Abstract
The posttranscriptional regulatory element (PRE) is considered to enhance hepatitis B virus (HBV) gene expression by facilitating the nuclear export of intronless viral subgenomic RNAs. Its role in the RNA metabolism of the viral pregenomic RNA (pgRNA) is currently unknown. We identified a positively cis-acting splicing regulatory element (SRE-1) and present two lines of evidence for its functionality. Firstly, in a heterologous context SRE-1 functionally substitutes for a retroviral bidirectional exonic splicing enhancer (ESE). As expected, SRE-1 is a splicing enhancer also in its natural viral sequence context, since deletion of SRE-1 reduces splicing of pgRNA in cell culture experiments. Secondly, we show that stimulation of HBV RNA splicing by the splicing factor PSF was repressed by the PRE. Analysis of a variety of PSF mutants indicated that RNA-binding and protein–protein interaction were required to enhance splicing. In addition, we show that the PRE contributed to pgRNA stability, but has little influence on its nuclear export. Herein, we report for the first time that the PRE harbors splicing stimulating and inhibiting regulatory elements controlling processing of the viral pregenome. We discuss a model in which the regulation of pgRNA splicing depends on cellular factors interacting with the PRE.
INTRODUCTION
RNA metabolism largely depends on the formation of ribonucleoprotein particle arbitrating RNA processing steps like splicing, nuclear export and the regulation of mRNA stability [reviewed in (1–3)]. Precise knowledge of human hepatitis B virus (HBV) gene expression requires the analysis of its RNA metabolism. This knowledge is supposed to be useful for the development of novel antiviral strategies based on the disturbance of accurate viral RNA processing. The HBV is a noncytopathic, hepatotropic virus with a 3.2 kb circular partial-double stranded DNA genome which serves as template for transcription of the pregenomic/precore mRNA and subgenomic RNAs [reviewed in (4–6)]. Although there are several promoters, all viral RNAs terminate at a common polyadenylation signal. HBV is distantly related to retroviruses, contains four open reading frames (ORFs)—core (C), reverse transcriptase-polymerase (P), surface (S) and X—which are all encoded on the positive strand. Subgenomic preS and S RNAs encode three viral surface proteins, all expressed from a single ORF, whereas the small 0.7 kb RNA encodes the X-protein. In addition to its function as template for reverse transcription (RT) into genomic minus strand DNA, the bicistronic pregenomic RNA (pgRNA) serves as mRNA for translation of the core and P-proteins. The precore mRNA initiates upstream of the pgRNA and the precore-region serves as the translation template for synthesis of hepatitis B e-antigen. The dual function of the pgRNA as pregenome and mRNA for more than one protein argues for accurate processing and stability of viral RNAs being of paramount importance for the hepadnaviral life cycle.
In contrast to the related duck HBV where splicing is important for infection and expression of the large surface protein (7) constitutive splicing of HBV is not required to produce viral proteins. However, splice variants of the HBV pgRNA have been detected in sera and in liver samples of infected patients as well as from transfected hepatoma cells (8–12), indicating that there is yet not recognized function of pgRNA splicing in HBV infection. In addition, it has been shown recently by RT–PCR analysis that preS/S mRNAs are spliced utilizing the 5′splice site (5′ss) at position 458 and the 3′splice site (3′ss) at position 1305 (13). Besides a variety of spliced genomes the major two splice products (SP) of the pgRNA are referred to as SP1 and SP2 (8,10). SP1 is generated by removing the intron flanked by the 5′ss at position 2448 and 3′ss at position 489, whereas SP2 is generated by utilizing an alternative 5′ss at position 2067 but the same 3′ss at position 489. SP1 is easily detectable by northern blotting and runs close to the S subgenomic RNA (about 2.1 kb) encoding the small surface protein. Earlier it was documented that about 30% of the pgRNA was spliced (14), raising the important question what are the functions of the fusion and aberrant proteins that are translated from the spliced viral transcripts (15–17). Furthermore, the question has to be addressed how HBV pgRNA splicing is regulated and limited to warrant expression of functional HBV proteins.
During a search for cis-regulatory elements within the HBV genome a genetic element was identified referred to as posttranscriptional regulatory element (PRE) (18–20). The PRE [nt 1217–1582, (21,22)] is located in the untranslated region of preS/S subgenomic RNAs, but in the polymerase and X-protein ORF of the pgRNA and X mRNA, respectively and facilitated the nucleocytoplasmic transport of unspliced preS/S subgenomic RNAs (18–20). Prediction of potential 5′ss and 3′ss within the preS/S subgenomic RNAs suggested that the PRE might also play a role in inhibiting preS/S RNA splicing (18). The PRE mediates the nuclear export of unspliced viral RNAs and it was concluded that ΔPRE-subgenomic RNAs are retained in the nucleus where they are subsequently degraded (18–20). The PRE is able to functionally substitute for the HIV-1 Rev Responsible Element and it was suggested that a CRM1-dependent nuclear export pathway (23) exports PRE-containing transcripts. However, a PRE-dependent increase in gene expression was not inhibited by Leptomycin B, an inhibitor of the CRM1-dependent export pathway or by over expression of ΔCAN, which competes with nucleoporins for binding to CRM1 (24–26). Recently, binding of the polypyrimidine tract-binding (PTB) protein to the 3′ end of the PRE was demonstrated and PTB was suspected to be involved in PRE-mediated nuclear export of unspliced subgenomic RNAs (27).
The PRE is located about 750 nt downstream of the 3′ss (nt 489) known to be utilized intensively in splicing of pgRNA and therefore has in this case an exonic position. In comparison to the HBV PRE (HPRE) the related woodchuck hepatitis virus PRE (WPRE) is even more potent to posttranscriptionally facilitate gene expression especially of heterologous transgenes. The functional difference between HPRE and WPRE probably depends on an additional sequence element referred to as WPREγ domain, which is absent in the HPRE (28). In addition, both PREs contain subelements referred to as PREα and PREβ domains, which are required for full activity (21,22). Interestingly, WPRE-dependent stimulation of gene expression could be inhibited partially by Leptomycin B or over expression of ΔCAN, indicating that the WPRE utilizes other and/or additional mechanism than the HPRE to boost posttranscriptionally gene expression (25). It was suggested that the WPRE enhances gene expression at different posttranscriptional levels like 3′ end processing, splicing and export and that all three subelements are required for full activity (25,29,30). Similar conclusions were made for the HPRE (31–33). Hence, comparison of the properties of HPRE and WPRE reveals significant functional, structural as well as mechanistically similarities but also distinct differences.
The current study was conducted to evaluate the function of the HPRE in its natural sequence context of the viral pgRNA. We discovered that the HPRE regulates HBV pgRNA splicing due to its positively cis-acting splicing regulatory element (SRE-1). Accordingly, deletion of the SRE-1 reduced splicing of pgRNA. We show a moderate additional role of the HPRE in stabilization of the pgRNA, and its dispensability for nuclear export of pgRNA. Furthermore, we report that the splicing factor PSF stimulates pgRNA splicing in an HPRE-dependent manner underscoring the splicing-regulatory function of the HPRE. Collectively, we report that the HPRE harbors an exonic splicing enhancer (ESE) required for pgRNA splicing. We discuss a model in which the HPRE regulates splicing of viral RNAs.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides were synthesized and purified as described previously (34). The oligonucleotides are as follows: 183: 5′-ATCCTGCAGGGCTTTAGGCTTTGATCCC; 1813: 5′-CAACAGAAATCCAACCTAGAGCTGCT; 1815: 5′-TCTTCCAGCCTCCCATCAGCGTTTGG; 1954: 5′-ATCGAATTCGCTTGTTTTGCTCGCAGCAGGTCTGGAGCAAACATTATCGGGACTG; 1955: 5′-ATCGAGCTCGGACAACAGAGTTATCAGTCCCGATAA; 2068: 5′-GATGAATTCTCCATACTGCGGAACTCCTAGCCGCTTGTTTTGCTCGC; 2213: 5′-ATCGAATTCGCTCCTCTGCCGATCCATACTGCGGAACT; 2217: 5′-TAAGTCTCTCAAGCGGTGGT; 2244: 5′-TAATACGACTCACTATAGGGCCT. The following oligonucleotides were commercially synthesized (Invitrogen): antisense primer HBV96-AS (5′-GAGATTCCCGAGATTGAGATCTTCTGC-3′; nt 2420–2447); sense primer HBV97-S (5′-TCCTCAACAACCAGCACGGGACC-3′; nt 493–515); antisense primer HBV64-AS (5′-CGAAAAGGTTCCACGCATG-3′; nt 1235–1253); sense primer HBV69-S (5′-GCACTTCGCTTCACCTCTGCACG-3′; nt 1583–1604); sense primer HBV63-S (5′-GCTTGTTTTGCTCGCAGC-3′; nt 1290–1307); HBV7-T7AS (5′-ccatcgattaatacgactcactatagGGTTGACATACTTTCCAAT-3′, HBV nt 996–979 in upper case letters); HBV1A (5′-TCATCCTCAGGCC-3′; HBV nt 3161–3173 in upper case letters); HBV44-T7AS (5′-ccatcgattaatacgactcactatagGGTGTAAATAGTGTCTAGTTTGG-3′; HBV nt 2734–2755 upper case letters); HBV43-S (5′-TCATCCTCAGGCC-3′; HBV nt 2021–2041); H2A-AS (5′-ggatcctaatacgactcactataggGAACATTGAGATTTCAGGC-3′, H2A nt 829–847 in upper case letters); H2A-S (5′-CAGCAGTGAGAATGAACGC-3′, H2A nt 19–37); GFP-AS (5′-ggatcctaatacgactcactataggGTCCATGCCGAGAGTGATCCC-3′); GFP-S (5′-CCTGGTCGAGCTGGACGGC-3′); glycerinaldehyde-3-phosphate dehydrogenase (GAPDH) intron-AS (5′-ggactagttaatacgactcactataGGGTGCGGTGGAGATCTG-3′); GAPDH intron-S (5′-CAAGGAGAGCTCAAGGTC-3′). T7 RNA polymerase promoter sequences are shown in boldface.
