Abstract
Prior to conjugative transfer of plasmids, one plasmid strand is cleaved in a site- and strand-specific manner by an enzyme called a relaxase or nickase. In F and related plasmids, an inverted repeat is located near the plasmid strand cleavage site, and others have proposed that the ability of this sequence to form a hairpin when in single-stranded form is important for transfer. Substitutions were introduced into a cloned F oriT region and their effects on plasmid transfer were assessed. For those substitutions that substantially reduced transfer, the results generally correlated with effects on in vitro binding of oligonucleotides to the F TraI relaxase domain rather than with predicted effects on hairpin formation. One substitution shown previously to dramatically reduce both plasmid transfer and in vitro binding to a 17-base oligonucleotide had little apparent effect on binding to a 30-base oligonucleotide that contained the hairpin region. Results from subsequent experiments strongly suggest that the relaxase domain can bind to hairpin oligonucleotides in two distinct manners with different sequence specificities, and that the protein binds the oligonucleotides at the same or overlapping sites.
INTRODUCTION
Bacterial conjugation is the unidirectional transfer of a single strand of a conjugative plasmid from donor bacterium to recipient. Prior to transfer, one plasmid strand, called the transfer strand, is cleaved in a reaction catalyzed by a protein named a relaxase or nickase. These proteins cut DNA in a sequence-specific manner within the origin of transfer (oriT) at a site called nic (1–3). Relaxases use an active site tyrosyl hydroxyl for nucleophilic attack on the backbone phosphorus at nic (4–7). The reaction forms a long-lived intermediate in which protein and DNA are joined by a phosphotyrosyl linkage (2,4,8). This linkage probably endures through transfer, with the attached relaxase ligating the ends of the plasmid in the recipient cell (9,10). Relaxases readily bind and cleave single-stranded, but not double-stranded, oligonucleotides in vitro. This leads to the suggestion that the role of accessory proteins that are required for efficient transfer is to bind oriT near nic and assist cleavage by generating the single-stranded DNA (ssDNA) conformation that relaxases are capable of binding. Plasmid oriT regions contain not only binding sites for relaxases and accessory proteins, but they also include features such as inverted repeats and A-tracts (11). The complexity of oriT regions suggests that the transfer process requires multiple recognition and regulatory steps.
Results from our previous in vitro and in vivo experiments indicate that TraI, the relaxase from conjugative plasmid F, recognizes its oriT site with a remarkable level of sequence specificity (12). In these initial experiments, we focused on a 22-base region around nic. Using a fluorescently-labeled single-stranded oligonucleotide to measure in vitro binding affinity and specificity of the TraI relaxase domain (TraI36), we found that single base changes could reduce binding affinity by over 5000-fold. A subset of these substitutions was introduced into a cloned F oriT, and substitutions that substantially decreased in vitro binding also significantly reduced plasmid transfer efficiency. The crystal structure of the F TraI36 variant Y16F, which is deficient in ssDNA cleavage, bound to oriT ssDNA [TraI36(Y16F):ssDNA] helps explain the specificity of TraI36 recognition. DNA bases that define much of the binding specificity are involved in intricate intramolecular and intermolecular interactions (13).
In our initial binding experiments and in our crystal structure, the oligonucleotides used did not extend far enough 5′ to nic to include all of a region that includes a perfect inverted repeat. This segment, the 3′ end of which is 10 bases 5′ to nic, has a 2-base spacer between the 8-base repeated sequences. Deonier and coworkers (14) demonstrated that altering the sequence within the inverted repeat of F oriT can affect termination, and sometimes also initiation, of the transfer process. The crystal structure of the TrwC relaxase domain from plasmid R388 was determined with bound ssDNA that included a 5′ segment corresponding to a 5 bp perfect inverted repeat (15). In the TrwC structure, this region forms a hairpin, and several protein–DNA contacts are formed between TrwC and the hairpin. The TrwC structure and in vitro binding experiments clearly indicate that the protein binds well to a partial duplex oligonucleotide (15). What is not clear, however, is whether during transfer the protein binds a hairpin or some other partial duplex.
The TraI36 and TrwC relaxase domains are similar in structure (Figure 1) (13). The region in TrwC that interacts with the hairpin corresponds to a region in TraI36 that is poorly ordered. This disordered TraI36 region is potentially available to interact with DNA. We undertook a series of experiments to determine whether substitutions in a cloned F oriT that would alter hairpin stability would affect transfer, and whether the presence of this hairpin would affect the binding of an oligonucleotide to the protein. We did observe effects on transfer efficiency from substitutions in the inverted repeat, but they were consistent with effects due to loss of specific protein–DNA interactions rather than to destabilizing a hairpin. In vitro binding studies confirmed that these base substitutions reduced binding, but also suggested that the protein can bind DNA in two distinct manners with different specificities.
Figure 1.
Comparison of structures of TraI and TrwC relaxase domains with bound ssDNA. Depicted are (left) TraI36(Y16F) (PDB ID 2A0I) (13) and (right) TrwC (PDB ID 1QX0) (15). The proteins are shown as blue (TraI36) or green (TrwC) cartoons in which α-helices are depicted as ribbons and β-sheets as arrows. DNA is depicted as pink (TraI36) or yellow (TrwC) sticks. The circles highlight an area occupied in the TrwC structure by a 5′ DNA hairpin and a two-strand β-sheet but in which there is no DNA and largely unstructured protein in TraI36.
