Short abstract
G-protein-coupled receptors (GPCRs) are the largest and most diverse family of transmembrane receptors. A pylogenetic analysis shows the overall relationship of 277 human GPCRs. An emphasis on receptors with no identified natural ligand may prove valuable for identification of the ligands of 'orphan' receptors.
Abstract
Background
G-protein-coupled receptors (GPCRs) are the largest and most diverse family of transmembrane receptors. They respond to a wide range of stimuli, including small peptides, lipid analogs, amino-acid derivatives, and sensory stimuli such as light, taste and odor, and transmit signals to the interior of the cell through interaction with heterotrimeric G proteins. A large number of putative GPCRs have no identified natural ligand. We hypothesized that a more complete knowledge of the phylogenetic relationship of these orphan receptors to receptors with known ligands could facilitate ligand identification, as related receptors often have ligands with similar structural features.
Results
A database search excluding olfactory and gustatory receptors was used to compile a list of accession numbers and synonyms of 81 orphan and 196 human GPCRs with known ligands. Of these, 241 sequences belonging to the rhodopsin receptor-like family A were aligned and a tentative phylogenetic tree constructed by neighbor joining. This tree and local alignment tools were used to define 19 subgroups of family A small enough for more accurate maximum-likelihood analyses. The secretin receptor-like family B and metabotropic glutamate receptor-like family C were directly subjected to these methods.
Conclusions
Our trees show the overall relationship of 277 GPCRs with emphasis on orphan receptors. Support values are given for each branch. This approach may prove valuable for identification of the natural ligands of orphan receptors as their relation to receptors with known ligands becomes more evident.
Background
G-protein-coupled receptors (GPCRs) are the largest and most diverse family of transmembrane receptors. They respond to a wide range of stimuli including small peptides, lipid analogs, amino-acid derivatives, and sensory stimuli such as light, taste and odor [1], and transmit signals to the interior of the cell through interaction with heterotrimeric G proteins. Certain amino-acid residues of this receptor family are well conserved and approaches exploiting this, such as low-stringency hybridization and degenerate PCR, have been used to clone new members of this large superfamily [2,3,4]. Many of these putative receptors share GPCR structural motifs, but still lack a defined physiologically relevant ligand. One strategy to identify the natural ligand of these so-called orphan receptors uses changes in second-messenger activation in cells stably expressing the receptor in response to tissue extracts expected to contain the natural ligand [5]. In a second step, these extracts are tested and fractionated to purity, before being analyzed by mass spectrometry. This strategy led to the identification of several novel bioactive peptides or peptide families (for review see [6]). The identification of these natural ligands is likely to give further insight into the physiological role of these receptors and advance the design of pharmacologically active receptor agonists or antagonists. This is of particular interest, as GPCRs are the most targeted protein superfamily in pharmaceutical research [7]. Better prediction of the presumed chemical class or structure of the ligand facilitates the identification of orphan receptors by the strategy described above, as the ligand purification process can be tailored more specifically to the assumed class of substances.
Phylogenetic analysis of receptor relationships has already been used to elucidate the chemical nature of receptor ligands. The identification of sphingosine 1-phosphate as the ligand for the GPCR EDG-1 led to the prediction that EDG-3, EDG-5, EDG-6 and EDG-8 have the same ligand [8,9,10,11]. In contrast, phylogenetically distinct members of the EDG cluster - EDG-2, EDG-4 and EDG-7 - are receptors for the similar but distinct ligand lysophosphatidic acid (LPA) [12,13,14]. Neuromedin U, a potent neuropeptide that causes contraction of smooth muscle, was correctly predicted phylogenetically to be the ligand of the orphan GPCR FM3 (NMUR) [15]. Not only the ligand, but also the pharmacology of a novel receptor for histamine, was predicted and confirmed through phylogeny [16]. GPR86, related to the ADP receptor P2Y12, was similarly recently shown to bind ADP [17], and UDP-glucose, a molecule involved in carbohydrate biosynthesis, was shown to be the ligand for the related receptor KIAA0001 [18].
Mammalian GPCRs were previously classified by phylogeny into three families [19,20]: the rhodopsin receptor-like family (A), the secretin receptor-like receptor family (B) and the metabotropic glutamate receptor family (C). These results were generated by neighbor joining, a fast distance-based method suited for large datasets, but influenced by methodological flaws that can in part be overcome by methods not generally applied previously.
In this work, we compiled an exhaustive list that includes all available synonyms and accession numbers of 196 human GPCRs with known ligands and 84 human orphan receptors. The 241 sequences belonging to family A were aligned, and a tentative tree constructed by neighbor joining with 1,000 bootstrap steps. Subgroups of family A defined by this tree and sequences from families B and C were then used for more accurate phylogenetic analysis by state-of-the-art techniques. From this analysis, we tried to predict possible ligands for orphan receptors.
Results and discussion
We set out to define the phylogenetic relationship of human GPCRs by state-of-the-art tools, assuming that the identification of cognate ligands of orphan receptors will be facilitated by a more complete knowledge of their relationship within the large and diverse superfamily.
Database mining and multiple sequence alignment
Most receptors were identified by different groups; therefore, many confusing names and synonyms exist. We adhered to SWISS-PROT names where possible, and compiled a list including all available synonyms and accession numbers of 196 human GPCRs with known ligands and 84 human orphan receptors (Table 1 shows all receptors mentioned in this work; the complete list is supplied as an additional data file). Gustatory and olfactory receptors were omitted. Multiple protein sequences were aligned and the extremely variable amino termini upstream of the first transmembrane domain and carboxyl termini downstream of the seventh transmembrane domain were deleted to avoid length heterogeneity (see Figure 1). The deleted regions contained no significant sequence conservation.
Table 1.
List of example receptor names, accession numbers and abbreviations
| Receptor | Group | Accession no. | Names and synonyms |
| Human GPCR - Family A | |||
| ADMR | A02 | O15218 | Adrenomedullin receptor, Am-R |
| APJ | A03 | P35414 | Apelin receptor, Apj, Agtrl1 |
| CML1 | A08 | Q99788 | Chemokine receptor-like 1, Dez, Chemr23, Ch23, Cmklr1 |
| CML2 | A02 | Q99527 | Chemokine receptor-like 2, flow-induced endothelial G protein-coupled receptor, Feg-1, Gpr30, Cmkrl2, Dry12, Cepr |
| DUFF | A02 | Q16570 | Duffy antigen, Fy glycoprotein, glycoprotein D, Gpfy, Fy, Gpd, Darc |