Plasmid constructs and mutagenesis
The HBV expression plasmid pCH-9/3091 (kind gift from H. Schaller ZMBH, Heidelberg) contains a more than full-length HBV genome (subtype ayw) in which synthesis of the pgRNA is under control of a cytomegalovirus (CMV) promoter. Different nucleotide deletions were introduced into the plasmids pCH-9/3091 and ptetHBV [kindly provided by Dr Christoph Seeger, (35)] according to site directed mutagenesis method (Stratagene, USA) using proofreading Pwo DNA Polymerase (Roche, Germany). To introduce the different nucleotide deletions the following oligonucleotides were used: for mutant pHBV-ΔPRE and ptetHBV-ΔPRE HBV64-AS and HBV69-S; for mutant pHBV-Δintron and pHBV-ΔintronΔPRE HBV96AS and HBV97S, for mutant pHBV-ΔSRE HBV64-AS and HBV63-S; PCR products were purified with the QIAquick PCR purification Kit (Qiagen, Germany), phosphorylated at the 5′ end with 10 U T4 polynucleotide kinase (Roche) subsequently ethanol precipitated and resuspended in an appropriate volume of H2O. PCR products were ligated with the Rapid DNA Ligation Kit (Roche) following the manufacturer's instructions. Finally, the ligated DNA was digested with 10 U DpnI (New England Biolabs, USA) for 1 h at 37°C with appropriate buffers, transformed into Escherichia coli strain DH5α. Correct mutagenesis was confirmed by sequencing using the Taq-cycle sequencing protocol with IRD-800 labeled primers (MWG Biotech, Ebersberg, Germany) and a Licor automated sequencing device (MWG Biotech). Cloning of the HIV-1 subgenomic env expression vectors SV-env/GAR and SV-env/HIV#18 (used as a control) have been described recently (36). The bidirectional SF2/ASF- and SRp40-dependent splicing enhancer regulates human immunodeficiency virus type 1 rev, env, vpu and nef gene expression (36). For the construction of SV-env/SRE-3 (HBV nt 1290–1350) the GAR ESE flanked by an EcoRI and SacI restriction site was substituted for the EcoRI–SacI adaptor #1954/#1955. For SV-env/SRE-2 (HBV nt 1267–1350) the GAR ESE was substituted with a PCR amplified fragment using primer pair #2068/#183 and SV-env/SRE-2 as template. For SV-env/SRE-1 (HBV nt 1254–1350) the GAR ESE was substituted with a PCR amplified fragment using primer pair #2213/#183 and SV-env/SRE-2 as template. Cloning of pT7cytCoxII used as a probe to detect human mitochondrial cytochrome c oxidase II (GenBank accession no. NC 001807) has been described previously (37). All sequences either inserted by oligonucleotide adaptors or PCR were confirmed by DNA sequencing.
Cell culture and transfection procedure
Huh-7 human hepatoma cells or HeLa-T4+ cells (38) were grown as monolayers in DMEM supplemented with 10% fetal calf serum (FCS). All plasmids used for transfection were purified by ion exchange chromatography using the Maxiprep kit (Qiagen). Transfection was performed using FuGENE™ 6 Transfection Reagent (Roche) according to the manufacturer's protocol.
In vitro transcription for northern blot analysis
Plasmid pCH-9/3091 was used for the production of DNA templates for the generation of HBV antisense transcripts (probe 1 and probe 2, Figure 1A). The following two primers were used to generate the template for probe 1 by PCR: the antisense primer HBV7-T7AS and the sense primer HBV1A and for probe 2 the antisense primer HBV44-T7AS and the sense primer HBV43-S. cytochrome c oxidase II-specific antisense RNA-probe was generated by in vitro transcription with T7 RNA polymerase (Promega, USA) using the EcoRI linearized plasmid pT7cytCoxII (37). Plasmid pWA 175 (containing the Histone H2A/a gene, kindly provided by W. Albig, Göttingen, Germany) was used for the production of DNA templates for the generation of histone 2A antisense transcripts. Two primers were used to generate the template for the H2A probe by PCR: the antisense primer H2A-AS and the sense primer H2A-S. Plasmid pEGFP-N1 was used for the production of DNA templates for the generation of GFP antisense transcripts. Two primers were used to generate the template for the GFP probe by PCR: the antisense primer GFP-AS and the sense primer GFP-S. DNA template used for the in vitro transcription to generate antisense RNA probes specific for the GAPDH intron B was raised by PCR on genomic DNA using antisense primer GAPDH intron-AS and the sense primer GAPDH intron-S. PCRs for the generation of the in vitro transcription templates were performed with the mixture containing 80 pmol of each primer in 1× PCR buffer, 0.2 mM of GTP, ATP, TTP and CTP, and 2.5 U of Taq DNA polymerase (Roche). The PCR products were purified by size exclusion using Microspin G-25 columns (Amersham Bioscience, Germany) according to the manufacturer's protocol, ethanol precipitated and used as templates for in vitro transcription. Transcription reactions were carried out with 0.5–1.0 µg linearized plasmid or PCR product in a final volume of 20 µl in transcription buffer (Promega) containing 0.31 mM ATP, CTP, GTP, 0.25 µM UTP and 5.0 µM [α-32P]UTP (800 Ci/mmol) (Hartmann Analytic, Germany), 5 mM DTT and 20 U RNasin (Promega). The reaction was started by addition of 20 U T7 RNA polymerase (Promega). After incubation for 45 min at 37°C, another 20 U of T7 RNA polymerase were added, and the reaction was continued for 45 min at 37°C. The reaction was terminated by adding 10 µg of yeast tRNA and 1 U of DNase I (Promega) and incubated for 15 min at 37°C. Unincorporated nucleotides were removed by using Microspin G-25 columns.
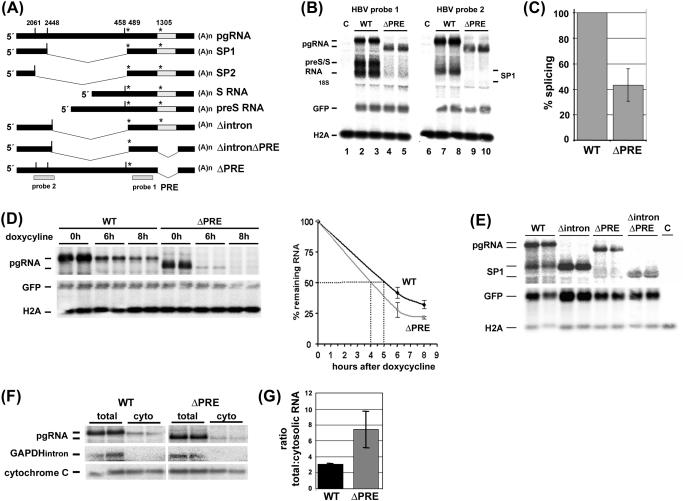
Figure 1.
