and 68 kDa with strong homologies to the p66-component from Xenopus laevis. Although p66β has been characterized as being a component of the human Mi-2/NuRD complex, the association of p66α and p66β within the repressor complex had until now not been elucidated. In order to investigate the organization of both p66-paralogs within the repressor complex, the expression of one p66-protein was knocked down and the influence on the repressing activity of the paralogous partner was determined. HeLa cells were transfected with Gal-p66α, Gal-p66β or the Gal4 DNA binding domain alone together with an appropriate reporter gene. Protein expression of p66α or p66β was specifically reduced by cotransfecting pSil-αX or pSil-βX and reporter gene activity was determined. As illustrated in Figure 5, reducing the expression of p66α or p66α influences repression by the Gal-fused paralog. Transfection of pSil-αX resulted in an expected decrease of Gal-p66α mediated repression by more than 14-fold; however, repression by p66β was also decreased by a factor of 2.4, compared with a non-targeting knock-down construct (pSil-neg). Similarly, knock-down of p66β not only decreased Gal-p66β mediated repression about 3-fold, but also relieved Gal-p66α mediated repression by more than 4-fold. This effect could be explained by a functional interplay between p66α and p66β, consistent with the idea that both proteins function as a part of the same complex.
Figure 5.
p66α or p66β requires the paralogous p66 partner for repression. HeLa cells were transfected with plasmids coding for the Gal-DNA binding domain, Gal-p66α or Gal-p66β together with pSil-neg, pSil-αX or pSil-βX. Cell extracts were analyzed for reporter gene activity; fold repression was determined relative to the Gal-DNA binding domain; significant changes relative to pSil-neg are indicated by asterisk.
Acetylation-sensitive association of p66α and p66β with histone tails
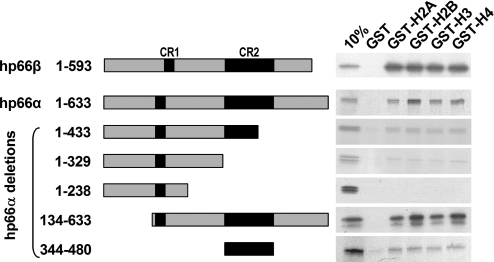
Post-translational modifications of histone tails are known to influence chromatin structure and gene expression (33). Deacetylation of histone tails by the MeCP1 complex causes chromatin condensation and transcriptional repression (34). MBD2, the DNA binding component of MeCP1, has been shown to associate with heterochromatin and to largely colocalize with p66α and p66β (25). To address the question whether chromatin association of the MeCP1 complex might also be due to a direct interaction of the p66-proteins with histones, GST-fused histone tails of H2A, H2B, H3 and H4 were used in an in vitro pull-down assay with radioactively labeled p66-constructs. In vitro translated p66α and p66β showed a strong affinity for all histone tails tested (Figure 6). C-terminal deletions of p66α that removed a part of or all of conserved region 2 (CR2) showed a reduced affinity for all histone tails tested. However, an N-terminal deletion of p66α or CR2 alone still retained the capacity to interact with histones. Although the CR2-domain is sufficient to mediate histone-tail interaction, the overall affinity is reduced compared with the full-length protein. This suggests that either a second interaction domain exists outside of CR2, or that other regions of the protein stabilize the association of the CR2-domain with histones.
Figure 6.
Both p66α and p66β interact with histone tails in vitro. GST and GST-histone tails were purified with glutathione–Sepharose beads and analyzed by SDS–PAGE to normalize protein amounts. Equivalent amounts of GST fusion proteins were incubated with [35S]methionine-labeled in vitro translated p66-proteins, as indicated left of the figure. The bound proteins were eluted with SDS sample buffer, fractionated on SDS–PAGE and visualized by fluorography.