| EDG1 | A13 | P21453 | Endothelial differentiation, Sphingosine 1-phosphate receptor, Lp-B1 |
| EDG2 | A13 | Q92633 | Endothelial differentiation, lysophosphatidic acid receptor, Lp-A1, Vzg-1 |
| EDG3 | A13 | Q99500 | Endothelial differentiation, lysosphingolipid receptor, Lp-B3 |
| EDG4 | A13 | NM_004720 | Endothelial differentiation, lysophosphatidic acid receptor, Lp-A2 |
| EDG5 | A13 | NP_004221 | Endothelial differentiation, sphingolipid receptor, Lp-B2, H218, Agr16 |
| EDG6 | A13 | AJ000479 | Endothelial differentiation, lysosphingolipid receptor, Lp-C1 |
| EDG7 | A13 | NP_036284 | Endothelial differentiation, lysophosphatidic acid receptor, Lp-A3 |
| EDG8 | A13 | NP_110387 | Endothelial differentiation, sphingosine 1-phosphate receptor, Lp-B4 |
| ETBR-LP2 | A07 | Y16280 | Endothelin B receptor-like protein-2, Etbrlp2, Ebp2, Cns2 |
| FSHR | A10 | P23945 | Follicle stimulating hormone receptor, Fsh-R, follitropin receptor |
| GPR | A06 | NM_007223 | G protein-coupled receptor |
| GPR1 | A08 | P46091 | G protein-coupled receptor Gpr1 |
| GPR3 | A13 | P46089 | G protein-coupled receptor, Acca orphan receptor |
| GPR6 | A13 | P46095 | G protein-coupled receptor 6 |
| GPR7 | A04 | P48145 | G protein-coupled receptor 7 |
| GPR8 | A04 | P48146 | G protein-coupled receptor 8 |
| GPR25 | A03 | NM_005298 | G protein-coupled receptor 25 |
| GPR27 | A18 | NM_018971 | G protein-coupled receptor 27, Sreb1 |
| GPR34 | A12 | NM_005300 | G protein-coupled receptor, Gpry |
| GPR35 | A15 | NM_005301 | G protein-coupled receptor 35 |
| GPR37 | A07 | NM_005302 | G protein-coupled receptor 37, Endothelin receptor type B-like, Cns1 |
| GPR39 | A07 | O43194 | G protein-coupled receptor Gpr39 |
| GPR40 | A11 | O14842 | G protein-coupled receptor Gpr40 |
| GPR41 | A11 | O14843 | G protein-coupled receptor Gpr41, Hia-R |
| GPR42 | A11 | O15529 | G protein-coupled receptor Gpr42 |
| GPR43 | A11 | O15552 | G protein-coupled receptor Gpr43 |
| GPR44 | A08 | AAD21055 | G protein-coupled receptor 44 |
| GPR44 | A08 | AAD21055 | G protein-coupled receptor 44 |
| GPR48 | A10 | NM_018490 | G protein-coupled receptor 48 |
| GPR49 | A10 | NM_003667 | G protein-coupled receptor 49, Hg38, G protein-coupled receptor 67, Fex |
| GPR52 | A18 | Q9Y2T5 | G protein-coupled receptor Gpr52 |
| GPR55 | A15 | NM_005683 | G protein-coupled receptor 55 |
| GPR57 | A17 | NM_014627 | G protein-coupled receptor 57 |
| GPR58 | A17 | NM_014626 | G protein-coupled receptor 58 |
| GPR61 | A18 | AF317652 | G protein-coupled receptor 61 |
| GPR62 | A18 | AF317653 | G protein-coupled receptor 62 |
| GPR63 | A18 | AF317654 | G protein-coupled receptor 63 |
| GPR72 | A09 | NM_016540 | G protein-coupled receptor 72, Jp05 |
| GPR73 | A09 | AAE24084 | G protein-coupled receptor 73 |
| GPR75 | A09 | NM_006794 | G protein-coupled receptor 75 |
| GPR80 | A11 | AF411109 | G protein-coupled receptor 80 |
| GPR81 | A11 | AF411110 | G protein-coupled receptor 81 |
| GPR85 | A18 | NM_018970 | G protein-coupled receptor 85, Sreb2 |
| GPR86 | A12 | NP_076403 | Adp receptor |
| GPR87 | A12 | NM_023915 | G protein-coupled receptor 87 |
| GPR88 | A18 | NM_022049 | G protein-coupled receptor 88 |
| GPR91 | A11 | NM_033050 | G protein-coupled receptor 91 |
| GPR101 | A18 | NM_054021 | G protein-coupled receptor 101 |
| GPR102 | A17 | NM_053278 | G protein-coupled receptor 102 |
| GPR103 | A06 | AF411117 | G protein-coupled receptor 103 |
| GPRC | A13 | P47775 | Gpr12 |
| GPRF | A03 | P49685 | Gpr15, Bob |
| GPRJ | A09 | Q15760 | Gpr19, Gpr-Nga |
| GPRL | A18 | Q99679 | Gpr21 |
| GPRM | A06 | Q99680 | Gpr22 |
| GPRV | A11 | O00270 | Gpr31 |
| GPRW | A08 | O75388 | Gpr32 |
| HM74 | A11 | P49019 | G protein-coupled receptor Hm74 |
| KI01 | A12 | Q15391 | Udp-Glucose receptor, Kiaa0001 |
| LSHR | A10 | P22888 | Lutropin-choriogonadotropic hormone receptor, Lh/Cg-R, Lsh-R, luteinizing hormone receptor, Lhcgr, Lhrhr, Lcgr |
| MAS | A08 | P04201 | Mas proto-oncogene, Mas1 |
| ML1A | A09 | P48039 | Melatonin receptor Type 1a, Mel-1a-R, Mtnr1a |
| ML1B | A09 | P49286 | Melatonin receptor Type 1b, Mel-1b-R, Mtnr1b |
| ML1X | A09 | Q13585 | Melatonin-related receptor, H9, Gpr50 |
| MRG | A08 | P35410 | Mas-related G protein-coupled receptor |
| NMU1R | A07 | AF272362 | Neuromedin U receptor 1, Nmur1, Gpr66, Fm-3 |
| NTR1 | A07 | P30989 | Neurotensin receptor Type 1, Nt-R-1, Ntsr1, Ntrr |
| NTR2 | A07 | O95665 | Neurotensin receptor Type 2, Nt-R-2, levocabastine-sensitive neurotensin receptor, Ntr2 receptor, Ntsr2 |
| NY1R | A09 | P25929 | Neuropeptide Y receptor Type 1, Npy1-R, Npy1r, Npyr, Npyy1 |
| NY2R | A09 | P49146 | Neuropeptide Y receptor Type 2, Npy2-R, Npy2r |
| NY4R | A09 | P50391 | Neuropeptide Y receptor Type 4, Npy4-R, Pancreatic Polypeptide receptor 1, Pp1, Ppyr1, |
| Npy4r | |||
| P2Y5 | A15 | P43657 | P2y purinoceptor 5, P2y5, purinergic receptor 5, P2ry5, 6h1 |
| P2Y7 | A05 | Q15722 | P2y purinoceptor 7, P2y7, Leukotriene B4 receptor, Chemoattractant receptor-like 1, P2ry7, P2y7, Gpr16, Cmkrl1, Ltb4r |
| P2Y9 | A15 | Q99677 | P2y purinoceptor 9, P2y9, purinergic receptor 9, Gpr23, P2ry9 |
| P2Y10 | A15 | AF000545 | Putative purinergic receptor P2y10 |
| P2Y12 | A12 | AF313449 | Adp receptor, Sp1999 |
| PAFR | A12 | P25105 | Platelet Activating Factor receptor, Paf-R, Ptafr |
| PNR | A17 | AF021818 | Putative neurotransmitter receptor |
| PSP24 | A18 | U92642 | High-affinity lysophosphatidic acid receptor homolog, Gpr45 |
| RDC1 | A02 | P25106 | G protein-coupled receptor Rdc1 homolog |
| RE2 | A18 | AF091890 | G protein-coupled receptor Re2 |
| SALPR | A05 | NM_016568 | Somatostatin and angiotensin-like peptide receptor, Loc51289 |
| SREB3 | A18 | NM_018969 | Super conserved receptor expressed in brain 3 |
| TM7SF1 | A01 | AF027826 | Putative seven pass transmembrane protein |
| TSHR | A10 | P16473 | Thyroid stimulating hormone receptor, thyrotropin receptor, Tsh-R |
| Human GPCR - Family B | |||
| EMR1 | B | Q14246 | Cell surface glycoprotein emr1, Emr1 hormone receptor |
| EMR2 | B | AF114491 | Egf-like module Emr2 |
| EMR3 | B | AF239764 | Egf-like module-containing mucin-like receptor Emr3 |
| BAI1 | B | O14514 | Brain-specific angiogenesis inhibitor 1 |
| BAI2 | B | O60241 | Brain-specific angiogenesis inhibitor 2 |
| BAI3 | B | O60242 | Brain-specific angiogenesis inhibitor 3, Kiaa0550 |
| GPR56 | B | NM_005682 | G protein-coupled receptor 56 |
| Human GPCR - Family C | |||
| GPRC5B | C | NM_016235 | G PROTEIN-COUPLED RECEPTOR, FAMILY C, GROUP 5, MEMBER B, GPRC5B |
| GPRC5C | C | NM_018653 | G protein-coupled receptor, family C, group 5, member C, Gprc5c |
| GPRC5D | C | NM_018654 | G protein-coupled receptor, family C, group 5, member D, Gprc5d |
A complete list is supplied as additional data file. Orphan receptors are shown in bold.
Figure 1.
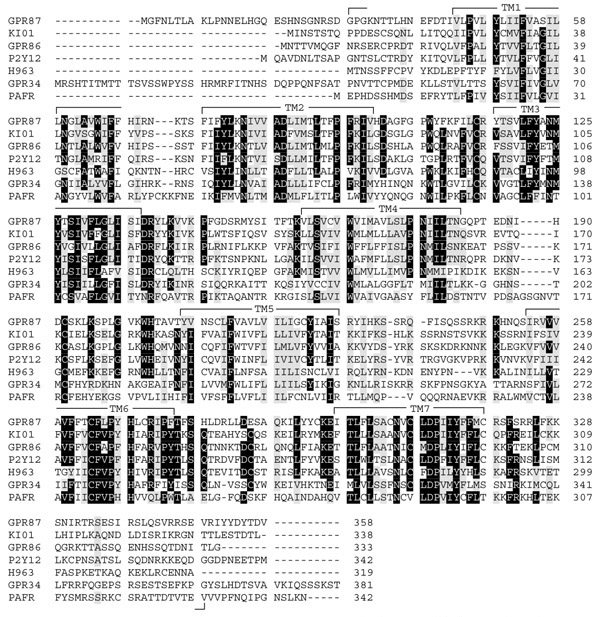
An example multiple sequence alignment of seven receptors. Protein sequences of GPR87, KI01, GPR86, P2Y12, H963, GPR34 and PAFR belonging to subgroup 12 were aligned with ClustalX and modified by deleting the extremely variable amino termini upstream of the first transmembrane domain and carboxyl termini downstream of the seventh transmembrane domain as indicated. Identical amino-acid residues in all aligned sequences are shaded in black and similar residues in gray. Transmembrane (TM) domains identified by the TMpred program are indicated.