The PRE is required for HBV pgRNA splicing. (A) Schematic drawing of pgRNA, the major spliced RNAs (SP1 and SP2), the subgenomic preS/S RNAs and the pgRNA mutants Δintron, ΔintronΔPRE and ΔPRE. The HBV probes to detect pgRNA and subgenomic RNAs (probe 1), pgRNA and SP1 RNA species (probe 2) are depicted. Grey box = PRE; vertical dash, 5′ splice donor; *, 3′ splice acceptor. (B) Deletion of the PRE reduces the levels of the major splice product SP1 and of preS/S mRNA. HuH-7 cells were cotransfected with each 1.5 µg of plasmid tetHBV-Wild-type (WT) or tetHBV-ΔPRE (ΔPRE, deletion of nt 1254–1582), 0.5 µg of the transactivator expressing plasmid pUHD-TA and 0.07 µg pEGFP-N1 to monitor transfection efficiency. Forty-eight hours thereafter RNA was prepared and 10 µg total RNA was analyzed by northern blotting. The pgRNA and the subgenomic HBV RNAs (preS and S RNA) were detected using HBV probe 1. A second aliquot of RNA from the same preparation was loaded on to a second gel and analyzed by northern blotting using HBV probe 2 detecting pgRNA and the major spliced RNA (SP1). To normalize RNA loading and transfection efficiency the same blots were hybridized with histone H2A (H2A) and GFP mRNA (GFP) specific probes. M, total RNA prepared from untransfected cells. (C) Deletion of the PRE reduces splicing. Six independent experiments performed in duplicates were quantified by phosphoimaging and HBV RNA levels were normalized against RNA loading and transfection efficiency. The percentage of splicing reflects the ratio between the signal intensity of the SP1 RNA and the pgRNA. The HBV wild-type SP1: pgRNA ratio was set as 100%. The SD is indicated (± SD; n = 6). (D) The PRE contributes to pgRNA stability. HuH7 cells were cotransfected with each 1.5 µg of plasmid tetHBV-WT (WT) or tetHBV-ΔPRE (ΔPRE), 0.5 µg of the transactivator expressing plasmid pUHD-TA and 0.07 µg pEGFP-N1 to monitor transfection efficiency. Forty-hours thereafter, cells were harvested (time point 0 h) or doxycycline was added and cells were harvested at 6 or 8 h thereafter. RNA was prepared and 10 µg total RNA was analyzed by northern blotting using HBV probe 2 to detected pgRNA. To normalize RNA loading and transfection efficiency the same blots were hybridized with a histone H2A (H2A) and a GFP mRNA (GFP) specific probe. Two independent experiments performed in duplicates were analyzed, quantified by phosphorimaging and HBV RNA levels were normalized against RNA loading and transfection efficiency. The SDs are indicated (±; n = 2). (E) ΔintronΔPRE pgRNA is stably expressed. HuH7 cells were cotransfected with each 1.5 µg of plasmid tetHBV-WT (WT), tetHBV-Δintron (Δintron, deletion of nt 2446–2496), tetHBV-ΔPRE (ΔPRE; deletion of nt 1254–1582), tetHBV-ΔintronΔPRE (ΔintronΔPRE, deletion of nt 2446–2496 and of nt 1254–1582), 0.5 µg of the transactivator expressing plasmid pUHD-TA and 0.07 µg pEGFP-N1 to monitor transfection efficiency. Forty-eight hours thereafter RNA was prepared and 10 µg total RNA was analyzed by northern blotting using HBV probe 2 to detected pgRNA, GFP and H2A probes to confirm transfection efficiency and RNA loading, respectively. C, mock transfection. (F) Cytosolic accumulation of pgRNA does not depend on the PRE. HuH7 cells were cotransfected in duplicates with each 1.5 µg of plasmid tetHBV-WT (WT) or tetHBV-ΔPRE (ΔPRE), 0.5 µg of the transactivator expressing plasmid pUHD-TA and 0.07 µg pEGFP-N1 to monitor transfection efficiency. Forty hours thereafter, cells were harvested and total RNA (total) and cytosolic RNA (cyto) was prepared as described in Materials and Methods. RNAs were analyzed by northern blotting using HBV probe 2 to detected HBV pgRNAs. Contamination of the cytosolic RNA fraction with pre-mRNAs was monitored by reprobing the membrane with a probe directed against a GAPDH intron (GAPDHintron) and the quality of the cytosolic RNA preparation was monitored by detection of the cytochrome c Oxidase II mRNA. RNAs of two independent transfections were loaded and analyzed. One representative northern blot is shown. For quantification, the northern blot membranes were reprobed with a Histone H2A probe (data not shown) and used for normalization of RNA loading. Each transfection was quantified independently, the mean was calculated and the SD is indicated (±, n = 2). (G) Ratio between total and cytosolic WT and ΔPRE pgRNA was calculated and blotted.
Northern blot analysis
Huh-7 cells were harvested for 48 h or as indicated after transfection. Total RNA was prepared using TriPure Isolation Reagent (Roche) according to the manufacturer's protocol. For preparation of cytosolic RNA transfected Huh-7 cells were washed twice with ice cold phosphate-buffered saline (PBS), harvested in lysis buffer [10 mM Tris–HCl (pH 8.0), 10 mM KCl, 10 mM MgCl2 and 0.2% NP40] supplemented with 200 U of RNasin/1 ml lysis buffer. The slurry was transferred to 2 ml tubes and placed on ice for 10 min, slurry was pipetted up and down to support cell lysis and cell lysis was controlled by light microscopy. Nuclei and cell debris were pelleted by centrifugation (3 min 1000 g at 4°C) and supernatants were centrifuged again (3 min 1000 g at 4°C). Final supernatants were transferred to new 2 ml tubes and cytosolic RNA was extracted using TriPure Isolation Reagent (Roche) according to the manufacturer's protocol. RNA was separated on a 1.2% agarose–formaldehyde gel. RNA was blotted on to a nylon membrane (Osmonics, USA) and hybridized with 32P-labeled in vitro transcribed RNA probes overnight at 68°C. To standardize transfection efficiency and RNA loading, northern blots were hybridized with 32P-labeled in vitro transcribed antisense GFP probe and antisense GAPDH or antisense histone 2A probes, respectively. Blots were exposed to Fuji imaging screens, and signals were quantified by Fujix BAS 2000 bio-imaging analyzer (Fuji, Japan) and by TINA software (Raytest, Germany).
Determination of pregenomic HBV-RNA half-life using the Tet-Off-System
Huh-7 cells were transfected with 1.5 µg ptetHBV-WT or ptetHBV-ΔPRE and 0.5 µg pUHD-TA, expressing tTA, the tetracycline-controlled transactivator. Synthesis of pgRNA was blocked by addition of 500 ng/ml doxycycline to the culture medium 40 h after transfection. Cells were harvested directly upon addition of doxycycline or 6 and 8 h thereafter. Total RNA was prepared and analyzed by northern blot analysis.
RT–PCR assay
A total of 2.5 × 105 HeLa-T4+ cells were seeded into wells of a 6-well plate (Greiner) and grown as monolayers in 2.5 ml DMEM supplemented with 10% FCS for 24 h. For RT–PCR, cells were transfected with 1.5 g of the respective SV-env plasmid and 0.5 µg of pXGH5 (39) to control transfection efficiency, and total RNA was isolated after 30 h. Total RNA was isolated using a modified guanidinium isothiocyanate protocol (40). Cells were washed twice with 2 ml of PBS and cell lysis was performed with 500 µl of buffer D [4 M guanidinium isothiocyanate, 25 mM sodium citrate (pH 7.0), 0.5% N-lauroyl-sarkosine, sodium salt and 0.4% dextran blue]. The following solutions are added with vortexing in between: 7.2 µl of 2-mercaptoethanol, 50 µl of 2 M sodium acetate (pH 4), 500 µl of phenol and 100 µl of a chloroform-isoamyl alcohol mixture (24:1). After addition of the latter, the mixture was vigorously shaken for 15 s, placed on ice for 15 min and phases were separated by centrifugation (10 600 g) for 20 min at 4°C. RNA was precipitated in 1 vol of isopropanol overnight. After centrifugation (10 600 g) RNA was washed twice with 70% ethanol and dissolved in 16 µl of DMDC-ddH2O. Before RT, 8 µl of RNA samples were subjected to DNase I digestion using 10 U DNase I (Roche) with 50 mM Tris (pH 7.5) and 10 mM MgCl2 in a total volume of 10 µl at room temperature for 1 h. After DNase I inactivation at 80°C for 10 min, 4.5 µl of the DNase digested RNA samples were reversed transcribed with 200 U SuperScript III RNase H-Reverse Transcriptase (Invitrogen) according to the manufacturer's protocol using 0.375 mM oligo(dT)15 (Roche) as primer. PCR was carried out with 1.25 U AmpliTaq (Applied Biosystems) in a total volume of 50 µl according to the manufacturer's protocol in a Robocycler Gradient 96 Temperature Cycler (Stratagene, USA). All primers were used at a final concentration of 0.2 µM. Spliced RNA was detected with primer pair #2244/#2217. PCR products were separated on 3% Metaphor-agarose gels and stained with ethidium bromide. For the detection of hGH mRNA in the RT–PCR assay primer pair #1813/#1815 was used.