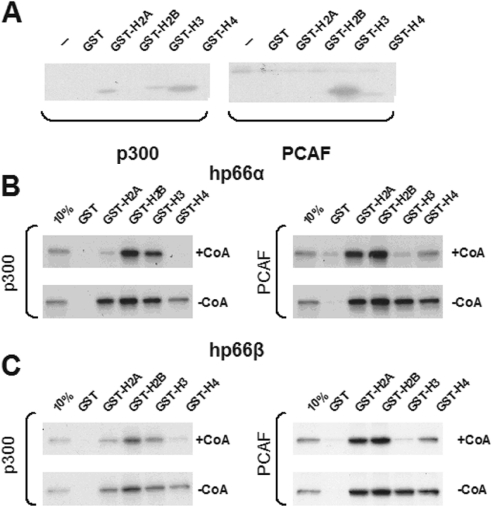
Since the GST-histone tails used in these experiments had been purified from bacteria, and therefore contained no post-translational modifications, the histone-acetyltransferases (HATs) p300 and PCAF were used to differentially modify the N-terminal domains of the histones. In order to do so, the acetyltransferase domains of p300 and PCAF fused to GST were bacterially expressed and incubated with individual histone tails. Radioactive labeled acetyl-CoA was used as an acetyl-donor. Figure 7A depicts the specificity and intensities with which the different enzymes acetylated the individual substrates. While GST-p300 preferentially modified the N-terminal region of histone H4 followed by H2A and H3, GST-PCAF was autoacetylated (35) (seen in all lanes at top of the figure) and acetylated mainly the histone tails of H3 and to a lesser extent H4.
Figure 7.
Acetylation of histone tails reduces the association with both p66-proteins in vitro. (A) The histone acetyltransferase domains of p300 and of PCAF were bacterially expressed and incubated with individual GST-histone tails together with radioactive acetyl-CoA. Glutathione–Sepharose purified GST-histone tails were eluted with SDS sample buffer, fractionated on SDS–PAGE and visualized by fluorography. p300 acetylated primarily H2A, H3 and H4 histone tails, whereas PCAF acetylated primarily the tails of histone H3, and also of H4. Acetylation by p300 or PCAF reduced association of p66α (B) or of p66β (C) with acetylated histone tails. Acetylated GST-histone tails as in (A), but incubated with non-radioactive acetyl-CoA (+CoA) or non-acetylated histone tails (−CoA), were incubated with [35S]methionine-labeled in vitro translated p66-proteins. All samples were purified by glutathione–Sepharose, fractionated on SDS–PAGE and visualized by autoradiography.
We next investigated whether this modification altered the binding capacity to p66α and p66β. Using a modified GST pull-down assay, one set of histone-substrates was acetylated by GST-p300 or GST-PCAF and another set remained unmodified. Both sets were subsequently bound to Sepharose beads and incubated with [35S]labeled p66α or p66β. Acetylation by p300 resulted in a considerable reduction of retained p66α or p66β on the N-terminal domains of histone H4, H2A and to a lesser degree H3 (Figure 7B and C). The association of both p66-paralogs with the histone tails was also decreased upon acetylation by the HAT-domain of PCAF. Corresponding to the acetylation specificity of this acetyltransferase, the affinity for histone H3 was reduced to a greater extent than for H4, whereas the unmodified histone tails all bound similarly. We also found an association of both p66-proteins with PCAF. Upon auto-acetylation of E.coli expressed PCAF, the interaction with either p66 is not reduced, rather slightly increased (see Supplementary Data). This suggests that p66 loss of binding upon acetylation of histone tails is likely not due to an unspecific charge effect of the acetyl groups since in the case of PCAF acetylation the reverse effect is seen. Thus, the affinities of p66α and p66β for the N-terminal histone regions are dependent on the modification state; acetylation of histone tails specifically reduces the association with both p66-proteins.
Nuclear distribution of p66α and p66β depends on CR2 and MBD2
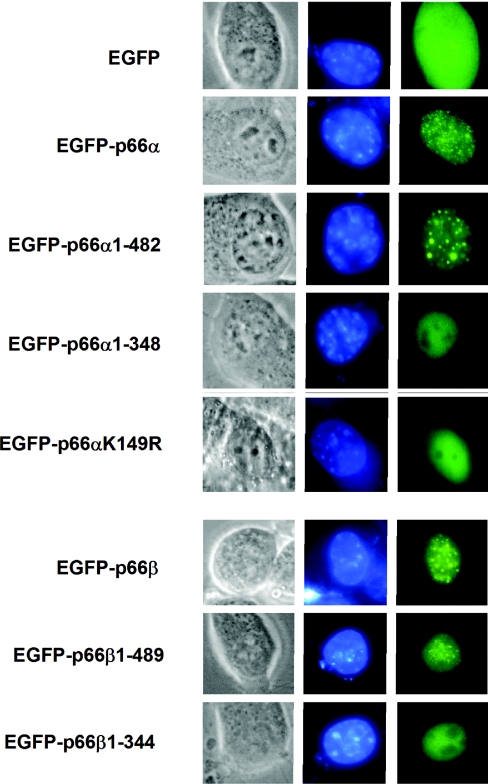
We have shown an in vitro association of p66α and p66β with the N-terminal regions of histone tails and have identified the conserved region 2 as a potential interaction domain. To substantiate these findings in vivo, we constructed several GFP-fused deletion constructs of both p66-paralogs and analyzed their localization in NIH 3T3 cells. Full-length p66-GFP fusions as well as C-terminal deletions of up to the CR2-domain of p66α (p66α 1–482) or p66β (p66β 1–489) show a speckled distribution in the nucleus (Figure 8). However, deletion of the CR2-domain (p66α 1–348, p66β 1–344) resulted in an overall loss of the speckled pattern and a uniform localization within the nucleus. The wild-type distribution of both paralogs is therefore dependent on CR2.