MATERIALS AND METHODS
Bacterial strains, plasmids, plasmid mutagenesis and oligonucleotides
Escherichia coli strains ER2738 [F′ proA+B+ lacIq Δ(lacZ)M15 zzf::Tn10(TetR)/fhuA2 glnV Δ(lac-proAB) thi-1 Δ(hsdS-mcrB)5] and TB1 [F− ara Δ (lac-proAB) [φ80dlac Δ(lacZ)M15] rpsL(StrR) thi hsdR] and plasmid pACYC177 were obtained from New England BioLabs. E.coli strain XL-1 Blue [F′ proAB lacIq lacZΔM15 Tn10 (TetR)/recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac] was purchased from Stratagene. Plasmid pACYC-ForiT was engineered by PCR amplifying bp 1–530 of ForiT [numbering according to Frost et al. (11)] from the ER2738 F′ plasmid and cloning the product into the DraIII and BamHI sites of pACYC177. Nucleotide substitutions were introduced into pACYC-ForiT using the QuikChange Kit (Stratagene). Sequences of cloning and mutagenesis oligonucleotides are available upon request. Mutagenesis results were confirmed by DNA sequencing and plasmid variants were transformed into ER2738.
Oligonucleotides were synthesized by Integrated DNA Technologies. Oligonucleotides used for binding and cleavage studies included WT Hairpin d(GCAAAAACTTGTTTTTGCGTGGGGTGT|GGT), G144′C Hairpin d(GCAAAAACTTGTTTTTGCGTGGGCTGT|GGT), G151′C/C150′G Hairpin d(GCAAAAACTTGTTTTTCGGTGGGGTGT|GGT), G167′C/C166′G Hairpin d(CGAAAAACTTGTTTTTGCGTGGGGTGT|GGT), G167′C/C166′G/G151′C/C150′G Hairpin d(CGAAAAACTTGTTTTTCGGTGGGGTGT|GGT), No Hairpin d(CGTTTTTGTTGTTTTTGCGTGGGGTGT|GGT), Hairpin + Random d(GCAAAAACTTGTTTTTGCCACCCCACACCA), WT 17mer d(TTTGCGTGGGGTGT|GGT), G144′C 17mer d(TTTGCGTGGGCTGT|GGT), G151′C/C150′G 17mer d(TTTCGGTGGGGTGT|GGT) and Hairpin Only d(GCAAAAACTTGTTTTTGC), where differences from the wild-type sequence are underlined and ‘|’ denotes nic, the site of cleavage by TraI. For binding studies in which binding is followed by changes in fluorescence intensity and anisotropy, oligonucleotides were synthesized with a 3′-carboxytetramethylrhodamine (TAMRA) label. For nicking assays, oligonucleotides were 5′ end-labeled with [γ-32P]ATP (GE Healthcare) using T4 Polynucleotide Kinase according to the manufacturer's (New England BioLabs) instructions. Unincorporated nucleotides were separated from oligonucleotide using Sephadex G-50 Fine Quick Spin Columns (Roche).
Plasmid mobilization assay
ER2738 containing wild-type or variant pACYC-ForiT, or pACYC177 as a negative control, were used as the donors and TB1 as recipients in mobilization assays. TB1 were grown in Luria–Burtani broth (LB) with streptomycin (50 µg/ml) (LB-strep) and ER2738/pACYC-ForiT in LB with tetracycline (25 µg/ml) and ampicillin (100 µg/ml) (LB-tet-amp) overnight at 37°C with shaking. Cultures were diluted 1:200 in LB-tet-amp (donors) or LB-strep (recipients) and grown to an OD600 = 0.5–0.6. Donor and recipient cells were pelleted by centrifugation, the supernatant removed, and the pellet resuspended in an equal volume of phosphate-buffered saline (PBS) (16). Donors and recipients were mixed at a 1:9 ratio and incubated at 37°C with no shaking. After 2 min, cells were vortexed vigorously for 10 s and immediately placed on ice. After 10 min on ice, serial dilutions were made into ice-cold PBS. Of each dilution 100 µl were plated onto selective agar (LB-amp to select for donors; LB-strep-amp to select for transconjugates). Viability of recipient TB1 cells was confirmed by plating on LB-strep. Plates were incubated overnight at 37°C. Mobilization efficiency was calculated as the number of transconjugate colonies divided by the number of donor colonies. The efficiencies are reported as an average of 3–21 assays with the SE of the measurement. Mobilization efficiencies were considered statistically significantly different from wild type if a two-sided Student's t-test gave P < 0.05.
Affinity measurements
Affinity measurements using changes in fluorescence intensity and anisotropy of 3′-TAMRA-labeled oligonucleotides were performed as described (12) except a high salt buffer [20 mM Tris–HCl (pH 7.5), 0.1 mM EDTA and 300 mM NaCl] was used. Dissociation rate constants were measured by first combining 4 nM TAMRA-labeled oligonucleotide with TraI36. Protein concentrations, which were chosen based on the affinity of TraI36 for the oligonucleotide, were 40 nM (WT 17mer), 85 nM (G151′C/C150′G Hairpin), 15 nM (G144′C Hairpin) or 10 nM (WT Hairpin). After a 10 min incubation, excess unlabeled oligonucleotide (400 nM final concentration) was added and fluorescence intensity was recorded. The unlabeled competitor oligonucleotide was the same as the labeled oligonucleotide except for G151′C/C150′G Hairpin (WT Hairpin used as competitor), and for experiments in which the ability of G144′C Hairpin to compete with WT 17mer for binding was examined.
Oligonucleotide cleavage assay
The ability of TraI36 to cleave an oriT oligonucleotide was measured by combining 9 µl of reaction buffer [300 mM NaCl, 50 mM Tris (pH 7.5) and 20 mM MgCl2], 3 µl of radiolabeled oligonucleotide (4 nM final concentration) and 3 µl of TraI36 (ranging from 1 nM to 1 µM, final concentration). The reactions were incubated at 25°C for 30 min and were stopped by addition of SDS (0.1% final concentration). Samples were incubated for an additional 10 min at 25°C and then mixed with an equal volume of loading buffer [80% formamide (v/v) in 225 mM Tris–Borate and 5 mM EDTA]. Samples were heated to 90°C for 5 min, snap cooled on ice and run on a 16 (Hairpin oligonucleotides) or 20% (17mer oligonucleotides) polyacrylamide-urea gel at 300 V for 2.5 h. Gels were placed in a phosphorimager cassette overnight and exposed using a SF PhosphorImager (Molecular Dynamics).