Western blot analysis
A total of 3.0 × 105 HeLa-T4+ cells were seeded into wells of a 6-well plate (Greiner, Germany) and grown as monolayers in 2.5 ml DMEM supplemented with 10% FCS for 24 h. Cells were cotransfected with 1.0 µg of the respective SV-env plasmid, 1 µg of SVcrev and 1.0 µg pGL3 (Promega) to monitor transfection efficiency. The medium was changed 24 h after transfection and cells were scraped into the medium 24 h later, sedimented at 12 000 g for 14 s, washed in 1 ml PBS and suspended in 0.5 ml PBS. An aliquot of 70 µl was analyzed for luciferase activity (luciferase assay system, Promega) and protein concentration (Bradford Protein Assay, Bio-Rad). The remaining volume of the second PBS washing step was centrifuged and suspended in 200 µl of SDS–PAGE sample buffer (41). Each sample was adjusted to equal amounts of luciferase activity and protein amount by adding extracts of mock-transfected cells. Samples were heated for 10 min at 95°C and subjected to electrophoresis on a 7% SDS–polyacrylamide gel and transferred to a PVDF-membrane (Immobilon™ P, pore size 0.45 µm; Millipore) by electroblotting with 500 mA in transfer-buffer (200 mM glycine, 25 mM Tris base and 20% methanol) for 1 h. Blots were blocked in PBS with 10% skim milk powder (Oxoid), 0.1% Tween-20, for 15 min at RT. Protein detection was performed in PBS, 5% skim milk powder, 0.1% Tween-20, with a monoclonal mouse-anti-gp120 antibody (87–133/026, 1:5000, kindly provided by Dade Behring) for 1 h, washed four times with PBS, incubated in PBS, 10% skim milk powder, 0.1% Tween-20 with a sheep-anti-mouse antibody conjugated with horseradish peroxidase (HRP) (NA 931, Amersham Bioscience, 1:1000) for 1 h, washed five times with PBS, rinsed with water and visualized by a chemiluminescence detection system (ECL™-system and ECL™ hyperfilm, Amersham Bioscience; Super Signal® ultra, Pierce).
RESULTS
The PRE modulates splicing of HBV pgRNA
So far, no information is available on the role of the PRE in pgRNA processing. To gain insight into the function of the PRE in the posttranscriptional control of pgRNA metabolism we deleted the PRE [Figure 1A; ΔPRE, deletion of nt 1254–1582, Galibert nomenclature, (42)] within the HBV expression plasmid pHBV-wild-type (WT) and ptetHBV-WT and monitored splicing, stability and nuclear export of WT and ΔPRE pgRNA. To study pgRNA processing DNA of WT or mutant HBV expression plasmids were transiently transfected into Huh-7 cells and total RNA was prepared 48 h thereafter. HBV RNA level, extend of splicing and nuclear export was analyzed and quantified as described in Materials and Methods. As shown in Figure 1B, pgRNA transcribed from plasmid ptetHBV-ΔPRE migrates faster than the WT pgRNA (pgRNA, cf. lanes 2, 3 with 4, 5) due to the deletion of the PRE and was expressed at slightly reduced levels. Furthermore, as previously published (18–20) deletion of the PRE abolished expression of subgenomic preS/S RNAs (Figure 1B, cf. lanes 2, 3 with 4, 5). The major splice product of the pgRNA, referred to as SP1, was hardly detected (Figure 1B, SP1, cf. lanes 7, 8 with 9, 10), indicating that deletion of the PRE strongly reduced pgRNA splicing. Quantification of five independent experiments revealed reduction in pgRNA splicing by about 60% (Figure 1C). To also visualize whether the expression of the second prominent pgRNA splice product SP2 was PRE-dependent, RT–PCR analysis was applied. RT–PCR analysis also revealed that splice product SP2 was strongly reduced when the PRE was deleted from the pgRNA (data not shown). Thus, production of SP1 and SP2 spliced pgRNAs depend on the PRE, indicating that the PRE contains element(s) mediating the usage of upstream located splice sites.
Since it was observed that ΔPRE pgRNA levels were slightly reduced when the PRE was deleted (Figure 1B, cf. lanes 2, 3 with 4, 5), we next measured the half-life of WT and ΔPRE pgRNA. To determine and to compare the half-life of WT and ΔPRE pgRNAs, the tetracycline expression system was used and transfection experiments with ptetHBV-WT or ptetHBV-ΔPRE plasmids were accomplished. Huh-7 cells were transfected either with ptetHBV-WT or ptetHBV-ΔPRE plasmid DNA, 40 h thereafter doxycycline was added to block further transcription and RNA was harvested 6 and 8 h afterwards. The half-life of both, WT and ΔPRE pgRNAs were determined in duplicates in two independent experiments. Wild-type pgRNA comprised a half-life of ∼5 h [Figure 1D; (35)] whereas the half-life of ΔPRE-pgRNA was reduced by about 1 h (Figure 1D). We conclude that the PRE to some extent contributes to pgRNA stability.
Since the half-life of the pgRNA was slightly reduced in the absence of the PRE we addressed the question of whether lack of SP1 RNA detection in the ΔPRE context might be due to an altered stability of the spliced SP1-ΔPRE RNA. We thus deleted the intron sequence (referred to as Δintron, nt 2446–2496, Figure 1A and E) and created the double mutant ΔintronΔPRE (Figure 1A and E). Huh-7 cells were transfected either with ptetHBV-WT, ptetHBV-Δintron, ptetHBV-ΔPRE or ptetHBV-ΔintronΔPRE plasmid DNA, 48 h thereafter RNA was harvested. HBV RNA levels were determined in duplicates in two independent transfections experiments and RNA was analyzed by northern blotting. The experiment shows that the ΔintronΔPRE pgRNA is easily detectable although at somewhat lower levels than Δintron pgRNA, indicating that the PRE contributes to the stabilization of WT-pgRNA and SP1 RNA. However, the strong expression of SP1-ΔPRE pgRNA excludes the possibility that SP1 RNA is highly unstable and therefore hardly detectable when the PRE is deleted. This experiment strongly supports the assumptions that the PRE is required for HBV RNA splicing to occur.
Originally, the PRE was identified as a nuclear export element of unspliced subgenomic preS/S RNAs. Therefore, we next asked whether the cytosolic accumulation of pgRNA depended on the PRE. Hence, Huh-7 cells were transfected in duplicates with either pHBV-WT or pHBV-ΔPRE plasmid DNA and total as well as cytosolic RNAs were prepared 48 h later. Fractionation was monitored using a probe detecting an intronic sequence of nuclear pre-GAPDH mRNA which is present in total RNA but absent in the cytosolic RNA fraction (Figure 1F, cf. total with cyto) and using a probe detecting the mitochondrial cytochrome c oxidase II mRNA which is present in both, total RNA and the cytosolic RNA fraction (Figure 1F, cf. total with cyto). The representative northern blot analysis in Figure 1F shows comparable levels of total and cytosolic WT and ΔPRE pgRNAs (Figure 1F). Quantification revealed that the ratio between total and cytosolic WT ΔPRE pgRNA was increased compared to the ratio of total to cytosolic WT pgRNA, indicating a minor contribution of the PRE to RNA instability and/or export (Figure 1G). The experiment also indicates that the nuclear export of unspliced pgRNA did not largely depend on the PRE. Taken together, the characterization of the ΔPRE-pgRNA revealed that the PRE is important for pgRNA splicing and contributes to pgRNA stability. However, PRE is not crucial for nuclear export of viral pgRNA. Data suggest major functional differences of the PRE in the pgRNA and in the preS/S RNA sequence context.
The PRE covers an ESE
The most frequent used 3′ss within the pgRNA is located about 750 nt (nt 489) upstream of the PRE (Figure 1A). We assumed that the PRE regulates splicing of the pgRNA via one or more splicing regulatory regions situated within the PRE recruiting trans-acting factors required for 3′ss definition. The assumption was based on the finding that RNA levels of the splice products SP1 and SP2 (using the same 3′ss) were drastically reduced when the PRE was deleted (see above). For this reason, we considered that the PRE might contain a SRE-1 recognized by SR proteins, a family of splicing regulatory proteins mostly increasing utilization of either 5′ss or 3′ss [reviewed in (43)]. We used the ESEfinder program (44) to predict potential binding sites for the SR proteins SF2/ASF, SC35, SRp55 and SRp40. Analysis of the PRE sequence (nt 1253–1583) led to the prediction of numerous putative SR protein-binding sites within the PRE with one cluster between nt 1253 and 1290 at its 5′ end (Figure 2A).
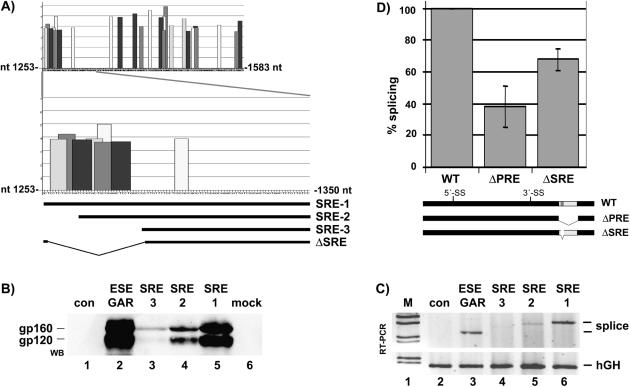
Figure 2.