Figure 8.
Nuclear distribution of p66α and p66β depends on CR2 and K149 for p66α. NIH3T3 cells were transfected with the indicated constructs coding for various EGFP-fused p66-proteins. Cells were stained with ‘Hoechst DNA stain’ and phase contrast images (left), Hoechst stain fluorescence (middle) and EGFP fluorescence (right) was visualized.
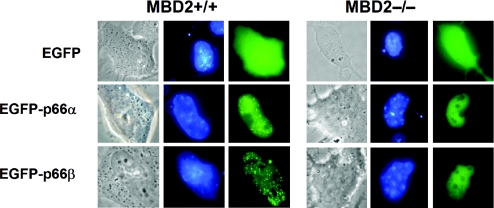
Because the CR1 domain of p66α and p66β has already been shown to interact with MBD2 and a point mutation in the CR1 domain of p66α (p66α K149R) disrupts MBD2-binding (see above), we used this mutant to test the MBD2-dependent distribution of p66α. Transfection of a GFP-p66α K149R construct into NIH 3T3 cells resulted in a diffuse nuclear localization. In order to provide further evidence that the subnuclear localization of p66α and p66β depends on the presence of MBD2, we compared the localization of GFP-p66α and GFP-p66β in MBD-containing fibroblasts versus MBD2−/− fibroblast cells (Figure 9). Similar to the previously described distribution in HEK 293 cells, fibroblasts containing MBD2 displayed a speckled nuclear localization of both GFP-p66-proteins. Interestingly, MBD2 knock-out cells lacked the speckled pattern but displayed a diffuse nuclear distribution. Taken together, these results are in agreement with the notion of a dual mechanism that is responsible for the localization of p66α and p66β. The interaction with MBD2 via the CR1-domain as well as the presence of the CR2 simultaneously determine the distribution of the p66-paralogs.
Figure 9.
Nuclear distribution of p66α and p66β depends on the presence of MBD2. MBD2+/+ wild-type or MBD2−/− knock-out fibroblast cells were transfected with constructs coding for the indicated EGFP-fused p66-proteins. Images were taken as in Figure 8.
DISCUSSION
Regulation of gene expression is largely influenced by DNA methylation, histone modifications and chromatin structure. The repressor complex Mi-2/NuRD is thought to be targeted to methylated DNA via MBD2 thereby generating the MeCP1 complex. The MeCP1 complex is composed of 10 subunits including the methyl binding proteins MBD2 and MBD3, the histone deacetylases HDAC1 and HDAC2, as well as two proteins of 66 and 68 kDa. Although these p66/68 components were originally hypothesized to be a single protein possibly differing in a post-translational modification, we have previously demonstrated that two homologous but distinct p66α and p66β proteins exist in humans (25).
Here we show that, when tethered to DNA, transcriptional repression by p66α or p66β does not require the methyl binding protein MBD2. Overexpression of each p66-protein augments repression by MBD2 and knock-down of either p66-paralogs reduces repression by MBD2. Combined knock-down of both p66-proteins decreases the repression by MBD2 further. However, since repression by MBD2 is also mediated by proteins other than the Mi-2/NuRD complex, a complete relief of repression would not be expected using this approach. These results indicate that the repressive function of MBD2 is mediated by the paralogous p66-proteins and that this repression might depend on a synergistic function with both p66α and p66β being present simultaneously in a single complex.
The CR1-region of p66β has previously been shown to interact with MBD2 and to be responsible for the integration of p66β within the MeCP1 complex (32). By mutating CR1 of p66α, we identified a critical amino acid essential for the interaction of p66α with MBD2 in vivo. Our observation that this point mutant was unable to enhance MBD2-mediated repression is in agreement with its inability to associate with MBD2. Additionally, transfection of a GFP-fused mutant resulted in a loss of the speckled nuclear localization observed with the wild-type protein. Therefore, as is the case with p66β, the CR1 of p66α is required for the association of p66α with MBD2. An analysis of the distribution of GFP-p66-proteins in the absence of MBD2, which revealed a diffuse nuclear distribution as well, further supports this view. MBD2 is therefore essential for the correct nuclear localization of p66α as well as of p66β, and very likely recruits both proteins to their target sites via CR1.