Phylogenetic analysis
Because of the large number of sequences in family A, we had to use a combination of computational methods to accomplish the best possible description of their phylogenetic relationship. In a first step we used the distance-based neighbor-joining method as the only one computationally feasible. Neighbor joining has been shown to be efficient at recovering the correct tree topology [21], but is greatly influenced by methodological errors, for example, the sampling error [22]. This can in part be overcome by bootstrapping, a method of testing the reliability of a dataset by the creation of pseudoreplicate datasets by resampling. Bootstrapping assesses whether stochastic effects have influenced the distribution of amino acids [23]. In previous publications on this topic, bootstrapping has not been generally used.
We generated a neighbor-joining tree of family-A sequences, and considered tree branches to be confirmed if they were found in more than 500 of 1,000 bootstrap steps (Figure 2). The same branching pattern was found by least squares (data not shown) as implemented in FITCH [24], but it was not possible to compute enough bootstrap steps with the equipment used. The remaining sequences of unconfirmed branches were then assigned to existing branches according to results obtained with the local alignment tool BLASTP (see Additional data files) [25] to account for similarities in parts of the sequences not sufficient for repeated global alignment. The p-value was used as a measure of similarity.
Figure 2.
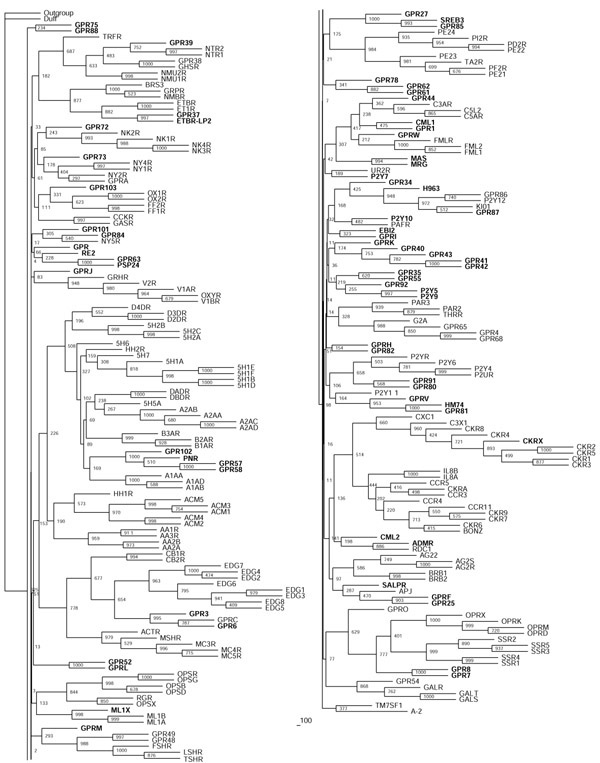
Neighbor-joining tree of the rhodopsin receptor-like family A inferred from the multiple sequence alignment using PHYLIP 3.6. Support values for each internal branch were obtained by 1,000 bootstrap steps, and are indicated. Pairwise distances were determined with PROTDIST and the JTT substitution frequency matrix. The tree was calculated with NEIGHBOR using standard parameters and rooted with the distant, though related, family-B receptor GPRC5B as the outgroup. The consensus tree of all bootstrapped sequences was obtained with CONSENSE. Orphan receptors are shown in bold. Scale bar indicates the branch length of 100 substitutions per site.
As this strategy still left four subgroups too large for detailed analyses, we recalculated neighbor-joining trees and in some cases least-square trees of these sequences to create subgroups A1 and 2, A4 and 5, A11 and 15 and A17 and 18. This approach finally resulted in 19 differently sized subgroups of family A (Table 2) that were further subjected to the more reliable maximum-likelihood and quartet-puzzling algorithms. Maximum-likelihood approaches calculate the probability of the observed data assuming that it has evolved in accordance with a chosen evolutionary model. Phylogenies are then inferred by finding trees and parameters that yield the highest likelihood. Maximum-likelihood approaches tend to outperform alternative methods such as parsimony or distance-based methods. The main advantage is the application of a well defined model of sequence evolution to a given dataset [26]. Maximum likelihood is the estimation method least affected by sampling error and tends to be robust to many violations of the assumptions in the evolutionary model. The methods are statistically well founded, evaluate different tree topologies and use all sequence information available [27,28]. Because of their smaller size, families B and C could be subjected to these methods without prior subgrouping. This resulted in 19 phylogenetic trees, comprising 241 receptors for family A (Figures 3,4,5,6), one tree from 23 sequences for family B and one tree from 14 sequences for family C (Figure 7). Family-A trees were rooted with the human family-B receptor GPRC5B and families B and C with family-A receptor 5H1A. The sequence used to root the tree (the outgroup) is supposed to be a distant, though related, sequence. In some of our groups, the phylogenetic trees could not be fully resolved. This could be due to either very similar or very distant sequences. In both cases the phylogenetic signal is too weak to resolve the tree [29]. Several receptors (for example, TM7SF1, DUFF, GPR, GPRM, GPR75, GPR88, MAS and MRG) were found to be only distantly related to other known receptors used in our analysis. A possible explanation could be the previously proposed convergent evolution of this large protein family, meaning that these receptors have acquired the compelling similarity in their overall structures as a result of functional need, not phylogenetic relationship. The lack of significant sequence similarity among the different GPCR families favors this assumption [30,31,32]. Other explanations for the lack of significant sequence similarities might be an extraordinary divergence (genetic drift) or technical problems of the sequence-analysis methods used in analyzing polytopic membrane proteins or large protein families [33].
Table 2.
Receptor subgroups derived from a combination of neighbor-joining and BLASTP results
| A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 |
| C3X1 | ADMR | AG22 | GPR7 | GALR | FF1R | BRS3 | C3AR | GPR72 | FSHR | GPR40 |
| CKR1 | BONZO | AG2R | GPR8 | GALS | FF2R | ET1R | C5AR | GPR73 | GPR48 | GPR41 |
| CKR2 | CCR11 | AG2S | OPRD | GALT | GASR | ETBR | C5L2 | (GPR75) | GPR49 | GPR42 |
| CKR3 | CCR3 | APJ | OPRK | GPR54 | (GPR) | ETBR-LP2 | CML1 | GPRA | LSHR | GPR43 |
| CKR4 | CCR4 | BRB1 | OPRM | GPRO | GPR103 | GHSR | FML1 | GPRJ | TSHR | GPR80 |
| CKR5 | CCR5 | BRB2 | OPRX | P2Y7 | (GPRM) | GPR37 | FML2 | ML1A | GPR81 | |
| CKR8 | CKR6 | GPR25 | SSR1 | SALPR | GRHR | GPR38 | FMLR | ML1B | GPR82 | |
| CKRX | CKR7 | GPRF | SSR2 | UR2R | OX1R | GPR39 | GPR1 | ML1X | GPR91 | |
| CXC1 | CKR9 | SSR3 | OX2R | GRPR | GPR44 | NK1R | GPRV | |||
| (TM7SF1) | CKRA | SSR4 | OXYR | NMBR | GPRW | NK2R | HM74 | |||
| CML2 | SSR5 | V1AR | NMU1R | (MAS) | NK3R | P2UR | ||||
| (DUFF) | V1BR | NMU2R | (MRG) | NK4R | P2Y11 | |||||
| IL8A | V2R | NTR1 | NY1R | P2Y4 | ||||||
| IL8B | NTR2 | NY2R | P2Y6 | |||||||
| RDC1 | TRFR | NY4R | P2YR | |||||||
| NY5R | ||||||||||
| A12 | A13 | A14 | A15 | A16 | A17 | A18 | A19 | B | C | |
| GPR34 | ACTR | PD2R | EBI2 | OPSB | 5H2A | AA1R | 5H1A | BAI1 | CASR | |
| GPR86 | CB1R | PE21 | G2A | OPSD | 5H2B | AA2A | 5H1B | BAI2 | GBR1 | |
| GPR87 | CB2R | PE22 | GPR35 | OPSG | 5H2C | AA2B | 5H1D | BAI3 | GBR2 | |
| H963 | EDG1 | PE23 | GPR4 | OPSR | 5H6 | AA3R | 5H1E | CALR | GPRC5B | |
| KI01 | EDG2 | PE24 | GPR55 | OPSX | A1AA | ACM1 | 5H1F | CD97 | GPRC5C | |
| P2Y12 | EDG3 | PF2R | GPR65 | RGR | A1AB | ACM2 | 5H5A | CGRR | GPRC5D | |
| PAFR | EDG4 | PI2R | GPR68 | A1AD | ACM3 | 5H7 | CRF1 | MGR1 | ||
| EDG5 | TA2R | GPR92 | A2AA | ACM4 | CRF2 | MGR2 | ||||
| EDG6 | GPRH | A2AB | ACM5 | EMR1 | MGR3 | |||||
| EDG7 | GPRI | A2AC | GPR101 | EMR2 | MGR4 | |||||
| EDG8 | GPRK | A2AD | GPR27 | EMR3 | MGR5 | |||||
| GPR3 | P2Y10 | B1AR | GPR52 | GIPR | MGR6 | |||||
| GPR6 | P2Y5 | B2AR | GPR61 | GLPR | MGR7 | |||||
| GPRC | P2Y9 | B3AR | GPR62 | GLR | MGR8 | |||||
| MC3R | PAR2 | D2DR | GPR63 | GPL2 | ||||||
| MC4R | PAR3 | D3DR | GPR78 | GPR56 | ||||||
| MC5R | THRR | D4DR | GPR84 | GRFR | ||||||
| MSHR | DADR | GPR85 | PACR | |||||||
| DBDR | (GPR88) | PTR2 | ||||||||
| GPR102 | GPRL | PTRR | ||||||||
| GPR57 | HH1R | SCRC | ||||||||
| GPR58 | PSP24 | VIPR | ||||||||
| HH2R | RE2 | VIPS | ||||||||
| PNR | SREB3 | |||||||||
Very distantly related receptors that are possibly not phylogenetically related are shown in brackets. Orphan receptors are shown in bold.