The PRE contains a splicing enhancer referred to as SRE-1. (A) Prediction of a cluster of SR protein-binding sites at the 5′ end of the PRE. The ESEfinder program (44) predicts potential binding sites for SF2/ASF, SC35, SRp55 and SRp40. The PRE sequence (nt 1253–1583) was analyzed by the ESEfinder and to increase stringency the threshold was set to 3 for all predicted binding sites. The 5′ end of the PRE (nt 1253–1350) containing a cluster of potential SR protein-binding sites is enlarged. SRE-1, SRE-2 and SRE-3 depicts the sequence cloned into the HIV glycoprotein reporter plasmid and analyzed for enhancer activity. ΔSRE illustrate a deletion (nt 1254–1289) introduced into the HBV expression plasmid pHBV-ΔSRE used in Figure 2D. (B–D) The 5′ end of the PRE contains a splicing enhancer. (B) The PRE sequences SRE-1, SRE-2 and SRE-3 were cloned into the HIV glycoprotein reporter plasmid SV-env. HeLa-T4+ cells were co-transfected with different HIV glycoprotein reporter plasmids containing no enhancer (con), the HIV GAR ESE (36) or the SRE-1, SRE-2 or SRE-3 sequences. The HIV Rev protein was co-expressed. Protein extracts were prepared 48 h later and analyzed by western blotting (WB) as described in Materials and Methods. As shown in (B) the SRE-1 element and the GAR ESE increase strongly gp160 and gp120 expression, whereas SRE-2 and SRE-3 were less efficient. (C) HeLa-T4+ cells were cotransfected with different HIV glycoprotein reporter plasmids containing no enhancer (con), the HIV GAR ESE (36) or the SRE-1, SRE-2 or SRE-3 sequences. The HIV Rev protein was not co-expressed. Total RNA was prepared 30 h later and analyzed by RT–PCR as described in Materials and Methods. SRE-1 and the GAR-ESE strongly stimulate splicing (splice) whereas SRE-2 and SRE-3 were less efficient. The human growth hormone gene (hGH) was used as a loading control. M, molecular weight marker. (D) HuH-7 cells were cotransfected with each 1.5 µg of plasmid tetHBV-WT (WT), tetHBV-ΔPRE (ΔPRE), tetHBV-ΔSRE (ΔSRE; nt 1254–1289) and analyzed as described in Figure 1B and Materials and Methods. Three independent experiments performed in duplicates were quantified by phosphoimaging and HBV RNA levels were normalized against RNA loading and transfection efficiency. The percentage of splicing reflects the ratio between the signal intensity of the SP1 RNA and the pgRNA. The HBV wild-type SP1: pgRNA ratio was set as 100%. The SD is indicated (±; n = 3–6). SRE, splicing regulatory element; ESE, exonic splicing enhancer; 5′ss/3′ss, splice donor/splice acceptor site.
To analyze whether this cluster covers a functional ESE we were interested in a heterologous ESE-dependent reporter system that not only allowed verifying ESE-dependent splicing but also additionally ESE-dependent protection for RNA degradation. Such a reporter turned out to be the HIV-1 subgenomic env-reporter, which we have extensively characterized during the past (36,45–47). In the presence of the cis-acting GAR ESE U1 snRNA is recruited to the HIV-1 5′ss (SD4) and protects the RNA from nuclear degradation before splicing. In the presence of the retroviral HIV-1 Rev protein the protected, unspliced message is exported into the cytoplasm and is translated into the envelope glycoprotein. In the absence of Rev U1 snRNA binding initiates spliceosome formation, resulting in removal of the intron and transportation of the spliced transcript into the cytoplasm. Since the HIV-1 envelope glycoprotein mediates syncytium formation when expressed at the surface of e.g. HeLa-T4+ cells, ESE-dependent U1 snRNA binding can even be visualized microscopically by syncytia. Using this HIV-1 env reporter we substituted the GAR ESE for the HBV SRE sequences (SRE-1 nt 1254–1350; SRE-2 nt 1267–1350; SRE-3 nt 1290–1350) and analyzed their activity to support U1 snRNA binding and to enhance splicing.
Firstly, we co-transfected HeLa-T4+ cells with the HIV-1 Rev expression plasmid SVcrev and different gp160-reporter gene constructs either containing a negative control sequence (Figure 2B, con; carrying a fragment of similar length as the GAR ESE but avoid of any ESE), or the bidirectional GAR ESE (Figure 2B, ESE GAR), or HBV SRE-1, SRE-2 or SRE-3 sequence elements (Figure 2B). As demonstrated by western blot analysis, mock-transfected cells (Figure 2B, lane 6) or cells transfected with SV-env/HIV#18 (lane 1) did not allow glycoprotein detection. In contrast, the GAR ESE and the SRE-1 (Figure 2B, lanes 2 and 5) led to a strong increase in gp160/gp120 expression. Gp160/gp120 protein expression was moderately or weakly enhanced with SRE-2 and SRE-3, respectively (Figure 2B, lanes 3 and 4). From this analysis it is apparent that the strongest effect was only seen when the sequence covering the cluster of predicted SR protein-binding sites was included. Shortening of SRE-1 led to a stepwise reduction in gp160/gp120 protein expression (SRE-1 > SRE-2 > SRE-3). Data suggest that SRE-1 mediates efficiently U1 RNA recruitment to the HIV-1 5′ss. The functionality of SRE-1 was further shown by its ability to induce syncytium formation in HeLa-T4+ cells (data not shown). Thus, results imply that the predicted cluster of SR protein-binding sites at the 5′ end of the SRE-1 acts as an ESE.
Next, the same experiment was repeated in the absence of HIV-1 Rev in order to monitor ESE-dependent splicing. Again GAR ESE and the SRE-1 strongly induced splicing whereas SRE-2 and SRE-3 were less efficient (Figure 2C, cf. lanes 3, 6 with 4, 5). The result is in line with the western blot analysis and syncytium formation assay emphasizing that the predicted SR cluster at the 5′ end of the SRE-1 acts like an ESE even in a heterologous sequence context.
To investigate whether deletion of only the predicted cluster of SR protein-binding sites (Figure 2A; ΔSRE; deletion of nt 1254–1289) affects pgRNA splicing transfection experiments were performed with pHBV-WT, pHBV-ΔPRE or pHBV-ΔSRE plasmid DNA and the ratio between SP1 and pgRNA was analyzed by northern blotting. Quantification of three independent experiments revealed a reduction in splicing by about 60% when the PRE (Figure 2D, ΔPRE) and by about 30% when only the cluster of potential SR protein-binding sites were deleted (Figure 2D, ΔSRE). The result supports the assumption that deletion of a splicing enhancer should decrease splicing efficiency. However, this result also demonstrates that the predicted cluster of SR protein-binding sites was not the only determinant for splicing regulation of pgRNA indicating that additional cis-acting elements in the PRE might be involved in splicing regulation. Such additional elements could either act positively in supporting SRE-1 activity but also in counteracting SRE-1, so that the net outcome of pgRNA splicing is a result of positive and negative cis-acting SREs-1, similar to the 3′ss regulation of e.g. HIV-1 SA7 (48).
The PTB associated splicing factor PSF-enhances HBV RNA splicing in a PRE-dependent manner
The PRE-dependent modulation of splicing efficiency observed raised the question of whether a splicing factor stimulates HBV RNA splicing in a PRE-dependent manner. To address this question, we chose the PTB associated splicing factor PSF (49) because the splicing factor PTB is known to bind to the 3′ end of the PRE (27) and thus may recruit PSF to the PRE. In order to examine the possible PRE-dependent modulatory role of PSF in pgRNA splicing, Huh-7 cells were cotransfected with plasmid pHBV-WT and expression vectors for GFP-PSF-WT or GFP. RNA analysis by northern blotting revealed a PSF-dependent increase in SP1 RNA level and a slight increase in HBV pgRNA levels (Figure 3B, cf. lanes 2, 3 with 4, 5). The quantitative evaluation of the ratio between SP1 showed GFP-PSF-dependent enhancement of pgRNA splicing by about 40% (Figure 3C, WT). In contrast, in cells cotransfected with GFP-PSF-WT and pHBV-ΔPRE expression plasmids the ratio between SP1 and ΔPRE pgRNA increased to about 110% (Figure 3C, ΔPRE) if compared to cotransfection with the GFP-expression vector alone (Figure 3, panels B and C). Since WT pgRNA splicing was only increased with PSF by about 40% we conclude that the PRE counteracts PSF-dependent splicing. The findings support the view that the HPRE contains one or more SREs-1 and argues for a region within the PRE, which represses PSF-dependent splicing.