RESULTS
Mobilization efficiencies of oriT variants
The DNA sequence of the F factor oriT near the TraI binding site includes a perfect inverted repeat (Figure 2). The repeated sequence (located on the transfer strand 5′ to nic) is 8 bp long, with a 2 bp spacer between the sequences. Within the repeated sequence is a 5 bp A-tract. We examined the effects of substitutions in these regions on transfer by engineering variants in an F oriT region cloned into pACYC177 and measuring the ability of these variant plasmids to be mobilized between strains (Figure 2). Transfer of this plasmid is referred to as mobilization because all proteins required for transfer are provided in trans from an F′ within the donor. A plasmid containing the wild-type F oriT (pACYC-ForiT) is mobilized with an average efficiency of 5 × 10−4 while pACYC177 with no oriT has no detectable transfer (efficiency <2 × 10−6; <0.6% of pACYC-ForiT). G144′ is known to be important for F transfer (17). The G144′C substitution, shown previously to dramatically reduce mobilization when introduced into F oriT and significantly reduce binding when introduced into an oligonucleotide (12), also reduces mobilization below the detectable limit.
Figure 2.
Relative mobilization efficiencies of F oriT variants. The mobilization efficiencies of oriT variants are shown (right) with the sequences of the transfer strands in the region of nic (center). The location of the inverted repeat is shown as arrows above the wild-type sequence. Sequence differences of the variants relative to the wild-type sequence are indicated by underlining and shaded boxes. Vertical lines indicate the position of nic. Mobilization efficiencies are given as percents of the average mobilization of the plasmid containing the wild-type oriT. Error bars represent the SE of the measurement. n indicates the number of measurements. Values that significantly differ (P < 0.05) from the wild-type efficiency as determined by a two-sided Student's t-test are indicated by asterisks. Variants with mobilization efficiencies below a detectable level have the average lower limit of transfer efficiency indicated within the bar.
To test whether the A-tracts are functionally important and whether a single base substitution within the inverted repeat would affect transfer, we substituted a T for the middle A in each A-tract individually (T154′A, A163′T). Neither variant differed significantly from the wild-type oriT in mobilization efficiency. The double variant (A163′T/T154′A), which should disrupt both A-tracts but maintain the base complementarity within the hairpin, also showed no significant difference from the wild-type mobilization efficiency.
To disrupt the inverted repeat further, we converted the TTGC sequence (153′–150′) at the base of the putative hairpin to AACG. The mobilization efficiency of this variant was below detection. The analogous substitution of CGTT for GCAA at 167′–164′, however, caused a small (3-fold) but statistically significant increase in mobilization efficiency. A variant with the combined 153′–150′ and 167′–164′ substitutions had a mobilization efficiency below detection. The increased mobilization caused by the 167′–164′ substitutions, and the failure of the 167′–164′ substitutions to detectably compensate for the 153′–150′ substitutions suggests that the reduced mobilization of the 153′–150′ variant does not result from disruption of a hairpin. Because both the 153′–150′ and the 167′–164′/153′–150′ variants had undetectable mobilization, we cannot say whether the 167′–164′ substitutions compensated partially for the 153′–150′ substitutions.
The T153′A/T152′A substitutions play a role in the reduced affinity of the 153′–150′ variant. The T153′A/T152′A variant mobilized with 17% of wild-type efficiency, a statistically significant reduction. The A165′T/A164′T variant transferred similarly to wild type. The combined T153′A/T152′A and A165′T/A164′T substitutions demonstrated 11% of wild-type mobilization, again a statistically significant reduction.
The C150′G and G151′C substitutions did not individually reduce transfer significantly, but the G151′C/C150′G variant had only 0.8% of wild-type mobilization. The G167′C/C166′G variant demonstrated wild-type mobilization, while the G167′C/C166′G/G151′C/C150′G variant had 3% wild-type mobilization. While it is possible that the 4-fold increase in mobilization efficiency of the G167′C/C166′G/G151′C/C150′G variant over the G151′C/C150′G variant reflects a small improvement due to compensatory substitutions that would stabilize a hairpin, the improvement in efficiency is not significant, at least with the number of measurements (n = 3 for each).
Binding affinities of oriT variants
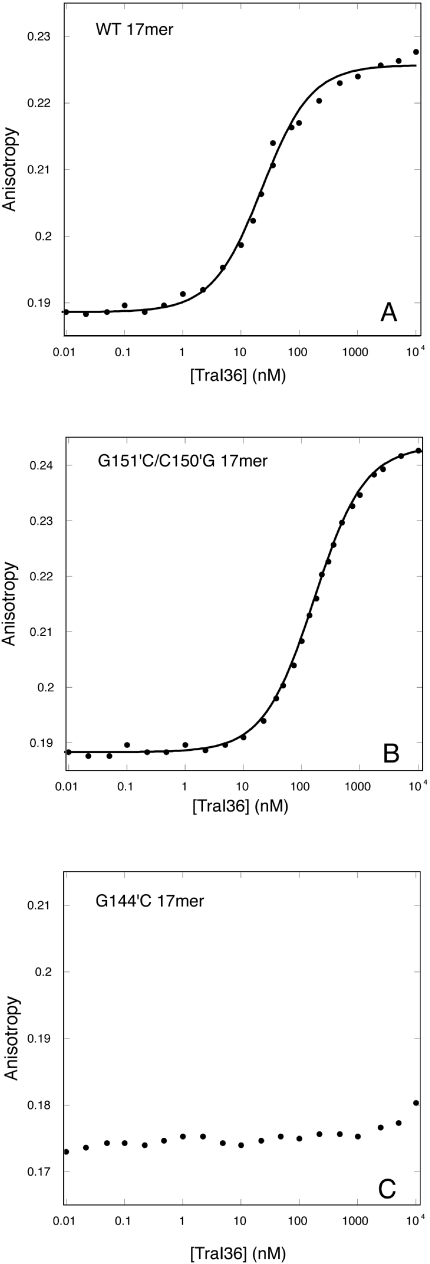
We measured binding of oligonucleotides to TraI36 to determine whether the reduced mobilization efficiencies of oriT sequence variants might correlate with reduced affinities of TraI36 for these sequences, and to ascertain the effect of the hairpin on binding (Table 1 and Figures 3 and 4). As expected based on earlier measurements (12,18), binding of a 17-base oligonucleotide that extends 14 bases 5′ to nic and 3 bases 3′ to nic (WT 17mer) binds to TraI36 with a KD of 39 nM, while TraI36 demonstrates little or no affinity for a similar oligonucleotide containing the G144′C substitution (G144′C 17mer). The binding affinity for the variant oligonucleotide was too low for an accurate measurement, but is reduced by at least three orders of magnitude. The G151′C/C150′G 17mer variant oligonucleotide binds TraI36 with a KD of 390 nM, a 10-fold reduction in affinity relative to WT 17mer.