Figure 3.
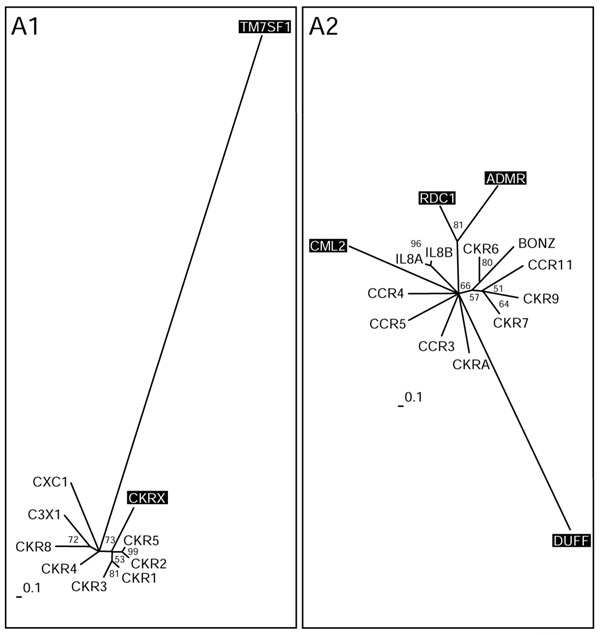
Chemokine receptors (subgroups A1 and A2). Phylogenetic trees of the subgroups were inferred using Puzzle 5.0 corrected by the JTT substitution frequency matrix. Quartet-puzzling support percentage values from 10,000 puzzling steps are shown. The scale bars indicate a maximum likelihood branch length of 0.1 inferred substitutions per site. Orphan receptors are shaded.
Figure 4.
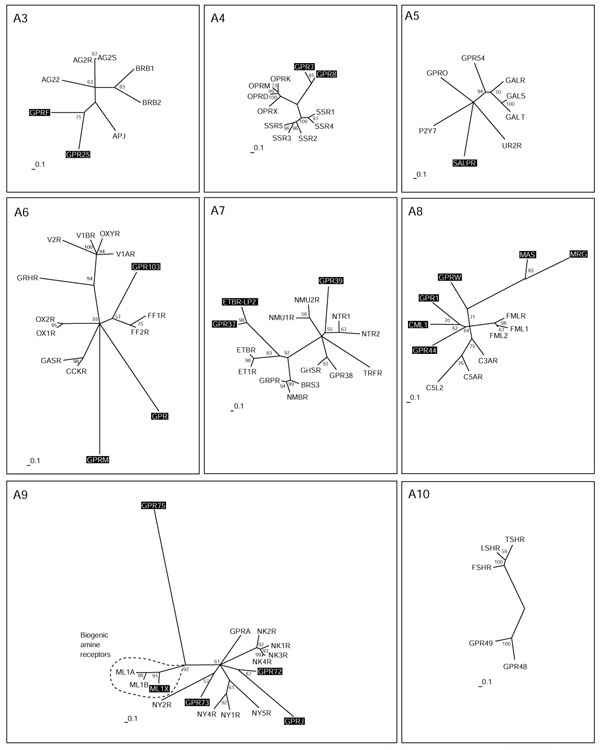
Peptide receptors (subgroups A3-A10). The scale bar indicates a maximum likelihood branch length of 0.1 inferred substitutions per site. Orphan receptors are shaded. For method see Figure 2
Figure 5.
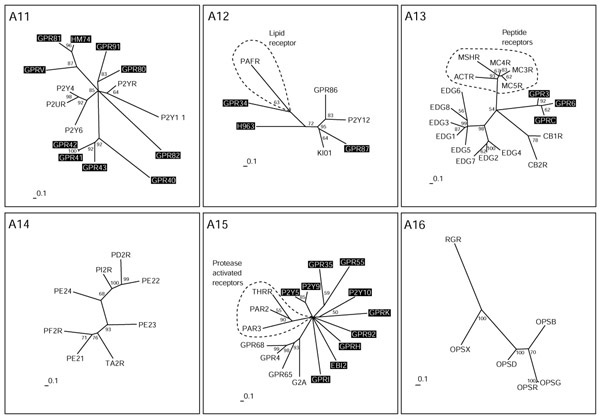
Nucleotide and lipid receptors (subgroups A11-A16). The scale bar indicates a maximum-likelihood branch length of 0.1 inferred substitutions per site. Orphan receptors are shaded. For method see Figure 2.
Figure 6.
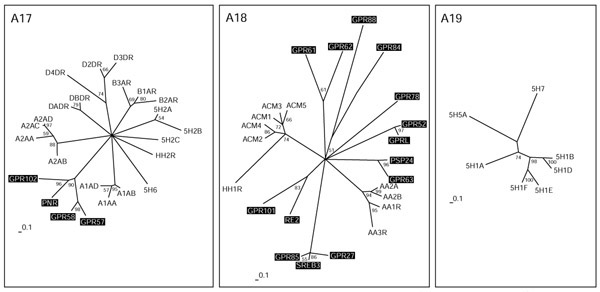
Biogenic amine receptors (subgroups A17-A19). The scale bar indicates a maximum-likelihood branch length of 0.1 inferred substitutions per site. Orphan receptors are shaded. For method see Figure 2.
Figure 7.
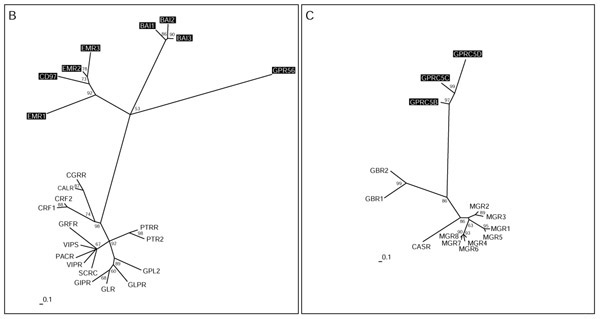
Families B and C of the G-protein-coupled receptors (GPRCs). Phylogenetic trees of families B and C were inferred using Puzzle 5.0 corrected by the JTT substitution frequency matrix. Quartet-puzzling support percentage values from 10,000 puzzling steps are shown. The scale bar indicates a maximum likelihood branch length of 0.1 inferred substitutions per site. Orphan receptors are shaded.
Receptor family A subgroups
In contrast to the subfamilies presented in GPCRDB [34], a database widely used in the field, our grouping shows the orphan receptors within their respective subgroup and their relationship to receptors with known ligands. In addition, our method sometimes resulted in subgroups with members whose ligands belong to different substance classes. These results are discussed in more detail below.
Chemokine receptors
Groups A1 and A2 comprise the chemokine receptors (Figure 3). The chemokine ligand superfamily is defined by four conserved cysteines that form two disulfide bonds, and can be structurally subdivided into two major branches based on the spacing of the first cysteine pair. Chemokines in which these residues are adjacent form the CC subfamily (corresponding to the SWISS-PROT CKR nomenclature used here), and those separated by a single amino acid comprise the CXC subfamily (here CCR and IL8R; for a review see [35]). We had to divide the whole subfamily into two groups to perform a detailed phylogenetic analysis. This sub-grouping produced the same dichotomy, as suggested by the two-ligand motifs, as another example of the parallel evolution of receptors and ligands. Similar results describing this parallel evolution were found previously using a different computational approach [36].