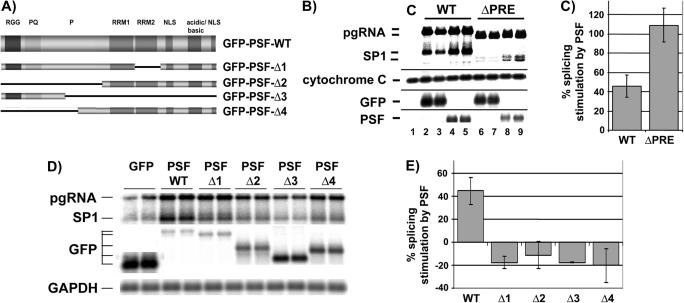
Figure 3.
PSF-dependent stimulation of HBV RNA splicing is repressed by the PRE. (A) GFP-PSF expression vectors used in this study (79). WT, wild-type; pgRNA, pregenomic RNA; SP1, major splice; PSF, PTB associated splicing factor; RRM, RNA recognition motif; NLS nuclear localization signal; RGG, Arginine-glycine-glycine tripeptide rich region; PQ, proline-glutamine rich domain; P, proline-rich domain; M, non-transfected cells. (B) PSF stimulates HBV splicing in a PRE-dependent manner. HuH-7 cells were cotransfected with each 1.5 µg of plasmid tetHBV-wild-type (WT) or tetHBV-ΔPRE (ΔPRE) and 1.0 µg GFP-PSF-WT or 1.0 µg empty GFP vector and 0.5 µg of the transactivator expressing plasmid pUHD-TA. Forty-eight hours thereafter RNA was prepared and 10 µg total RNA was analyzed by northern blotting. The pgRNA and the SP1 RNAs were detected using HBV probe 2. To normalize RNA loading and to monitor GFP-PSF or GFP expression the same blots were hybridized with cytochrome c oxidase II [(Cytochrome (C) and GFP mRNA (GFP and PSF)] specific probes. C, total RNA prepared from untransfected cells. A representative experiment is shown. (C) Quantification of PSF-dependent stimulation of WT and ΔPRE pgRNA splicing. The percentage of splicing stimulation reflects the ratio between the signal intensity of the SP1 RNA and the pgRNA. The HBV wild-type SP1: pgRNA ratio in presence of only GFP was set as 100%, therefore the graph shows the PSF-dependent stimulation of pgRNA splicing. Three independent experiments performed in duplicates were analyzed and quantified. The SD is indicated (±; n = 3). (D) The RRM2 and the N-terminal region of PSF are required to stimulate HBV RNA splicing. Wild-type HBV expression plasmids were cotransfected with GFP or with GFP-PSF-WT, GFP-PSF-Δ1, GFP-PSF-Δ2, GFP-PSF-Δ3 or GFP-PSF-Δ4 expression vectors into HuH-7 cells. Forty-eight hours later RNA was prepared and analyzed by northern blotting for pgRNA and spliced pgRNA (SP1) using HBV probe 2. Expression of GFP and different GFP-PSF plasmids was verified by hybridization with a GFP specific probe. To normalize RNA loading the same blot was hybridized with a GAPDH specific probe. (E) Quantification reveals that only wild-type GFP-PSF is able to stimulate HBV RNA splicing. The percentage of splicing stimulation reflects the ratio between the signal intensity of the SP1 RNA and the pgRNA. The HBV wild-type SP1: pgRNA ratio in presence of only GFP was set as 100%, therefore the graph shows the PSF-dependent stimulation of pgRNA splicing. Three independent experiments performed in duplicates were analyzed and quantified. The SD is indicated (±; n = 3).
In order to map putative domains of PSF required for stimulation of HBV splicing, expression plasmids encoding GFP-tagged mutants of PSF (Figure 3A) were cotransfected with plasmid pSM2. The HBV expression plasmid pSM2 contains an EcoRI head to tail dimer of an HBV genome and synthesis of all viral RNAs are driven by authentic HBV promoters only. Northern blot analysis of RNA of the transfected cells and quantification of the signals revealed that GFP-PSF-WT increased pgRNA splicing (Figure 3D and E). In contrast, none of the PSF mutant proteins enhanced splicing of pgRNA but rather reduced it slightly (Figure 3E). We conclude that the RNA recognition motif 2 (RRM2; Δ1 in Figure 3A, D and E) and the N-terminal part of PSF (Δ4 in Figure 3A, D and E) of PSF are required to increase pgRNA splicing. The results support the previous assumption that the RRM2 is required for RNA-binding and the proline-glutamine rich domain in the N-terminal region represents a protein–protein interface (49). Therefore, a likely prerequisite of the splicing modulatory function of PSF in HBV splicing is its direct interaction with HBV RNA via its RRM2. Furthermore, the data suggest that PSF requires the interaction with splicing-regulatory factors via its N-terminal region to modulate splicing of pgRNA.
DISCUSSION
Splicing licensed bulk mRNAs for nuclear export, whereas a splicing-independent pathway exports intronless mRNAs. A variety of viruses requiring the temporal or constitutive expression of unspliced mRNAs, carrying specific export elements to warrant export of unspliced RNAs [for review see (50–52)]. Even though HBV mRNAs contain a number of frequently used 5′ss and 3′ss (8–11,14,16,53), functional implication of splicing in the viral replication cycle is still not understood yet. The assumption is of interest since viral proteins are thought to be encoded only by unspliced RNAs. To ensure translation of the unspliced mRNAs, utilization of all splice sites must be tightly controlled. Therefore, it seems reasonable that HBV RNAs carry cis-acting elements, e.g. the PRE, that guarantees the export of unspliced viral RNAs and/or that represses splicing (18,19). However, here we describe that the PRE consists of distinct modular elements, which positively and negatively act on 3′ss usage of the pgRNA. The newly identified SRE-1 is located upstream to the PRE α and β domain that is required for full nuclear export activity of the PRE (21,22). Additionally, we show that the SRE-1 could functionally substitute for the bidirectional ESE in a heterologous context.
The observation that deletion of the cluster of predicted SR binding sites at the 5′ end of the PRE significantly reduced pgRNA splicing, although not as efficient as compared to the deletion of the whole PRE (Figure 2D), argues for additional SREs-1 in the PRE. Interestingly, it has been shown that PTB interacts with the 3′ end of the PRE, suggesting that PTB is involved in the export of unspliced subgenomic RNAs (27). However, PTB is known to repress splicing by interfering with splice site definition (54–61) and therefore might effect pgRNA splicing by binding to the PRE. In previous PRE investigations the new identified SRE-1 might have been overlooked due to the peculiarity of the widely used HIV-1 Rev-dependent CAT reporter pDM138 (20,21). In this reporter construct the bacterial CAT ORF is substituted for intronic HIV-1 gp120 sequences, which is flanked by the weak HIV-1 3′ss SA7 (62). Hence, the unspliced CAT message is exported to the cytoplasm only if an export element, such as the PRE, overcomes the nucleocytoplasmic export of the spliced message (20). As shown by Yen and coworkers (27) mutation of the predicted PTB binding site 2 within the PRE led to a decrease in CAT activity. The finding indicates an impeded export of the unspliced message and thus supports the hypothesis that the PRE serves as an export element. However, if PTB binding represses splicing of pDM138 by binding to the PRE, mutation of the PTB binding site would strengthen the SRE-1 leading to efficient utilization of the HIV-1 3′ss SA7. Therefore, the CAT ORF would be removed, the spliced RNA exported and consequently a reduction in CAT activity determined. However, pDM138 cytoplasmic RNA levels have been assessed only by a CAT probe (intronic sequence) and thus it remains unclear whether the spliced message has increased within the cytoplasm due to the PTB mutation. Hence, the PRE might have sequence context dependent functions: located within the pgRNA, PRE regulates splicing and located within the subgenomic RNA, PRE mediates the export of unspliced RNAs. In addition, factors interacting with the PRE eventually support both functions, e.g. PTB represses pgRNA splicing and facilitates the export of unspliced subgenomic RNAs (27,63).