Table 1.
Apparent dissociation constants for 3′-TAMRA-labeled ssDNA oligonucleotides from F TraI36
| Oligonucleotide | Average KD ± SE (nM)a | nb |
|---|---|---|
| WT 17mer | 39 ± 0.8 | 2 |
| G151′C/C150′G 17mer | 390 ± 9 | 3 |
| G144′C 17mer | >10 000c | 3 |
| WT Hairpin | 6.8 ± 1.2d | 7 |
| G151′C/C150′G Hairpin | 76 ± 3.4 | 4 |
| G167′C/C166′G Hairpin | 12 ± 1.8d | 4 |
| G167′C/C166′G/G151′C/C150′G Hairpin | 78 ± 2.0 | 4 |
| G144′C Hairpin | 7.6 ± 0.8d | 9 |
| No Hairpin | 16 ± 1.0 | 3 |
| Hairpin + Random | >10 000c | 2 |
aDissociation constants were estimated by a simultaneous fit of fluorescence intensity and anisotropy data using SPECTRABIND (23). Values from multiple experiments were averaged and listed with the SE of the population.
bNumber of measurements averaged.
cThe poor binding and insufficient upper baseline results in poor fits.
dOwing to suboptimal fits of the two-state model to the data, these values can only be considered estimates of the KD.
Figure 3.
Reduced affinity of TraI36 for G151′C/C150′G and G144′C 17-base oligonucleotides. TraI36 protein was titrated into 4 nM 3′-TAMRA-labeled WT (A), G151′C/C150′G (B) or G144′C (C) 17-base oligonucleotides. Anisotropy and intensity (not shown) data were simultaneously fit with a 1:1 (protein:DNA) binding model using SPECTRABIND (23,24). Data points are shown as closed circles and the fit as solid lines.
Figure 4.
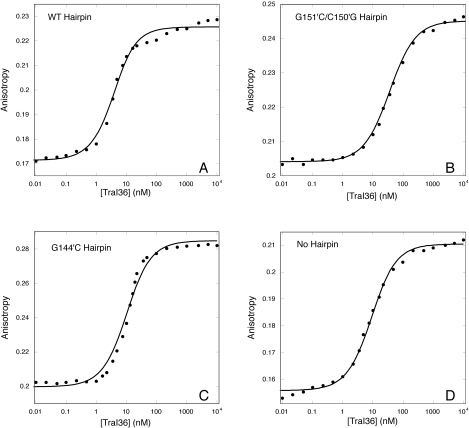
Reduced affinity of TraI36 for G151′C/C150′G but not G144′C 30-base hairpin oligonucleotides. TraI36 protein was titrated into 4 nM 3′-TAMRA-labeled 30-base WT Hairpin (A), G151′C/C150′G Hairpin (B), G144′C Hairpin (C) or No Hairpin (D) oligonucleotides. Anisotropy and intensity (not shown) data were simultaneously fit with a 1:1 (protein:DNA) binding model using SPECTRABIND (23,24). Data points are shown as closed circles and the fit as solid lines. Note the poor fit to the WT Hairpin and G144′C Hairpin data, and the flat upper baseline of the G144′C Hairpin titration.
A 30-base oligonuclotide that contains the hairpin 5′ to nic and extends 3 bases 3′ to nic (WT Hairpin) binds TraI36 with a several-fold increased affinity over the affinity for WT 17mer (Table 1 and Figure 4). The TrwC relaxase domain also binds with increased affinity to a hairpin-containing oligonucleotide (15). Using a simple binding model with a 1:1 protein:DNA stoichiometry to estimate a binding constant from the data yielded uncharacteristically poor fits, however. The data at the beginning of the transition fell below the fitted curve, while the data beyond the inflection tended to lie above the curve. Subsequent experimentation (see below) suggests that the poor fit may be due in part to nonequilibrium measurements for the hairpin oligonucleotides resulting from markedly slowed rates of association and dissociation. Although the WT Hairpin curve is clearly shifted to lower TraI36 concentrations, indicating a higher affinity interaction, the suboptimal quality of the fits for the hairpin oligonucleotides means that the affinities calculated for these oligonucleotides can only be considered estimates. The estimated KD of TraI36 for WT Hairpin is 7 nM.
The WT Hairpin curve also has an upper baseline with a pronounced positive slope. We reported previously a sloped upper baseline in binding curves when TraI36 binds 22- and 17-base oligonucleotides at lower salt concentrations, and attributed it to nonspecific binding by the protein (12). A sloped baseline is also present in the WT 17mer titration shown here (Figure 3) but is less pronounced than in the WT Hairpin data. The smaller apparent slope may result from the shorter upper baseline due to the lower affinity for the WT 17mer relative to the WT Hairpin.