Group A1 mainly comprises the CC family. We hypothesize that the orphan receptor CKRX, which constitutes a separate branch related to CKR1, 2, 3 and 5, might also bind a CC ligand. In contrast, TM7SF1 in this group seems to be only distantly, if at all, related to family-A receptors. It was grouped according to BLASTP results, where a misleading local alignment of approximately 20 amino acids placed it in the vicinity of the chemokine receptors. Group A2 is more heterogeneous and comprises receptors for CC and CXC ligands, as well as an orphan receptor (ADMR) previously thought to bind the peptide adrenomedullin. Adrenomedullin has now been shown to bind a family-B receptor and is discussed further below. The orphan receptor RDC1 in group A2 was first believed to be a receptor for vasointestinal peptide VIP [37], a notion not supported by phylogeny and later dismissed by experimental data [38]. Our results place it closer to the ADMR receptor than to the typical chemokine receptors. CML2 is a typical, but distant, member of the chemokine receptor family. The DUFF receptor (the Duffy antigen) is also very distantly related and was only grouped into A2 by BLASTP results.
Peptide receptors
Group A3 consists of receptors for the small peptides angiotensin (8 amino acids), bradykinin (9 amino acids) and apelin (Figure 4). Four forms of apelin (12, 13, 17 and 36 amino acids) have been described, but only those of 12 and 13 amino acids bind in nanomolar concentrations [39]. The orphan receptors GPRF and GPR25 in this group are related as closely to the apelin receptor APJ as to the angiotensin or bradykinin receptors, and might also bind small peptides. GPRF acts as a co-receptor for the human immunodeficiency virus (HIV) [40], like the APJ receptor [41], which further hints at structural homology of the two ligands. Opioid and somatostatin receptors make up group A4. Both somatostatin and opioid peptides are derived from the processing of larger precursors. The somatostatins are cyclic peptides of 14 and 28 amino acids. The opioid precursors preproenkephalin, preprodynorphin, prepro-opiomelanocortin and prepronociceptin display a strikingly similar general organization and a conserved amino-terminal region that contains six cysteines, probably involved in disulfide bond formation.
The processed neuropeptides, in contrast, are less similar to each other. It could be speculated that the receptors first bound the precursors themselves, and that the diversity derived from processing is evolutionarily new. Processing prepronociceptin gives rise to two evolutionarily conserved peptides besides orphanin FQ, the ligand for OPRX. It has not been reported whether these peptides bind to the orphan receptors GPR7 and GPR8, which constitute a new branch related to the opioid receptors.
In group A5 we find three receptors that bind the 30-amino-acid peptide galanin, and related to these the GPR54 receptor, which is activated by the 54-, 14-, and 13-amino-acid peptides derived from the product of KiSS-1, a metastasis suppressor gene for melanoma cells. These kisspeptins all share a common RF-amide caboxyl terminus. Although only distantly related to each other, both GPRO (melanin-concentrating hormone) and UR2R (urotensin II peptide) bind cyclic peptides originally isolated from fish. Similarly distant is the orphan receptor SALPR, which shares sequence similarity with somatostatin (A4) and angiotensin (A3) receptors, but subgrouping of groups A4 and 5 by neighbor joining led to its placement in group 5. SALPR does not bind somatostatin or angiotensin ligands [42], but could bind another cyclic peptide. The P2Y7 receptor in group A5 does not bind nucleotides [43], as suggested by the name, but was published as a receptor for the lipid leukotriene 84 [44], a notion not supported by phylogeny. In addition, two new leukotriene receptors - CLT1 and CLT2 - have been cloned and characterized during the preparation of this manuscript [45,46] and were found to be unrelated to P2Y7.
Group A6 is again composed solely of receptors for peptide ligands. The orphan receptor GPR103 is related to the neuropeptide FF receptors that bind two amidated mammalian neuropeptides - NPAF (A-18-F-amide) and NPFF (F-8-F-amide), also known as morphine-modulating peptides. These peptides, which may also be the ligand for GPR103, are members of a large family of neuropeptides related to the molluscan cardioexcitatory neuropeptide (FMRF-amide, Phe-Met-Arg-Phe-amide). The orphan receptors GPRM and GPR in group A6 are most probably also peptide receptors, but are only very distantly related to the others and show no relationship to receptors with known ligands. Group A7 is also composed of receptors for peptide ligands: neuromedin, neurotensin, motilin, endothelin, bombesin and the releasing hormones for growth hormone and thyrotropin. GPR39 might bind a small peptide ligand like the closely related neurotensin receptors NTR1 and 2, which binds a 13-amino-acid peptide derived from a larger precursor protein. GPR37 and ETBR-LP2 are related to each other and branch off the endothelin receptors that bind characteristic bicyclic peptides of 21 amino acids containing four cysteines linked by two disulfide bonds.
Group A8 has two branches with receptors with known ligands. These receptors bind the structurally diverse but functionally related chemotactic substances N-formylmethionyl and the anaphylatoxic complement factors. The N-formylmethionyl ligands are small hydrophilic peptides of bacterial origin, but recently a number of new peptide agonists have been identified that selectively activate the high-affinity fMLF receptor FPR and/or its low-affinity variant FPRL1. These agonists include peptide domains derived from the envelope proteins of HIV type 1 and at least three amyloidogenic polypeptides, the human acute-phase protein serum amyloid A, the 42-amino-acid form of beta-amyloid peptide and a 21-amino-acid fragment of the human prion protein. Furthermore, a cleavage fragment of neutrophil granule-derived bactericidal cathelicidin, LL-37, is also a chemotactic agonist for FPRL1 (for a review see [47]). The complement factors C3a and C5a are large but highly hydrophilic proteins with a mainly alpha-helical structure held together by three disulfide bridges. C5a is rapidly desarginated to the less potent derivative C5adR74, which is the ligand for the C5L2 receptor. The orphan receptors GPR1, CML1 and GPR44 all cluster, and constitute a separate branch as distant as the other two branches. No prediction of the possible structure of the ligands for these receptors can be derived from this tree, but maybe they will function as chemotactic peptides. This could at least hint at leukocytes or inflamed tissue as a possible source for these ligands. The receptor GPRW constitutes its own branch, not as distant to the main group as the MAS oncogene product and the related receptor MRG, which are only very distantly related to the group.
All receptors in group A9 with known ligands bind peptides, except for a side branch consisting of receptors for the biogenic amine melatonin. The orphan receptor ML1X is closely related to melatonin receptors ML1A and B, but apparently does not bind melatonin [48]. GPR73 is related to the neuropeptide Y (NPY) receptor NY2R which mainly binds the pancreatic peptide YY of 36 amino acids, and these two are placed together on a branch distinct from the NPY receptors NY4R and NY1R. GPR73 does not bind the NPY ligand family [49], but possibly a similar large peptide ligand. The orphan receptors GPR72 and GPRJ constitute a new subgroup that most probably bind related peptide ligands. GPR72 does not bind a NPY ligand [49]. GPR75 is only very distantly related to the whole A9 group. The receptors for the glycoprotein hormones thyroid-stimulating hormone (TSH), luteinizing hormone (LSH) and follicle-stimulating hormone (FSH) make up Group A10. GPR48 and 49 are very similar in their overall structure, with long amino termini, but their relationship is also evident in the neighbor-joining tree constructed from alignments without amino and carboxyl termini. It has been recently shown that these receptors mediate the action of relaxin, a peptide hormone of the insulin-like growth factor family secreted by the corpus luteum during pregnancy [50].
Nucleotide and lipid receptors
The receptors with known ligands in group A11 are the P2Y receptors, which bind pyrimidine as well as purine nucleotides (Figure 5). Several orphan receptors constitute new clusters. GPR80 and GPR91 are distantly related to each other and relatively close to the P2Y receptors. GPR80 is the closest relative of the newly identified CLT2 receptor for leukotrienes as judged by BLASTP results. GPR81, HM74 and GPRV and GPR 40-43 belong to branches only distantly related to P2Y receptors. Within these potential new subfamilies, GPR41-43, GPR81 and HM74 are more closely related to each other than to GPR40 (for GPR41-43) and GPRV (for GPR81 and HM74).
In group A12, the platelet-activated receptor, a lipid receptor and receptors activated by nucleotides mingle, but are found on different side branches. The orphan receptor GPR87 is closely related to the receptor for UDP-glucose KI01 and to the ADP-binding receptors P2Y12 and GPR86. We assume that this receptor might also bind UDP-glucose or another modified nucleotide. GPR34 is distantly related to the platelet-activating factor (PAF) receptor; it was not activated by available lipid ligands [51], but might nevertheless bind a lipid ligand. Group A13 contains both peptide and lipid receptors but they make up different branches. The peptide branch binds peptides derived from the processing of pro-opiomelanocortin that gives rise to peptides of between 12 and 36 amino acids. The EDG and cannabinoid receptors constitute clusters, and one cluster distinct from the other three consists of the orphan receptors GPR3, GPR6 and GPRC, which have been grouped closer to the lipid EDG receptors in the overall neighbor-joining tree (Figure 2). This information helped to identify a phospholipid ligand for GPRC (H. Chica Schaller, personal communication).