The SRE-1 is a splicing enhancer located about 750 nt downstream of the intensively used 3′ss. The dsxRE splicing enhancer has been shown to be active over a comparable far distance of 500 nt (64). However, RNA loop formation could bridge the apparent distance to bring the SREs-1 in close proximity to the 3′ss (65). Recently, Loeb and coworkers (66) have shown for the pgRNA of DHBV that RNA duplex formation between region A and region B could bridge 1100 nt to bring the 5′ and 3′ss in close proximity resulting in suppression of splicing. In addition, it is likely that PTB might be involved in counteracting splice site selection of the pgRNA by formation of an RNA loop. Recently, it has been described that PTB led to exon exclusion by binding to intronic sequences and subsequent PTB multimerization leading to RNA loop formation unfavorable for recognition of the looped exon (54). We envision a model in which PTB binds to the 3′ end of the PRE and the polypyrimidine tract of the 3′ss forming an RNA loop critical for pgRNA splicing. In such a scenario, PTB would eventually compete for the PTB-associated splicing factor PSF that strongly stimulates pgRNA splicing only in the absence of the PRE (Figure 3B and C). PSF specifically interacts with the polypyrimidine tract close to the 3′ss (49) and probably replaces U2AF65 during the second catalytic step of splicing (67). Hence, overexpression of PSF would change the balance between PTB-dependent splicing inhibition and PSF-dependent splicing stimulation—further supported by trans-acting factors such as the SRE-1-binding SR proteins—leading to an increase in splicing as depicted in Figure 3C. Accordingly, deletion of the PRE would prevent RNA loop formation, abrogating PTB-mediated inhibition of splicing and thus explain an increase in PSF-dependent splicing. Formally, it might also be possible that overexpression of PSF recruits PTB from the PRE leading to a loss of inhibition. However, our analysis of GFP-PSF mutants PSFΔ1 and PSFΔ2 (Figure 3D and E) revealed that the RNA recognition motif 2 was required, suggesting that PSF interacts with pgRNA to stimulate splicing. Hence, if both PTB and PSF compete for the polypyrimidine tract at the 3′ss, exogenous PSF would replace PTB and stimulate splicing of pgRNA. Furthermore, lack of PSF function following deletion of the N-terminal part including the proline-glutamine rich sequence suggests that the protein–protein interaction for splicing stimulation is essential, too. The regions required for direct interaction between PTB and PSF is not mapped yet and because PSF is part of an even bigger complex, containing p54nrb, U1, U2 snRNPs plus a variety of additional proteins (67,68), PTB and PSF could interact temporarily (69).
In conclusion, we show for the first time that the PRE is a splicing regulatory element containing at least one splicing enhancer balancing pgRNA splicing. It seems likely that the PRE functions in a RNA sequence context dependent manner: situated in the pgRNA it regulates splicing, present in subgenomic RNAs it regulates intronless RNA export (18,20) and placed in heterologous sequence context it affects RNA processing events (31,32). In the future, it will be challenging to investigate the fundamental importance of the PRE in the control of pgRNA splicing and its function in the nuclear export of subgenomic RNAs during viral infection. The functional modulation of SR proteins (70–76) and there function in nuclear RNA export (77,78) are important aspects for HBV RNA processing.
Acknowledgments
We are grateful to Jim Patton, Dove Zipori for providing the GFP-PSF plasmids, Dade Behring for providing the monoclonal mouse-anti-gp120 antibody (87-133/026) and Richard Axel for providing HeLa-T4+ cells (through the MRC AIDS Directed Program Reagent Project) and we would like to thank Andreas Rang and Irene Dornreiter for critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft, HE 2814/3-2 (T.H.), SCHA 909/2-2 (H.S.), SCHA 909/1-2 (H.S.), WI 664/9-2 (H.W.), the Kompetentznetz Hepatitis (Hep-Net) 01 KI 0417 (H.W.), and the Stiftung für AIDS-Forschung, Düsseldorf (HS). The Heinrich-Pette-Institut is financially supported by the Bundesministerium für Gesundheit and Freie und Hansestadt Hamburg. Funding to pay the Open Access publication charges for this article was provided by the Heinrich-Pette-Institut.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dreyfuss G., Kim V.N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nature Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 2.Keene J.D. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl Acad. Sci. USA. 2001;98:7018–7024. doi: 10.1073/pnas.111145598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reed R. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 4.Chisari F.V., Ferrari C. Hepatitis B virus immunopathogenesis. Annu. Rev. Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 5.Schaller H., Fischer M. Transcriptional control of hepadnavirus gene expression. Curr. Top Microbiol. Immunol. 1991;168:21–39. doi: 10.1007/978-3-642-76015-0_2. [DOI] [PubMed] [Google Scholar]
- 6.Seeger C., Mason W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obert S., Zachmann-Brand B., Deindl E., Tucker W., Bartenschlager R., Schaller H. A splice hepadnavirus RNA that is essential for virus replication. EMBO J. 1996;15:2565–2574. [PMC free article] [PubMed] [Google Scholar]
- 8.Gunther S., Sommer G., Iwanska A., Will H. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology. 1997;238:363–371. doi: 10.1006/viro.1997.8863. [DOI] [PubMed] [Google Scholar]
- 9.Rosmorduc O., Petit M.A., Pol S., Capel F., Bortolotti F., Berthelot P., Brechot C., Kremsdorf D. In vivo and in vitro expression of defective hepatitis B virus particles generated by spliced hepatitis B virus RNA. Hepatology. 1995;22:10–19. [PubMed] [Google Scholar]
- 10.Sommer G., van Bommel F., Will H. Genotype-specific synthesis and secretion of spliced hepatitis B virus genomes in hepatoma cells. Virology. 2000;271:371–381. doi: 10.1006/viro.2000.0331. [DOI] [PubMed] [Google Scholar]
- 11.Su T.S., Lai C.J., Huang J.L., Lin L.H., Yauk Y.K., Chang C.M., Lo S.J., Han S.H. Hepatitis B virus transcript produced by RNA splicing. J. Virol. 1989;63:4011–4018. doi: 10.1128/jvi.63.9.4011-4018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terre S., Petit M.A., Brechot C. Defective hepatitis B virus particles are generated by packaging and reverse transcription of spliced viral RNAs in vivo. J. Virol. 1991;65:5539–5543. doi: 10.1128/jvi.65.10.5539-5543.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hass M., Hannoun C., Kalinina T., Sommer G., Manegold C., Gunther S. Functional analysis of hepatitis B virus reactivating in hepatitis B surface antigen-negative individuals. Hepatology. 2005;42:93–103. doi: 10.1002/hep.20748. [DOI] [PubMed] [Google Scholar]
- 14.Wu H.L., Chen P.J., Tu S.J., Lin M.H., Lai M.Y., Chen D.S. Characterization and genetic analysis of alternatively spliced transcripts of hepatitis B virus in infected human liver tissues and transfected HepG2 cells. J. Virol. 1991;65:1680–1686. doi: 10.1128/jvi.65.4.1680-1686.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang H.L., Jeng K.S., Hu C.P., Tsai C.H., Lo S.J., Chang C. Identification and characterization of a structural protein of hepatitis B virus: a polymerase and surface fusion protein encoded by a spliced RNA. Virology. 2000;275:398–410. doi: 10.1006/viro.2000.0478. [DOI] [PubMed] [Google Scholar]
- 16.Soussan P., Garreau F., Zylberberg H., Ferray C., Brechot C., Kremsdorf D. In vivo expression of a new hepatitis B virus protein encoded by a spliced RNA. J. Clin. Invest. 2000;105:55–60. doi: 10.1172/JCI8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soussan P., Tuveri R., Nalpas B., Garreau F., Zavala F., Masson A., Pol S., Brechot C., Kremsdorf D. The expression of hepatitis B spliced protein (HBSP) encoded by a spliced hepatitis B virus RNA is associated with viral replication and liver fibrosis. J. Hepatol. 2003;38:343–348. doi: 10.1016/s0168-8278(02)00422-1. [DOI] [PubMed] [Google Scholar]
- 18.Huang J., Liang T.J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol. Cell. Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Z.M., Yen T.S. Hepatitis B virus RNA element that facilitates accumulation of surface gene transcripts in the cytoplasm. J. Virol. 1994;68:3193–3199. doi: 10.1128/jvi.68.5.3193-3199.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z.M., Yen T.S. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol. Cell. Biol. 1995;15:3864–3869. doi: 10.1128/mcb.15.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donello J.E., Beeche A.A., Smith G.J., III, Lucero G.R., Hope T.J. The hepatitis B virus posttranscriptional regulatory element is composed of two subelements. J. Virol. 1996;70:4345–4351. doi: 10.1128/jvi.70.7.4345-4351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith G.J., III, Donello J.E., Luck R., Steger G., Hope T.J. The hepatitis B virus post-transcriptional regulatory element contains two conserved RNA stem–loops which are required for function. Nucleic Acids Res. 1998;26:4818–4827. doi: 10.1093/nar/26.21.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth J., Dobbelstein M. Export of hepatitis B virus RNA on a Rev-like pathway: inhibition by the regenerating liver inhibitory factor IkappaB alpha. J. Virol. 1997;71:8933–8939. doi: 10.1128/jvi.71.11.8933-8939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otero G.C., Harris M.E., Donello J.E., Hope T.J. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J. Virol. 1998;72:7593–7597. doi: 10.1128/jvi.72.9.7593-7597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popa I., Harris M.E., Donello J.E., Hope T.J. CRM1-dependent function of a cis-acting RNA export element. Mol. Cell. Biol. 2002;22:2057–2067. doi: 10.1128/MCB.22.7.2057-2067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zang W.Q., Yen T.S., Fieno A.M., Grant R.A. Distinct export pathway utilized by the hepatitis B virus posttranscriptional regulatory element. Identification of glyceraldehyde-3-phosphate dehydrogenase as a cellular protein that binds to the hepatitis B virus posttranscriptional regulatory element. Virology. 1999;259:299–304. [Google Scholar]