The G151′C/C150′G Hairpin, like the G151′C/C150′G 17mer, showed a 10-fold reduced estimated affinity for TraI36 relative to WT Hairpin. The presence of the hairpin increases affinity of the G151′C/C150′G Hairpin relative to the G151′C/C150′G 17mer linear oligonucleotide (Hairpin KD = 76 nM; 17mer KD = 390 nM). The G167′C/C166′G/G151′C/C150′G Hairpin binds TraI36 with an estimated KD of 78 nM. The data from both of these variants were well fitted with a two-state model. The G167′C/C166′G Hairpin binds with an estimated KD of 12 nM, but these data were not well fit by the model. The G167′C/C166′G Hairpin had an upper baseline slope similar to the wild-type hairpin. The slope for the G151′C/C150′G and G167′C/C166′G/G151′C/C150′G Hairpin variants did not have a pronounced upper baseline slope, but this may be because the shifted binding transitions shortened the upper baseline and obscured the slope.
A 30-base oligonucleotide that retains the wild-type G151′/C150′ sequence but has the 5′ 8 bases substituted to prevent formation of a hairpin (No Hairpin) binds with a KD = 16 nM, underscoring the relative importance of G151′/C150′ over the hairpin to the binding (Figure 4). The data for the No Hairpin oligonucleotide were well fit with a two-state model, but did show an upper baseline slope. A 30-base oligonucleotide that possessed the full hairpin sequence and a substituted sequence 3′ to the hairpin (Hairpin + Random) bound poorly to TraI36, with an increase in KD of at least three orders of magnitude. An 18-base oligonucleotide comprising only the hairpin sequence (Hairpin Only) also showed little or no binding to TraI36 (data not shown).
Unexpectedly, the G144′C substitution, which caused a greater than three orders of magnitude reduction in affinity when introduced into the 17mer oligonucleotide, caused no change in binding relative to wild type in the context of the 30-base hairpin oligonucleotide. Products of two separate oligonucleotide syntheses gave virtually identical results. While the G144′C data showed the same sharp transition as WT Hairpin, the variant oligonucleotide did not yield a sloped upper baseline.
Dissociation rate constants of oligonucleotides
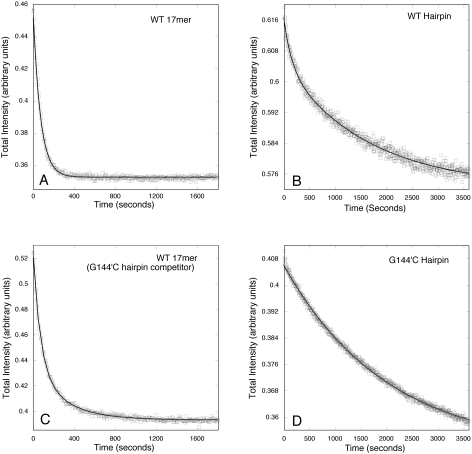
To further examine the interactions between TraI36 and oligonucleotides, we measured dissociation rate constants. Rate constants were measured by combining TraI36 and a labeled oligonucleotide, then adding a 100-fold excess of unlabeled oligonucleotide to prevent reassociation of labeled oligonucleotide after disassociation from TraI36. In most experiments, the unlabeled and labeled oligonucleotides had the same sequence. Dissociation of TAMRA-labeled WT 17mer oligonucleotide from TraI36 is described well by a single exponential curve. The average dissociation rate constant is 0.84 min−1 (Table 2 and Figure 5). In contrast, the WT Hairpin dissociation curve requires two exponentials for a good fit, with corresponding average rate constants of 0.42 min−1, accounting for 27% of the amplitude, and 0.026 min−1. The G151′C/C150′G Hairpin dissociation curve also requires two exponentials for an adequate fit, with average rate constants of 0.46 min−1 (90% of the amplitude) and 0.14 min−1. Similar results were obtained with the G167′C/C166′G/G151′C/C150′G Hairpin (data not shown), suggesting that the differences in results between WT Hairpin and G151′C/C150′G Hairpin are due to loss of base-specific interactions rather than disruption of the hairpin. In contrast, the G144′C Hairpin dissociation curve requires a single exponential for an adequate fit, with a corresponding rate constant of 0.034 min−1.
Table 2.
Dissociation rate constants of 3′-TAMRA-labeled ssDNA oligonucleotides from F TraI36
| Oligonucleotide | Competitor | k1 (min−1) | % Amp | k2 (min−1) | % Amp | na |
|---|---|---|---|---|---|---|
| WT 17-base | WT 17-base | 0.84 ± 0.0008 | 100 | 3 | ||
| WT Hairpin | WT Hairpin | 0.42 ± 0.16 | 27 | 0.026 ± 0.013 | 73 | 9 |
| G151′C/C150′G Hairpin | WT Hairpin | 0.46 ± 0.009 | 89 | 0.14 ± 0.04 | 11 | 3 |
| G144′C Hairpin | G144′C Hairpin | 0.034 ± 0.006 | 100 | 3 | ||
| WT 17-base | G144′C Hairpin | 0.67 ± 0.14 | 76 | 0.14 ± 0.05 | 24 | 3 |
aNumber of measurements averaged.
Figure 5.
Differing dissociation kinetics of oligonucleotides from TraI36. TraI36 was combined with 4 nM 5′-TAMRA-labeled WT 17-base (A and C), WT Hairpin (B) or G144′C Hairpin (D) oligonucleotide. After a 10 min incubation, excess unlabeled oligonucleotide was added and the decrease in fluorescence intensity of the oligonucleotide followed over time. The competitor oligonucleotide was an unlabeled version of the labeled binding oligonucleotide except in (D) in which the ability of G144′C Hairpin to compete with WT 17-base oligonucleotide for binding was tested.
The two rate constants observed for the dissociation of WT Hairpin from TraI36 suggest that there are at least two modes for the binding of the hairpin oligonucleotide to the protein. In contrast, the WT 17mer oligonucleotide and the G144′C Hairpin dissociation curves are consistent with a single bound population. To test whether the binding site of the 17-base oligonucleotides overlaps with the binding site for the Hairpin oligonucleotides, we combined TraI36 with the TAMRA-labeled WT 17mer oligonucleotide, equilibrated and then added an excess of unlabeled G144′C Hairpin. The G144′C Hairpin competed effectively with the WT 17mer for binding. This dissociation curve, however, required two exponentials for an adequate fit, with dissociation rate constants of 0.67 min−1 (76% amplitude) and 0.14 min−1. Excess unlabeled G144′C 17mer did not compete with WT 17mer for binding (data not shown).