The receptors in group A14 all bind ligands derived from arachidonic acid by the action of cyclooxygenase. These receptors for lipid-derived autacoids or prostanoids comprise receptors for the prostaglandins and thromboxanes. There are no orphan receptors in this group. Group A15 is a very heterogenous group composed of receptors for the lipids sphingosylphosphorylcholine (SPC), lysophosphatidylcholine (LPC) and psychosine, and receptors activated by proteases. GPR4 and GPR68 both bind SPC, like the EDG receptor branch consisting of the EDG1, 3, 6 and 8 receptors in A13, but are not closely related. Protease-activated receptors become activated by a part of the former amino terminus cleaved by the protease. The new amino terminus then functions as a tethered ligand and activates the receptor. This can be mimicked by very small peptides derived from this ligand; such receptors should therefore rather resemble peptide receptors. The orphans P2Y5, P2Y9 and P2Y10 receptors were not placed in group 11 and 12 like most P2Y receptors, but in group A15, supporting the fact that they were misnamed. P2Y5 and P2Y9 do not bind nucleotides [52,53], but this has not been shown yet for P2Y10. All other orphan receptors in this group, with the exception of GPR35 and GPR55 which cluster together, are as distantly related to each other as to the receptors with known ligands. Group A16 contains the opsins, receptors that are activated by isoprenoid ligands, and no orphan receptors.
Biogenic amine receptors
Some serotonin receptors and receptors for the biogenic amines adrenaline, dopamine and histamine are all placed on different branches in group A17 (Figure 6). An additional branch consists of the orphan receptors GPR102, PNR, GPR57 and GPR58, which are as distantly related to the others as, for example, is the alpha-adrenergic receptor branch. PNR and GPR58 expressed in COS cells did not bind various serotonin receptor-specific ligands [54]. Their ligands might be small molecules with similar properties. Group A18 is very heterogeneous and consists of receptors for the biogenic amines acetylcholine and adenosine, and the HH1R receptor for histamine, as well as many distantly related orphan GPCRs. GPR63 is closely related to the orphan receptor PSP24. The Xenopus laevis homolog of this receptor binds LPA [55]. GPR101 and RE2, GPRL and GPR52, and GPR61 and GPR62 constitute their own subgroups. In particular, the SREB1-3 cluster (GPR85, GPR27 and SREB3) makes up its own family, with only a distant relationship to other GPCRs in this group. No orphan receptors are found in group A19, which consists entirely of serotonin receptors distinct from those in A17.
During the preparation of this manuscript several new family-A receptors that could not be fitted into our analysis were identified. These comprise 15 new receptors distinct from the classical biogenic amine receptors that apparently bind the trace amines tyramine, β-phenylethylamine, tryptamine and octopamine [56]. In addition, a new subfamily of GPCRs related to the mas oncogene and uniquely expressed in small nociceptive sensory neurons were shown to be the receptors for a number of enkephalin fragments [57].
Receptor families B and C
Family B (Figure 7) was named after the secretin receptor. Yet proteins showing homology to this receptor make up only one of four distantly related subgroups. The receptors EMR1, EMR2 and EMR3, and the CD97 surface antigen, all have several epidermal growth factor (EGF)-like domains in the extracellular amino terminus. They constitute their own cluster only distantly related to the rest of the family. The same applies to the brain-specific angiogenesis inhibitor family BAI1-3. GPR56 was assigned to family B because it shows the typical signature [58], but is so far the only one of its kind. So far no non-protein ligand has been identified as a ligand for family-B receptors. Astonishingly, one family-B receptor, namely the CGRP receptor, requires coexpression with single transmembrane receptor activity-modifying proteins (RAMP1-3) for ligand binding and signal transduction [59]. Coexpression of different RAMPs results in binding of different cyclic peptide ligands such as adrenomedullin, amylin or the calcitonin gene-related peptide (for a review see [60]). This could further complicate the identification of the cognate ligands for these family-B orphan receptors, but we assume that they will also bind large peptide ligands. In family C (Figure 7), the metabotropic glutamate receptors MGR1-8 bind the small molecule glutamate, the CASR receptor senses extracellular calcium concentration, and receptors GBR1-2 bind the small molecule gamma-amino butyric acid (GABA). GPRC5B, C and D constitute their own subgroup with no closer relationship to the other members, but might also bind small molecules.
Conclusions
In this work, we calculated the phylogenetic distances of 277 human GPCRs and show the relationship of orphan receptors to receptors for known ligands with support values for each branch. We then grouped orphan receptors and receptors with known ligands into 19 subgroups that sometimes differ from previous classifications. Three subgroups are composed of receptors for ligands that belong to different substance classes; for example, in group A12, lipid receptors and receptors activated by nucleotides mingle, and in groups A13 and A15, peptide and lipid receptors. In both subgroups the receptors binding ligands of different substance classes make up different branches. We hope that this approach proves valuable for identifying the natural ligands of orphan receptors, as related receptors have previously been shown to have ligands with similar structural features.
Materials and methods
Sequence database mining
A database search excluding olfactory and gustatory receptors identified the amino-acid sequences of 281 human GPCRs. Only sequences annotated as GPCRs in the following databases were used: NCBI [61], SWISS-PROT [62], EMBL [63] and GPCRDB [34,64]. Receptors without published ligands in PubMed [65] were defined as orphan GPCRs.
Multiple sequence alignments
Multiple protein sequences were aligned with ClustalX 1.81 [66]. Pairwise alignment parameters were set as: slow/accurate alignment; gap opening penalty 10; gap extension penalty 0.10; protein weight matrix BLOSUM 30. Multiple alignment parameters were set as: gap opening penalty 10; gap extension penalty 0.05; delay divergent sequences 35%; protein weight matrix BLOSUM series [67]. The alignments were modified by deleting the extremely variable amino termini upstream of the first transmembrane domain and carboxyl termini downstream of the seventh transmembrane domain. Alignment editing and shading was done using BioEdit Sequence Alignment Editor [68] and GeneDoc Multiple Sequence Alignment Editor [69]. Transmembrane domains were identified using the TMpred program [70] and, where available, data from the original publication [71].
Clustering of subgroups
An overall phylogenetic tree of family A was inferred from the multiple sequence alignment with PHYLIP 3.6 [72]. Bootstrapping was performed 1,000 times using SEQBOOT to obtain support values for each internal branch. Pairwise distances were determined with PROTDIST and the JTT substitution frequency matrix [73]. Neighbor-joining phylogenetic trees [21] were calculated with NEIGHBOR using standard parameters. The human GPRC5B receptor belonging to family B was used as outgroup for family A. The out-group sequence is supposed to be a distant, though related, sequence and is used to root the tree. The majority-rule consensus trees of all bootstrapped sequences were obtained with the program CONSENSE. Representations of the calculated trees were constructed with TreeView [74]. Clusters with bootstrap values greater than 50% were defined as confirmed subgroups, and sequences with lower values added to these subgroups according to their sequence similarity in the alignment as judged by visual inspection and the results of pairwise local alignments with all other sequences by BLASTP [25]. The p-value was used as a measure of similarity.
Quartet-puzzling trees
Multiple protein sequence alignments of these new subgroups were created as described above. Phylogenetic trees were inferred from these alignments using Puzzle 5.0 [75] to calculate maximum-likelihood distances corrected by the JTT substitution-frequency matrix [73] with amino-acid usage estimated from the data, site-to-site rate variation modeled on a gamma distribution with eight rate categories plus invariant sites, and the shape parameter estimated from the data. The human GPRC5B receptor of family B was used as an outgroup for family A. The human 5H1A receptor of family A was used as an outgroup for families B and C (the outgroups are not shown in the figures here). Quartet-puzzling (QP) trees were constructed with the described settings and 10,000 puzzling steps to obtain support values (QP reliability) for each internal branch. The program Puzzle 5.0 was used in a parallelized version (ppuzzle) with a message-passing interface (MPI) implementation on a HP 9000 N-Class Enterprise Server Cluster consisting of five HP 9000 N-Class shared-memory multiprocessor systems with eight PA-RISC 8600 (552 MHz) processors each. Representations of the quartet-puzzling trees were constructed with TreeView [74].
Additional data files
Additional data files available with this paper include a data table with names, synonyms and accession numbers of all GPCRs, and the BLASTP results of all GPCRs (full-length sequences and sequences without amino or carboxyl termini).