- 27.Zang W.Q., Li B., Huang P.Y., Lai M.M., Yen T.S. Role of polypyrimidine tract binding protein in the function of the hepatitis B virus posttranscriptional regulatory element. J. Virol. 2001;75:10779–10786. doi: 10.1128/JVI.75.22.10779-10786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donello J.E., Loeb J.E., Hope T.J. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J. Virol. 1998;72:5085–5092. doi: 10.1128/jvi.72.6.5085-5092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schambach A., Wodrich H., Hildinger M., Bohne J., Krausslich H.G., Baum C. Context dependence of different modules for posttranscriptional enhancement of gene expression from retroviral vectors. Mol. Ther. 2000;2:435–445. doi: 10.1006/mthe.2000.0191. [DOI] [PubMed] [Google Scholar]
- 30.Zufferey R., Donello J.E., Trono D., Hope T.J. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guang S., Mertz J.E. Pre-mRNA processing enhancer (PPE) elements from intronless genes play additional roles in mRNA biogenesis than do ones from intron-containing genes. Nucleic Acids Res. 2005;33:2215–2226. doi: 10.1093/nar/gki506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y., Wimler K.M., Carmichael G.G. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 1999;18:1642–1652. doi: 10.1093/emboj/18.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu S., Cullen B.R. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9:618–630. doi: 10.1261/rna.5260303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaal H., Klein M., Gehrmann P., Adams O., Scheid A. Requirement of N-terminal amino acid residues of gp41 for human immunodeficiency virus type 1-mediated cell fusion. J. Virol. 1995;69:3308–3314. doi: 10.1128/jvi.69.6.3308-3314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehlers I., Horke S., Reumann K., Rang A., Grosse F., Will H., Heise T. Functional characterization of the interaction between human La and hepatitis B virus RNA. J. Biol. Chem. 2004;279:43437–43447. doi: 10.1074/jbc.M402227200. [DOI] [PubMed] [Google Scholar]
- 36.Caputi M., Freund M., Kammler S., Asang C., Schaal H. A bidirectional SF2/ASF- and SRp40-dependent splicing enhancer regulates human immunodeficiency virus type 1 rev, env, vpu, and nef gene expression. J. Virol. 2004;78:6517–6526. doi: 10.1128/JVI.78.12.6517-6526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krummheuer J., Lenz C., Kammler S., Scheid A., Schaal H. Influence of the small leader exons 2 and 3 on human immunodeficiency virus type 1 gene expression. Virology. 2001;286:276–289. doi: 10.1006/viro.2001.0974. [DOI] [PubMed] [Google Scholar]
- 38.Maddon P.J., Dalgleish A.G., McDougal J.S., Clapham P.R., Weiss R.A., Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 39.Selden R.F., Howie K.B., Rowe M.E., Goodman H.M., Moore D.D. Human growth hormone as a reporter gene in regulation studies employing transient gene expression. Mol. Cell. Biol. 1986;6:3173–3179. doi: 10.1128/mcb.6.9.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 41.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 42.Galibert F., Mandart E., Fitoussi F., Tiollais P., Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E.coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 43.Graveley B.R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freund M., Asang C., Kammler S., Konermann C., Krummheuer J., Hipp M., Meyer I., Gierling W., Theiss S., Preuss T., et al. A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res. 2003;31:6963–6975. doi: 10.1093/nar/gkg901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freund M., Hicks M.J., Konermann C., Otte M., Hertel K.J., Schaal H. Extended base pair complementarity between U1 snRNA and the 5′ splice site does not inhibit splicing in higher eukaryotes, but rather increases 5′ splice site recognition. Nucleic Acids Res. 2005;33:5112–5119. doi: 10.1093/nar/gki824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kammler S., Leurs C., Freund M., Krummheuer J., Seidel K., Tange T.O., Lund M.K., Kjems J., Scheid A., Schaal H. The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA. 2001;7:421–434. doi: 10.1017/s1355838201001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marchand V., Mereau A., Jacquenet S., Thomas D., Mougin A., Gattoni R., Stevenin J., Branlant C. A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J. Mol. Biol. 2002;323:629–652. doi: 10.1016/s0022-2836(02)00967-1. [DOI] [PubMed] [Google Scholar]
- 49.Patton J.G., Porro E.B., Galceran J., Tempst P., Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 50.Cullen B.R. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 2003;28:419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 51.Harris M.E., Hope T.J. RNA export: insights from viral models. Essays Biochem. 2000;36:115–127. doi: 10.1042/bse0360115. [DOI] [PubMed] [Google Scholar]
- 52.Sandri-Goldin R.M. Viral regulation of mRNA export. J. Virol. 2004;78:4389–4396. doi: 10.1128/JVI.78.9.4389-4396.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki T., Masui N., Kajino K., Saito I., Miyamura T. Detection and mapping of spliced RNA from a human hepatoma cell line transfected with the hepatitis B virus genome. Proc. Natl Acad. Sci. USA. 1989;86:8422–8426. doi: 10.1073/pnas.86.21.8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amir-Ahmady B., Boutz P.L., Markovtsov V., Phillips M.L., Black D.L. Exon repression by polypyrimidine tract binding protein. RNA. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carstens R.P., Wagner E.J., Garcia-Blanco M.A. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7388–7400. doi: 10.1128/mcb.20.19.7388-7400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Izquierdo J.M., Majos N., Bonnal S., Martinez C., Castelo R., Guigo R., Bilbao D., Valcarcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 57.Lin C.H., Patton J.G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 58.Shen H., Kan J.L., Ghigna C., Biamonti G., Green M.R. A single polypyrimidine tract binding protein (PTB) binding site mediates splicing inhibition at mouse IgM exons M1 and M2. RNA. 2004;10:787–794. doi: 10.1261/rna.5229704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spellman R., Rideau A., Matlin A., Gooding C., Robinson F., McGlincy N., Grellscheid S.N., Southby J., Wollerton M., Smith C.W. Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- 60.Wagner E.J., Garcia-Blanco M.A. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagner E.J., Garcia-Blanco M.A. RNAi-mediated PTB depletion leads to enhanced exon definition [Erratum (2002), Mol. Cell., 10, 1535.]. Mol. Cell. 10:943–949. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- 62.Hope T.J., McDonald D., Huang X.J., Low J., Parslow T.G. Mutational analysis of the human immunodeficiency virus type 1 Rev transactivator: essential residues near the amino terminus. J. Virol. 1990;64:5360–5366. doi: 10.1128/jvi.64.11.5360-5366.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li B., Yen T.S. Characterization of the nuclear export signal of polypyrimidine tract-binding protein. J. Biol. Chem. 2002;277:10306–10314. doi: 10.1074/jbc.M109686200. [DOI] [PubMed] [Google Scholar]
- 64.Tian M., Maniatis T. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 1994;8:1703–1712. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]
- 65.Graveley B.R., Hertel K.J., Maniatis T. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 1998;17:6747–6756. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loeb D.D., Mack A.A., Tian R. A secondary structure that contains the 5′ and 3′ splice sites suppresses splicing of duck hepatitis B virus pregenomic RNA. J. Virol. 2002;76:10195–10202. doi: 10.1128/JVI.76.20.10195-10202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gozani O., Patton J.G., Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kameoka S., Duque P., Konarska M.M. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 2004;23:1782–1791. doi: 10.1038/sj.emboj.7600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meissner M., Dechat T., Gerner C., Grimm R., Foisner R., Sauermann G. Differential nuclear localization and nuclear matrix association of the splicing factors PSF and PTB. J. Cell. Biochem. 2000;76:559–566. [PubMed] [Google Scholar]
- 70.Caceres J.F., Screaton G.R., Krainer A.R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fetzer S., Lauber J., Will C.L., Luhrmann R. The [U4/U6.U5] tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase. RNA. 1997;3:344–355. [PMC free article] [PubMed] [Google Scholar]
- 72.Gui J.F., Tronchere H., Chandler S.D., Fu X.D. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl Acad. Sci. USA. 1994;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jensen K.B., Dredge B.K., Stefani G., Zhong R., Buckanovich R.J., Okano H.J., Yang Y.Y., Darnell R.B. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 74.Kanopka A., Muhlemann O., Petersen-Mahrt S., Estmer C., Ohrmalm C., Akusjarvi G. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature. 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 75.Misteli T., Spector D.L. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol. Biol. Cell. 1996;7:1559–1572. doi: 10.1091/mbc.7.10.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao S.H., Manley J.L. Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 77.Gilbert W., Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol. Cell. 2004;13:201–212. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
- 78.Huang Y., Steitz J.A. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 79.Dye B.T., Patton J.G. An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp. Cell Res. 2001;263:131–144. doi: 10.1006/excr.2000.5097. [DOI] [PubMed] [Google Scholar]