Chromatin and histone tail association of Mi-2/NuRD has been documented (36,37). Histone H3 K4 methylation prevents NuRD binding, whereas H3 K9 methylation does not (37). Here, we demonstrated that both p66-paralogs bind directly to histone N-terminal tails in vitro, and that for p66α this association requires CR2. The localization of p66β has previously been shown to depend on CR2 (32). As discussed below, histone tail interaction may be important in mediating a two-interaction binding mode by DNA bound MBD2 as well as by histone tail interaction. In such a scenario, histone tail interaction may either be charge driven or may require specific binding surfaces or both. Interestingly, acetylation of histone N-termini by the histone acetyltransferases p300 or PCAF abrogated the association with the p66-proteins. The acetylation pattern by PCAF corresponds to reports by other groups in which the N-terminus of histone H3 (H3 K14) is preferentially modified over histone H4 (H4 K8) (38,39). Acetylation by p300 is known to differ from PCAF in that H2A is modified at lysine 5 (H2A K5), H3 at K14 and K18, and histone H4 at K5 and K8 (38,39). Although p300 has been reported to acetylate H2B as well, we were not able to detect acetylation of this histone. This might be explained by the fact that our experiments utilized the HAT-domain of the enzyme rather than the full-length enzyme. In any case, acetylation of the core histones (H2A, H2B, H3, H4) results in a decondensed chromatin structure and correlates with transcriptional activity. Again, loss of interaction upon acetylation of histone tails may be charge driven although acetylation per se does not reduce p66 binding as shown for the interaction with acteylated PCAF. A scenario in which a DNA bound repressor, that mediates chromatin histone deacetylation by initial unstable recruitment of a corepressor complex, has recently been shown for the SMRT/N-CoR complex (40). In this case, a stable two-interaction binding mediated by the DNA bound factor as well as by histone tails is observed only after deacetylation. Here, we suggest a similar two-interaction mode of the Mi-2/NuRD complex, which requires DNA binding by MBD2 as well as histone tail binding. The CR2-mediated speckled nuclear distribution may be caused by an unknown function of CR2 or histone tail binding of CR2 is involved in targeting of p66 as well. In such a scenario, a forward feedback loop can be envisaged, with a stable chromatin association taking place only after deacetylation of the histones has occurred. Such a stable Mi-2/NuRD binding might also facilitate a spreading of inactive deacetylated chromatin into active chromatin regions. Alternatively, histone association may play a role during remodeling rather during targeting of the complex to chromatin.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors would like to thank Sonja Sahner for excellent technical help, Drs C. Wu, A. Baniahmad, S. Berger and A. Bird for materials and the DFG for support (GRK 370). Funding to pay the Open Access publication charges for this article was provided by the DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bestor T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 2.Das P.M., Singal R. DNA methylation and cancer. J. Clin. Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg A.P., Cui H., Ohlsson R. DNA methylation and genomic imprinting: insights from cancer into epigenetic mechanisms. Semin. Cancer Biol. 2002;12:389–398. doi: 10.1016/s1044-579x(02)00059-7. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M., Herman J.G. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J. Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 5.Gaudet F., Hodgson J.G., Eden A., Jackson-Grusby L., Dausman J., Gray J.W., Leonhardt H., Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 6.Davey C., Pennings S., Allan J. CpG methylation remodels chromatin structure in vitro. J. Mol. Biol. 1997;267:276–288. doi: 10.1006/jmbi.1997.0899. [DOI] [PubMed] [Google Scholar]
- 7.Bednarik D.P., Duckett C., Kim S.U., Perez V.L., Griffis K., Guenthner P.C., Folks T.M. DNA CpG methylation inhibits binding of NF-kappa B proteins to the HIV- 1 long terminal repeat cognate DNA motifs. New Biol. 1991;3:969–976. [PubMed] [Google Scholar]
- 8.Kovesdi I., Reichel R., Nevins J.R. Role of an adenovirus E2 promoter binding factor in E1A-mediated coordinate gene control. Proc. Natl Acad. Sci. USA. 1987;84:2180–2184. doi: 10.1073/pnas.84.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voo K.S., Carlone D.L., Jacobsen B.M., Flodin A., Skalnik D.G. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol. Cell. Biol. 2000;20:2108–2121. doi: 10.1128/mcb.20.6.2108-2121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitz A., Short M., Ammerpohl O., Asbrand C., Nickel J., Renkawitz R. Cis-elements required for the demethylation of the mouse M-lysozyme downstream enhancer. J. Biol. Chem. 1997;272:20850–20856. doi: 10.1074/jbc.272.33.20850. [DOI] [PubMed] [Google Scholar]