Cleavage characteristics of oligonucleotide substrates
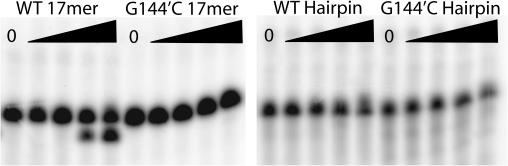
Various oligonucleotides were tested to determine whether the apparently altered binding specificities of oligonucleotides with and without the 5′ hairpin also differed in their abilities to be cleaved. Results from an oligonucleotide cleavage assay are shown in Figure 6. Consistent with the different affinities of TraI36 for the wild-type and G144′C 17-base linear oligonucleotides and with previous results (12), the wild-type oligonucleotide is readily cleaved in the presence of Mg2+ while the G144′C oligonucleotide is not cleaved under the same conditions. In contrast, neither WT Hairpin nor G144′C Hairpin is cleaved.
Figure 6.
Hairpin oligonucleotides are poor substrates for TraI36-mediated cleavage. The 5′-32P-labeled WT and G144′C 17mer (left panel) and WT and G144′C 30-base Hairpin (right panel) oligonucleotides were incubated with varying concentrations of TraI36. Oligonucleotide concentration was 4 nM, and for each reaction the TraI36 concentrations were (from left) 0 nM, 1 nM, 10 nM, 100 nM and 1 µM. After each reaction was stopped, the reaction mixtures were applied to a denaturing polyacrylamide gel. Cleavage activity is indicated by the appearance of a faster migrating band.
DISCUSSION
We examined the effect of altering F factor transfer strand sequences 5′ to nic, including an inverted repeat, on plasmid transfer. The results suggested that neither A-tracts in the indirect repeat nor formation of a hairpin by the sequences within the repeat are essential for F conjugative transfer. Instead, the reduced mobilization efficiency exhibited by some variants is more likely the result of reduced binding affinity of TraI for the variant plasmid sequence. In vitro measurements confirmed that at least within a limited set (WT, G151′C/C150′G and G144′C), the rank order of binding affinity for a 17-base oligonucleotide and of mobilization efficiency is maintained. Based on these results, it is plausible that the reduced transfer of G151′C/C150′G and G144′C variant plasmids results from a reduced affinity of TraI for the sequences.
Binding measurements of 30-base Hairpin oligonucleotides indicated that addition of a 5′ region to the 17-base oligonucleotide sequence improves affinity relative to the 17mer oligonucleotides. In addition, TraI36 binds the G144′C Hairpin oligonucleotide with an apparent affinity similar to that for WT Hairpin. The high-affinity binding of G144′C Hairpin comes despite the poor binding of G144′C 17mer and the dramatic reduction in plasmid mobilization caused by the G144′C substitution. The presence of the hairpin therefore improves oligonucleotide binding, and also alters the binding specificity.
Based on the G144′C 17mer and G144′C Hairpin results, and on subsequent experimentation, we conclude that TraI36 can bind with high affinity to the same or similar DNA sequences in two different conformations. Furthermore, these different oligonucleotide conformations can bind to the same or overlapping sites on TraI36. The measured dissociation rate constants are consistent with this model. The WT 17mer oligonucleotide dissociates from TraI36 with a single rate constant (0.84 min−1). The 30-base WT Hairpin oligonucleotide dissociates with two rate constants, one similar to the rate constant of the WT 17mer oligonucleotide (0.42 min−1), and one considerably smaller (0.026 min−1). The 30-base G144′C Hairpin oligonucleotide has a single rate constant (0.034 min−1) near the smaller rate constant of the WT Hairpin. Assuming that the two dissociation rate constants reflect dissociation of different oligonucleotide conformations, binding in the fast dissociation conformation (with the larger dissociation rate constant) requires the G144′ base, while the second (the slow dissociation conformation) does not. In the TraI36(Y16F):ssDNA structure, the G144′ base both interacts with the protein and participates in a noncanonical 3-base base-pairing interaction (13). The G144′C substitution would likely disrupt the local conformation, thereby destabilizing the complex and reducing binding affinity.
There is also sequence-specific recognition in the slow dissociation conformation. For G151′C/C150′G Hairpin, the slower dissociation rate constant (0.14 min−1) is several-fold faster than that for the slow dissociation conformation of the WT Hairpin while the larger rate constant (0.46 min−1) is similar to that of the fast dissociation conformation of the WT Hairpin. Results for G167′C/C166′G/G151′C/C150′G Hairpin are similar. Additional evidence that both conformations are bound sequence specifically is found in the poor binding by the Hairpin + Random oligonucleotide.
To examine whether the two binding conformations reflect binding to separate sites, we combined TraI36 with TAMRA-labeled WT 17mer, allowed binding to equilibrate and then added an excess of unlabeled G144′C Hairpin oligonucleotide. The G144′C Hairpin competed well with the WT 17mer, although the dissociation curve required two exponentials for an adequate fit. We are unsure what these two rate constants reflect. The ability of G144′C Hairpin and WT 17mer to compete for binding is consistent with the two sites overlapping each other. The extent to which they overlap is not clear, but the contacts that define the two modes of binding could be dramatically different given the distinct effects of the G144′C substitution on the two modes.