Supplementary Material
A data table with names, synonyms and accession numbers of all GPCRs
Names, synonyms and accession numbers of all GPCRs, and the BLASTP results of all GPCRs full-length sequences
Names, synonyms and accession numbers of all GPCRs, and the BLASTP results of all GPCRs sequences without amino or carboxyl termini
Acknowledgments
Acknowledgements
The DFG Graduiertenkolleg 255, the Dr Kurt und Irmgard Meister-Stiftung and the Hamburgische Wissenschaftliche Gesellschaft, supported this study. We appreciate the help of Chica Schaller in finding additional sequences and of Andreas Schuldei in reconfiguring ppuzzle and using MPI. Klaus Martens and his colleagues at the computing center of the Technical University Hamburg-Harburg provided an account at the HP N-Class Enterprise Server Cluster and helped us to use the software environment.
References
- Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- Libert F, Parmentier M, Lefort A, Dinsart C, Van Sande J, Maenhaut C, Simons MJ, Dumont JE, Vassart G. Selective amplification and cloning of four new members of the G protein-coupled receptor family. Science. 1989;244:569–572. doi: 10.1126/science.2541503. [DOI] [PubMed] [Google Scholar]
- Methner A, Hermey G, Schinke B, Hermans-Borgmeyer I. A novel G protein-coupled receptor with homology to neuropeptide and chemoattractant receptors expressed during bone development. Biochem Biophys Res Commun. 1997;233:336–342. doi: 10.1006/bbrc.1997.6455. [DOI] [PubMed] [Google Scholar]
- Lee D, George S, Evans J, Lynch K, O'Dowd B. Orphan G protein-coupled receptors in the CNS. Curr Opin Pharmacol. 2001;1:31–39. doi: 10.1016/s1471-4892(01)00003-0. [DOI] [PubMed] [Google Scholar]
- Reinscheid RK, Nothacker HP, Bourson A, Ardati A, Henningsen RA, Bunzow JR, Grandy DK, Langen H, Monsma FJ, Jr, Civelli O. Orphanin FQ: a neuropeptide that activates an opioidlike G protein-coupled receptor. Science. 1995;270:792–794. doi: 10.1126/science.270.5237.792. [DOI] [PubMed] [Google Scholar]
- Civelli O, Nothacker HP, Saito Y, Wang Z, Lin SH, Reinscheid RK. Novel neurotransmitters as natural ligands of orphan G-protein-coupled receptors. Trends Neurosci. 2001;24:230–237. doi: 10.1016/s0166-2236(00)01763-x. [DOI] [PubMed] [Google Scholar]
- Drews II. Drug discovery today - and tomorrow. Drug Discov Today. 2000;5:2–4. doi: 10.1016/s1359-6446(99)01428-2. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Van Brocklyn JR, Thangada S, Liu CH, Hand AR, Menzeleev R, Spiegel S, Hla T. Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science. 1998;279:1552–1555. doi: 10.1126/science.279.5356.1552. [DOI] [PubMed] [Google Scholar]
- Van Brocklyn JR, Tu Z, Edsall LC, Schmidt RR, Spiegel S. Sphingosine 1-phosphate-induced cell rounding and neurite retraction are mediated by the G protein-coupled receptor H218. J Biol Chem. 1999;274:4626–4632. doi: 10.1074/jbc.274.8.4626. [DOI] [PubMed] [Google Scholar]
- Van Brocklyn JR, Graler MH, Bernhardt G, Hobson JP, Lipp M, Spiegel S. Sphingosine-1-phosphate is a ligand for the G protein-coupled receptor EDG-6. Blood. 2000;95:2624–2629. [PubMed] [Google Scholar]
- Im DS, Heise CE, Ancellin N, O'Dowd BF, Shei GJ, Heavens RP, Rigby MR, Hla T, Mandala S, McAllister G, et al. Characterization of a novel sphingosine 1-phosphate receptor, Edg-8. J Biol Chem. 2000;275:14281–14286. doi: 10.1074/jbc.275.19.14281. [DOI] [PubMed] [Google Scholar]
- Hecht JH, Weiner JA, Post SR, Chun J. Ventricular zone gene-1 (vzg-1) encodes a lysophosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. J Cell Biol. 1996;135:1071–1083. doi: 10.1083/jcb.135.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Bleu T, Hallmark OG, Goetzl EJ. Characterization of a novel subtype of human G protein-coupled receptor for lysophosphatidic acid. J Biol Chem. 1998;273:7906–7910. doi: 10.1074/jbc.273.14.7906. [DOI] [PubMed] [Google Scholar]
- Im DS, Heise CE, Harding MA, George SR, O'Dowd BF, Theodorescu D, Lynch KR. Molecular cloning and characterization of a lysophosphatidic acid receptor, Edg-7, expressed in prostate. Mol Pharmacol. 2000;57:753–759. [PubMed] [Google Scholar]
- Szekeres PG, Muir AI, Spinage LD, Miller JE, Butler SI, Smith A, Rennie GI, Murdock PR, Fitzgerald LR, Wu H, et al. Neuromedin U is a potent agonist at the orphan G protein-coupled receptor FM3. J Biol Chem. 2000;275:20247–20250. doi: 10.1074/jbc.C000244200. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Michalovich D, Wu H, Tan KB, Dytko GM, Mannan IJ, Boyce R, Alston J, Tierney LA, Li X, et al. Cloning, expression, and pharmacological characterization of a novel human histamine receptor. Mol Pharmacol. 2001;59:434–441. doi: 10.1124/mol.59.3.434. [DOI] [PubMed] [Google Scholar]
- Communi D, Gonzalez NS, Detheux M, Brezillon S, Lannoy V, Parmentier M, Boeynaems JM. Identification of a novel human ADP receptor coupled to G(i). J Biol Chem. 2001;276:41479–41485. doi: 10.1074/jbc.M105912200. [DOI] [PubMed] [Google Scholar]
- Chambers JK, Macdonald LE, Sarau HM, Ames RS, Freeman K, Foley JJ, Zhu Y, McLaughlin MM, Murdock P, McMillan L, et al. A G protein-coupled receptor for UDP-glucose. J Biol Chem. 2000;275:10767–10771. doi: 10.1074/jbc.275.15.10767. [DOI] [PubMed] [Google Scholar]
- Attwood TK, Findlay JB. Fingerprinting G-protein-coupled receptors. Protein Eng. 1994;7:195–203. doi: 10.1093/protein/7.2.195. [DOI] [PubMed] [Google Scholar]
- Kolakowski LF., Jr GCRDb: a G-protein-coupled receptor database. Receptors Channels. 1994;2:1–7. [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Page RD, Holmes EC. Molecular Evolution: A Phylogenetic Approach. Oxford: Blackwell Science; 1999. pp. 218–225. [Google Scholar]
- Kumar S, Gadagkar SR. Efficiency of the neighbor-joining method in reconstructing deep and shallow evolutionary relationships in large phylogenies. J Mol Evol. 2000;51:544–553. doi: 10.1007/s002390010118. [DOI] [PubMed] [Google Scholar]
- Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Strimmer K, Goldman N, von Haeseler A. Bayesian Probabilities and Quartet Puzzling. Mol Biol Evol. 1997;14:210–211. [Google Scholar]
- Strimmer K, von Haeseler A. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–211. [Google Scholar]
- Strimmer K, Robertson DL. Inference and applications of molecular phylogenies: an introductory guide. In: Sansom C, Horton RM, editor. In The Internet for Molecular Biologists (Practical Approach Series) Oxford: Oxford University Press,; 2001. [Google Scholar]
- Pin JP, Joly C, Heinemann SF, Bockaert J. Domains involved in the specificity of G protein activation in phospholipase C-coupled metabotropic glutamate receptors. EMBO J. 1994;13:342–348. doi: 10.1002/j.1460-2075.1994.tb06267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst WC, Snyder LA, Schuster DI, Brosius J, Sealfon SC. Sequence alignment of the G-protein coupled receptor superfamily. DNA Cell Biol. 1992;11:1–20. doi: 10.1089/dna.1992.11.1. [DOI] [PubMed] [Google Scholar]
- Burbach JP, Meijer OC. The structure of neuropeptide receptors. Eur J Pharmacol. 1992;227:1–18. doi: 10.1016/0922-4106(92)90136-j. [DOI] [PubMed] [Google Scholar]
- Graul R, Sadee W. Evolutionary relationships among G protein-coupled receptors using a clustered database approach. AAPS PharmSci. 2001;3:E12. doi: 10.1208/ps030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn F, Weare J, Beukers MW, Horsch S, Bairoch A, Chen W, Edvardsen O, Campagne F, Vriend G. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res. 1998;26:275–279. doi: 10.1093/nar/26.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascieri MA, Springer MS. The chemokine/chemokine-receptor family: potential and progress for therapeutic intervention. Curr Opin Chem Biol. 2000;4:420–427. doi: 10.1016/s1367-5931(00)00113-7. [DOI] [PubMed] [Google Scholar]