- 11.Nickel J., Short M.L., Schmitz A., Eggert M., Renkawitz R. Methylation of the mouse M-lysozyme downstream enhancer inhibits heterotetrameric GABP binding. Nucleic Acids Res. 1995;23:4785–4792. doi: 10.1093/nar/23.23.4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iguchi-Ariga S.M., Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 13.Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 14.Ohlsson R., Renkawitz R., Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 15.Hendrich B., Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boeke J., Ammerpohl O., Kegel S., Moehren U., Renkawitz R. The minimal repression domain of MBD2b overlaps with the methyl-CpG-binding domain and binds directly to Sin3A. J. Biol. Chem. 2000;275:34963–34967. doi: 10.1074/jbc.M005929200. [DOI] [PubMed] [Google Scholar]
- 17.Fujita N., Shimotake N., Ohki I., Chiba T., Saya H., Shirakawa M., Nakao M. Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1. Mol. Cell. Biol. 2000;20:5107–5118. doi: 10.1128/mcb.20.14.5107-5118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Ng H.H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng H.H., Zhang Y., Hendrich B., Johnson C.A., Turner B.M., Erdjument-Bromage H., Tempst P., Reinberg D., Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nature Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 20.Nan X., Ng H.H., Johnson C.A., Laherty C.D., Turner B.M., Eisenman R.N., Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 21.Jones P.L., Veenstra G.J., Wade P.A., Vermaak D., Kass S.U., Landsberger N., Strouboulis J., Wolffe A.P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 22.Harikrishnan K.N., Chow M.Z., Baker E.K., Pal S., Bassal S., Brasacchio D., Wang L., Craig J.M., Jones P.L., Sif S., et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nature Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 23.Wade P.A., Gegonne A., Jones P.L., Ballestar E., Aubry F., Wolffe A.P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nature Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 24.Loyola A., Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Brackertz M., Boeke J., Zhang R., Renkawitz R. Two highly related p66-proteins comprise a new family of potent transcriptional repressors interacting with MBD2 and MBD3. J. Biol. Chem. 2002;277:40958–40966. doi: 10.1074/jbc.M207467200. [DOI] [PubMed] [Google Scholar]
- 26.Baniahmad A., Leng X., Burris T.P., Tsai S.Y., Tsai M.J., O'Malley B.W. The tau 4 activation domain of the thyroid hormone receptor is required for release of a putative corepressor(s) necessary for transcriptional silencing. Mol. Cell. Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georgel P.T., Tsukiyama T., Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L., Scolnick D.M., Trievel R.C., Zhang H.B., Marmorstein R., Halazonetis T.D., Berger S.L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendrich B., Guy J., Ramsahoye B., Wilson V.A., Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Standke G.J., Meier V.S., Groner B. Mammary gland factor activated by prolactin on mammary epithelial cells and acute-phase response factor activated by interleukin-6 in liver cells share DNA binding and transactivation potential. Mol. Endocrinol. 1994;8:469–477. doi: 10.1210/mend.8.4.7519723. [DOI] [PubMed] [Google Scholar]
- 32.Feng Q., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Zhang Y. Identification and functional characterization of the p66/p68 components of the MeCP1 complex. Mol. Cell. Biol. 2002;22:536–546. doi: 10.1128/MCB.22.2.536-546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berger S.L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 34.Feng Q., Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–832. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X.J. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Q., Zhang Y. The NuRD complex: linking histone modification to nucleosome remodeling. Curr. Top Microbiol. Immunol. 2003;274:269–290. doi: 10.1007/978-3-642-55747-7_10. [DOI] [PubMed] [Google Scholar]
- 37.Nishioka K., Chuikov S., Sarma K., Erdjument-Bromage H., Allis C.D., Tempst P., Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth S.Y., Denu J.M., Allis C.D. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 39.Schiltz R.L., Mizzen C.A., Vassilev A., Cook R.G., Allis C.D., Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 40.Yoon H.G., Choi Y., Cole P.A., Wong J. Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol. Cell. Biol. 2005;25:324–335. doi: 10.1128/MCB.25.1.324-335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.