The WT 17mer is readily cleaved by TraI36 in the presence of Mg2+, but we detected no cleavage of the WT Hairpin oligonucleotide. Because the fast dissociation conformation of the WT Hairpin has a dissociation rate constant similar to that of the 17mer and was not obviously populated in the G144′C Hairpin, we initially suspected that the fast dissociation conformation of the WT Hairpin resembles the binding conformation of WT 17mer. If this were so, however, we would have expected some cleavage product from the WT Hairpin oligonucleotide. A possible explanation for the absence of cleavage is that there are fundamental differences between the bound conformation of the WT 17mer and both binding modes of WT Hairpin. Another possibility is that TraI36 is competent to cleave the WT Hairpin oligonucleotide, but the altered binding characteristics somehow favor the ligation reaction over the cleavage reaction. As a result, cleaved oligonucleotide is likely to be ligated before release, reducing the amount of cleavage product.
The ability of TraI36 to interact in two distinct ways with DNA is not completely unexpected. The HUH superfamily of proteins includes TraI, other relaxases and mobilization proteins, and many replication initiator proteins involved in plasmid and viral rolling circle replication (19,20). The family is named for a metal-binding motif that contains a pair of His residues separated by a hydrophobic amino acid. Hickman et al. (21) have shown that the nuclease domain of the Rep protein of adeno-associated virus (AAV) can specifically interact both with a dsDNA Rep binding site, and with a hairpin structure that it cuts with sequence specificity. AAV Rep, however, interacts with the two forms of DNA with different faces of the protein. In contrast, TraI36 appears to bind DNA in two distinct ways, but the DNA apparently binds to the same or overlapping sites.
The fast and slow dissociating conformations of WT Hairpin and their different binding characteristics may partially explain the poor fits of a two-state model to the hairpin titration data. For WT Hairpin, there are two possible high-affinity binding interactions, but the different binding modes may cause different effects on fluorescence intensity and anisotropy. These factors, combined with the kinetics of association and dissociation could be affecting the curves. Binding of 4 nM WT 17mer and 40 nM TraI36 reaches equilibrium within 2 min, while the fluorescence intensity is still increasing, slowly but significantly, at 3 h in similar reactions using WT or G144′C Hairpin (data not shown). The binding model used in the fits assumes that the binding is in equilibrium at every step of the titration, and using this model with nonequilibrium data will certainly lead to inaccurate fits. Extending the equilibration time to ≥5 h at each step through the transition, however, did not yield data that produced significantly improved fits (data not shown). Longer equilibration times are impractical because signal drift increasingly affects the data.
Another potential complicating factor is that the hairpin oligonucleotides are capable of dimerizing. When binding an oligonucleotide dimer, TraI36 could cause a greater increase in both fluorescence intensity and anisotropy than if binding a monomer. The result would be an increased slope through the transition. The contribution of oligonucleotide dimers to the binding curves is probably small, though. Heating and snap cooling the labeled oligonucleotide prior to collecting titration data did not alter the results. Furthermore, evidence from nondenaturing gels (S. L. Williams and J. F. Schildbach, unpublished data) and from analytical ultracentrifugation (Chris Larkin, S. L. Williams and J. F. Schildbach, unpublished data) indicates that the large majority of the hairpin oligonucleotides is in a single conformation, presumably monomeric. This is not surprising given that even at the relatively high concentration used for the centrifugation experiments, hairpin formation should be greatly favored over dimerization because of the higher local concentration of the binding partners.
Finally, the protein may bind cooperatively to the oligonucleotide. The average protein:DNA stoichiometry, however, is 1.4:1 as experimentally determined using electrophoretic mobility shift assays (S.L. Williams and J.F. Schildbach, unpublished data). In addition, analytical ultracentrifugation experiments demonstrated that the protein alone is monomeric at high concentrations (7), and there is no evidence of oligomerization upon binding (Chris Larkin, S. L. Williams and J. F. Schildbach, unpublished data).
We still need to elucidate the biological implications of the two binding modes. The G144′C substitution reduces plasmid mobilization when introduced into an F oriT, and it apparently eliminates the fast dissociation conformation when introduced into the 30-base hairpin oligonucleotide. The G151′C/C150′G substitution reduces plasmid mobilization, and apparently predominantly affects the slow dissociation conformation of the hairpin. We do not know, however, what these binding conformations are in a structural sense, and when or if in transfer they occur. Work by Deonier and coworkers (14,22) demonstrated that initiation and termination of transfer have different oriT sequence requirements, and it is plausible that the biological function of TraI might require the protein to recognize different forms of DNA. The TraI protein interacts with the oriT sequence around nic prior to cleavage, as a stable intermediate after cleavage but prior to transfer, and after transfer as it ligates the ends of the plasmid. It is not difficult to imagine that the transfer strand around nic might be in a partial duplex prior to cleavage, and that the region 5′ to nic would be single stranded or in a hairpin immediately following transfer into the recipient. If so, TraI would be required to interact with these different DNA conformations. It is not surprising that the DNA recognition at these different stages could tolerate or even require different levels of sequence specificity.
Our results have definite implications for structural studies of relaxases. We assume that our TraI36(Y16F):ssDNA structure reflects the structure of the relaxase:ssDNA complex prior to ssDNA cleavage (13). Details of the structure match well with our biochemical data. Furthermore, as shown here and previously (12), DNA sequence substitutions that reduce in vitro binding correlate reasonably well with reduced mobilization efficiencies for plasmids containing those substitutions. Coll and coworkers (15) propose that the structure of the relaxase domain of R388 TrwC with bound ssDNA in the hairpin conformation reflects the structure of a relaxase:ssDNA complex after ssDNA cleavage. Certainly the high affinity of DNA for the hairpin construct and the complex series of intermolecular interactions present in the structure are consistent with the structure reflecting the state of TrwC at some step in transfer. The results we present here, however, underscore the complexity of relaxases and their DNA binding activities, and suggest caution is warranted when extrapolating from the structure to models of in vivo function.