- Goh C, Bogan A, Joachimiak M, Walther D, Cohen F. Co-evolution of proteins with their interaction partners. J Mol Biol. 2000;299:283–293. doi: 10.1006/jmbi.2000.3732. [DOI] [PubMed] [Google Scholar]
- Sreedharan SP, Robichon A, Peterson KE, Goetzl EJ. Cloning and expression of the human vasoactive intestinal peptide receptor. Proc Natl Acad Sci USA. 1991;88:4986–4990. doi: 10.1073/pnas.88.11.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Ishihara T, Robberecht P, Libert F, Parmentier M, Christophe J, Vassart G. RDC1 may not be VIP receptor. Trends Pharmacol Sci. 1992;13:102–103. doi: 10.1016/0165-6147(92)90037-7. [DOI] [PubMed] [Google Scholar]
- Tatemoto K, Hosoya M, Habata Y, Fujii R, Kakegawa T, Zou MX, Kawamata Y, Fukusumi S, Hinuma S, Kitada C, et al. Isolation and characterization of a novel endogenous peptide ligand for the human APJ receptor. Biochem Biophys Res Commun. 1998;251:471–476. doi: 10.1006/bbrc.1998.9489. [DOI] [PubMed] [Google Scholar]
- Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, et al. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf ME, Gerard N, et al. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Kamohara M, Sugimoto T, Hidaka K, Takasaki J, Saito T, Okada M, Yamaguchi T, Furuichi K. The novel G-protein coupled receptor SALPR shares sequence similarity with somatostatin and angiotensin receptors. Gene. 2000;248:183–189. doi: 10.1016/s0378-1119(00)00123-2. [DOI] [PubMed] [Google Scholar]
- Herold CL, Li Q, Schachter JB, Harden TK, Nicholas RA. Lack of nucleotide-promoted second messenger signaling responses in 1321N1 cells expressing the proposed P2Y receptor, p2y7. Biochem Biophys Res Commun. 1997;235:717–721. doi: 10.1006/bbrc.1997.6884. [DOI] [PubMed] [Google Scholar]
- Yokomizo T, Izumi T, Chang K, Takuwa Y, Shimizu T. A G-protein-coupled receptor for leukotriene B4 that mediates chemotaxis. Nature. 1997;387:620–624. doi: 10.1038/42506. [DOI] [PubMed] [Google Scholar]
- Lynch KR, O'Neill GP, Liu Q, Im DS, Sawyer N, Metters KM, Coulombe N, Abramovitz M, Figueroa DJ, Zeng Z, et al. Characterization of the human cysteinyl leukotriene CysLT1 receptor. Nature. 1999;399:789–793. doi: 10.1038/21658. [DOI] [PubMed] [Google Scholar]
- Takasaki J, Kamohara M, Matsumoto M, Saito T, Sugimoto T, Ohishi T, Ishii H, Ota T, Nishikawa T, Kawai Y, et al. The molecular characterization and tissue distribution of the human cysteinyl leukotriene CysLT(2) receptor. Biochem Biophys Res Commun. 2000;274:316–322. doi: 10.1006/bbrc.2000.3140. [DOI] [PubMed] [Google Scholar]
- Le Y, Yang Y, Cui Y, Yazawa H, Gong W, Qiu C, Wang JM. Receptors for chemotactic formyl peptides as pharmacological targets. Int Immunopharmacol. 2002;2:1–13. doi: 10.1016/s1567-5769(01)00150-3. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR, Ebisawa T, Mahle CD, Kolakowski LF., Jr Cloning of a melatonin-related receptor from human pituitary. FEBS Lett. 1996;386:219–224. doi: 10.1016/0014-5793(96)00437-1. [DOI] [PubMed] [Google Scholar]
- Parker R, Liu M, Eyre HJ, Copeland NG, Gilbert DJ, Crawford J, Sutherland GR, Jenkins NA, Herzog H. Y-receptor-like genes GPR72 and GPR73: molecular cloning, genomic organisation and assignment to human chromosome 11q21.1 and 2p14 and mouse chromosome 9 and 6. Biochim Biophys Acta. 2000;1491:369–375. doi: 10.1016/s0167-4781(00)00023-3. [DOI] [PubMed] [Google Scholar]
- Hsu S, Nakabayashi K, Nishi S, Kumagai J, Kudo M, Sherwood O, Hsueh A. Activation of orphan receptors by the hormone relaxin. Science. 2002;295:671–674. doi: 10.1126/science.1065654. [DOI] [PubMed] [Google Scholar]
- Schoneberg T, Schulz A, Grosse R, Schade R, Henklein P, Schultz G, Gudermann T. A novel subgroup of class I G-protein-coupled receptors. Biochim Biophys Acta. 1999;1446:57–70. doi: 10.1016/s0167-4781(99)00081-0. [DOI] [PubMed] [Google Scholar]
- Li Q, Schachter JB, Harden TK, Nicholas RA. The 6H1 orphan receptor, claimed to be the p2y5 receptor, does not mediate nucleotide-promoted second messenger responses. Biochem Biophys Res Commun. 1997;236:455–460. doi: 10.1006/bbrc.1997.6984. [DOI] [PubMed] [Google Scholar]
- Janssens R, Boeynaems JM, Godart M, Communi D. Cloning of a human heptahelical receptor closely related to the P2Y5 receptor. Biochem Biophys Res Commun. 1997;236:106–112. doi: 10.1006/bbrc.1997.6895. [DOI] [PubMed] [Google Scholar]
- Lee DK, Lynch KR, Nguyen T, Im DS, Cheng R, Saldivia VR, Liu Y, Liu IS, Heng HH, Seeman P, et al. Cloning and characterization of additional members of the G protein- coupled receptor family. Biochim Biophys Acta. 2000;1490:311–323. doi: 10.1016/s0167-4781(99)00241-9. [DOI] [PubMed] [Google Scholar]
- Guo Z, Liliom K, Fischer DJ, Bathurst IC, Tomei LD, Kiefer MC, Tigyi G. Molecular cloning of a high-affinity receptor for the growth factor-like lipid mediator lysophosphatidic acid from Xenopus oocytes. Proc Natl Acad Sci USA. 1996;93:14367–14372. doi: 10.1073/pnas.93.25.14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowsky B, Adham N, Jones K, Raddatz R, Artymyshyn R, Ogozalek K, Durkin M, Lakhlani PP, Bonini JA, Pathirana S, et al. Trace amines: identification of a family of mammalian G protein-coupled receptors. Proc Natl Acad Sci USA. 2001;98:8966–8971. doi: 10.1073/pnas.151105198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo P, Grazzini E, Groblewski T, O'Donnell D, Roy M, Zhang J, Hoffert C, Cao J, Schmidt R, Pelletier M, et al. Proenkephalin A gene products activate a new family of sensory neuron-specific GPCRs. Nat Neurosci. 2002;5:201–209. doi: 10.1038/nn815. [DOI] [PubMed] [Google Scholar]
- Liu M, Parker RM, Darby K, Eyre HJ, Copeland NG, Crawford J, Gilbert DJ, Sutherland GR, Jenkins NA, Herzog H. GPR56, a novel secretin-like human G-protein-coupled receptor gene. Genomics. 1999;55:296–305. doi: 10.1006/geno.1998.5644. [DOI] [PubMed] [Google Scholar]
- McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, Thompson N, Solari R, Lee MG, Foord SM. RAMPs regulate the transport and ligand specificity of the calcitonin- receptor-like receptor. Nature. 1998;393:333–339. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- Muff R, Born W, Fischer J. Adrenomedullin and related peptides: receptors and accessory proteins. Peptides. 2001;22:1765–1772. doi: 10.1016/s0196-9781(01)00515-0. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov
- The Swiss 7TM Search Tool http://www.expasy.ch/cgi-bin/search-7tm
- European Bioinformatics Institute http://www.ebi.ac.uk
- GPCRDB: Information system for G protein-coupled receptors (GPCRs) http://www.gpcr.org
- PubMed http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999:95–98. [Google Scholar]
- GeneDoc http://www.psc.edu/biomed/genedoc/
- Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- TMpred - Prediction of Transmembrane Regions and Orientation http://www.ch.embnet.org/software/TMPRED_form.html
- PHYLIP http://evolution.genetics.washington.edu/phylip.html
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A data table with names, synonyms and accession numbers of all GPCRs
Names, synonyms and accession numbers of all GPCRs, and the BLASTP results of all GPCRs full-length sequences
Names, synonyms and accession numbers of all GPCRs, and the BLASTP results of all GPCRs sequences without amino or carboxyl termini