Acknowledgments
We thank Christine DeGennaro for cloning the F oriT, Chris Larkin and Michalis Hadjithomas for assistance in analytical centrifugation data collection, Professors Ludwig Brand, Robert Schleif and Van Moudrianakis for use of equipment, and Dr Dima Toptygin for assistance with SPECTRABIND. We thank Chris Larkin, Professor Brand, Professor Beth Traxler, Professor Fernando de la Cruz and Dr Toptygin for helpful discussions and suggestions. This manuscript is based on work supported by National Institutes of Health grant GM61017 to J.F.S. Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health grant GM61017 to J.F.S.
Conflict of interest statement. None declared.
REFERENCES
- 1.Matson S.W., Morton B.S. Escherichia coli DNA helicase I catalyzes a site- and strand-specific nicking reaction at the F plasmid oriT. J. Biol. Chem. 1991;266:16232–16237. [PubMed] [Google Scholar]
- 2.Reygers U., Wessel R., Muller H., Hoffmann-Berling H. Endonuclease activity of Escherichia coli DNA helicase I directed against the transfer origin of the F factor. EMBO J. 1991;10:2689–2694. doi: 10.1002/j.1460-2075.1991.tb07812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pansegrau W., Balzer D., Kruft V., Lurz R., Lanka E. In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc. Natl Acad. Sci. USA. 1990;87:6555–6559. doi: 10.1073/pnas.87.17.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pansegrau W., Ziegelin G., Lanka E. Covalent association of the traI gene product of plasmid RP4 with the 5′-terminal nucleotide at the relaxation nick site. J. Biol. Chem. 1990;265:10637–10644. [PubMed] [Google Scholar]
- 5.Pansegrau W., Schroder W., Lanka E. Relaxase (TraI) of IncP alpha plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc. Natl Acad. Sci. USA. 1993;90:2925–2929. doi: 10.1073/pnas.90.7.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grandoso G., Avila P., Cayon A., Hernando M.A., Llosa M., de la Cruz F. Two active-site tyrosyl residues of protein TrwC act sequentially at the origin of transfer during plasmid R388 conjugation. J. Mol. Biol. 2000;295:1163–1172. doi: 10.1006/jmbi.1999.3425. [DOI] [PubMed] [Google Scholar]
- 7.Street L.M., Harley M.J., Stern J.C., Larkin C., Williams S.L., Miller D.L., Dohm J.A., Rodgers M.E., Schildbach J.F. Subdomain organization and catalytic residues of the F factor TraI relaxase domain. Biochim. Biophys. Acta. 2003;1646:86–99. doi: 10.1016/s1570-9639(02)00553-8. [DOI] [PubMed] [Google Scholar]
- 8.Matson S.W., Nelson W.C., Morton B.S. Characterization of the reaction product of the oriT nicking reaction catalyzed by Escherichia coli DNA helicase I. J. Bacteriol. 1993;175:2599–2606. doi: 10.1128/jb.175.9.2599-2606.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo Z.Q., Isberg R.R. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl Acad. Sci. USA. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Draper O., Cesar C.E., Machon C., de la Cruz F., Llosa M. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc. Natl Acad. Sci. USA. 2005;102:16385–16390. doi: 10.1073/pnas.0506081102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frost L.S., Ippen-Ihler K., Skurray R.A. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stern J.C., Schildbach J.F. DNA recognition by F Factor TraI36: Highly sequence-specific binding of single-stranded DNA. Biochemistry. 2001;40:11586–11595. doi: 10.1021/bi010877q. [DOI] [PubMed] [Google Scholar]
- 13.Larkin C., Datta S., Harley M.J., Anderson B.J., Ebie A., Hargreaves V., Schildbach J.F. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure (Camb) 2005;13:1533–1544. doi: 10.1016/j.str.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Luo Y., Gao Q., Deonier R.C. Boundaries of the nicking region for the F plasmid transfer origin, oriT. Mol. Microbiol. 1995;15:829–837. doi: 10.1111/j.1365-2958.1995.tb02353.x. [DOI] [PubMed] [Google Scholar]
- 15.Guasch A., Lucas M., Moncalian G., Cabezas M., Perez-Luque R., Gomis-Ruth F.X., de la Cruz F., Coll M. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nature Struct. Biol. 2003;10:1002–1010. doi: 10.1038/nsb1017. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J., Fritsch E.F., Maniatis F. Molecular Cloning: A Laboratory Manual 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Thompson R., Taylor L., Kelly K., Everett R., Willetts N. The F plasmid origin of transfer: DNA sequence of wild-type and mutant origins and location of origin-specific nicks. EMBO J. 1984;3:1175–1180. doi: 10.1002/j.1460-2075.1984.tb01947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stern J.C., Anderson B.J., Owens T.J., Schildbach J.F. Energetics of the sequence-specific binding of single-stranded DNA by the F factor relaxase domain. J. Biol. Chem. 2004;279:29155–29159. doi: 10.1074/jbc.M402965200. [DOI] [PubMed] [Google Scholar]
- 19.Ilyina T.V., Koonin E.V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin E.V., Ilyina T.V. Computer-assisted dissection of rolling circle DNA replication. BioSystems. 1993;30:241–268. doi: 10.1016/0303-2647(93)90074-m. [DOI] [PubMed] [Google Scholar]
- 21.Hickman A.B., Ronning D.R., Perez Z.N., Kotin R.M., Dyda F. The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol. Cell. 2004;13:403–414. doi: 10.1016/s1097-2765(04)00023-1. [DOI] [PubMed] [Google Scholar]
- 22.Gao Q., Luo Y., Deonier R.C. Initiation and termination of DNA transfer at F plasmid oriT. Mol. Microbiol. 1994;11:449–458. doi: 10.1111/j.1365-2958.1994.tb00326.x. [DOI] [PubMed] [Google Scholar]
- 23.Toptygin D., Brand L. Analysis of equilibrium binding data obtained by linear-response spectroscopic techniques. Anal. Biochem. 1995;224:330–338. doi: 10.1006/abio.1995.1048. [DOI] [PubMed] [Google Scholar]
- 24.Toptygin D., Brand L. Spectrabind User's Guide. Baltimore, MD: The Johns Hopkins University; 1995. [Google Scholar]