Abstract
From a forward genetic screen for phagocytosis mutants in Drosophila melanogaster, we identified a mutation that affects peptidoglycan recognition protein (PGRP) SC1a and impairs the ability to phagocytose the bacteria Staphylococcus aureus, but not Escherichia coli and Bacillus subtilis. Because of the differences in peptidoglycan peptide linkages in these bacteria, our data suggest that PGRP-SC1a is necessary for recognition of the Lys-type peptidoglycan typical of most Gram+ bacteria. PGRP-SC1a mutants also fail to activate the Toll/NF-κB signaling pathway and are compromised for survival after S. aureus infection. This mutant phenotype is the first found for an N-acetylmuramoyl-l-alanine amidase PGRP that cleaves peptidoglycan at the lactylamide bond between the glycan backbone and the crosslinking stem peptides. By generating transgenic rescue flies that express either wild-type or a noncatalytic cysteine–serine mutant PGRP-SC1a, we find that PGRP-SC1a amidase activity is not necessary for Toll signaling, but is essential for uptake of S. aureus into the host phagocytes and for survival after S. aureus infection. Furthermore, we find that the PGRP-SC1a amidase activity can be substituted by exogenous addition of free peptidoglycan, suggesting that the presence of peptidoglycan cleavage products is more important than the generation of cleaved peptidoglycan on the bacterial surface for PGRP-SC1a mediated phagocytosis.
Keywords: antimicrobial peptides, N-acetylmuramoyl-l-alanine amidase, pattern recognition receptor
Host receptors must recognize a microbe before any immune response can begin. After recognition, signaling pathways are activated and result in effector responses such as induction of antimicrobial peptides (AMPs), melanization, and phagocytosis, which are important for controlling the infection. Much is known about the signaling pathways important for the AMP response in Drosophila, but less is known about the regulation of the other two responses. In Drosophila, if phagocytosis is blocked in a mutant that is already impaired for the AMP response, a normally innocuous Escherichia coli infection becomes lethal (1). Phagocytosis is also likely to be a first line of defense, because the response involves specific interaction between microbe and host receptors and occurs within half an hour (1), whereas the induction of AMPs occurs over several hours and AMPs can act on diverse groups of microbes.
The peptidoglycan recognition proteins (PGRPs) are critical receptors in the Drosophila immune response that are required for the recognition of peptidoglycan, a component of bacterial cell walls (2, 3), and for subsequent activation of AMP gene expression (4–10). PGRPs were first characterized in the moths Bombyx mori and Trichoplusia ni (11–14) and proposed to be receptors that can trigger immune responses. PGRPs have also been identified in mammals, and mutant PGRP mice have been generated (15, 16), but the most comprehensive characterization of PGRPs has been performed in the genetic model organism Drosophila melanogaster.
Drosophila has 13 PGRP genes, six long (L) forms with four that are predicted to reside in the plasma membrane, and seven short forms (S) that are all predicted to be secreted (7, 8, 17). PGRPs share homology with N-acetylmuramoyl-l-alanine amidases, which cleave peptidoglycan at the lactylamide bond between the glycan backbone and the stem peptides (11). Some PGRPs, such as PGRP-LC, -LE -SA, and -SD, lack a critical cysteine in the catalytic pocket and are not able to cleave peptidoglycan (18). PGRP-LC, -LE, and -SA have been demonstrated to bind peptidoglycan and are necessary for expression of AMP genes (6, 10, 17, 19, 20), supporting the hypothesis that PGRPs directly recognize bacteria and activate immune responses. The identification of mutations in PGRP-SA (seml) and PGRP-LC (ird7 or totem) indicated that these genes are necessary for activation of the two signaling pathways regulating AMP gene expression, the Toll pathway that responds to Gram+ bacteria and fungi and the Imd pathway that responds to Gram– bacteria (4, 5, 8, 21). Drosophila uses PGRP-SA and PGRP-LC to distinguish between Gram+ and Gram– PGN for activation of the Toll and Imd signaling pathways (7). Gram+ PGN and Gram– PGN differ in the stem peptide portion; typical Gram+ bacteria have a lysine as the third amino acid, whereas Gram– bacteria and the Gram+ Bacillus have a diaminopimelic acid (DAP) in that position (22). Two other noncatalytic PGRPs, PGRP-LE and PGRP-SD, also play a role in activation of the Imd and Toll pathways, respectively (23, 24). PGRP-LC, PGRP-LE double mutants show a more dramatic phenotype to Bacillus and Gram– bacterial infection than either mutation alone, suggesting that PGRP-LE acts with PGRP-LC in the recognition of Gram– DAP-type peptidoglycan for the activation of the Imd pathway (24). PGRP-SD may be playing a similar role with PGRP-SA for activation of the Gram+/Toll pathway (23).
Catalytic PGRPs, such as PGRP-SC1a and -SC1b, include this cysteine residue in the active site, and are potent enzymes that cleave peptidoglycan between the N-acetylmuramic acid of the backbone and the l-alanine in the stem peptide (18). After digestion with PGRP-SC1b, staphylococcal peptidoglycan exhibits less activation of the AMP genes in a Drosophila blood cell line, so it was hypothesized that catalytic PGRPs may act as scavengers to limit an inflammatory response to free peptidoglycan (18). This may not be absolute, as PGRP-SC1b-digested Gram– peptidoglycan is still able to activate the AMP response in S2 cells, albeit at a higher dose (6). However, we wanted to examine the role of these genes in vivo.
In a genetic screen for phagocytosis mutants, we identified a novel mutant, picky. picky flies fail to phagocytose S. aureus particles but can phagocytose E. coli, Bacillus subtilis, and Saccharomyces cerevisiae zymosan particles. picky mutants have defects in the activation of the Toll pathway. picky maps to the location of the PGRP-SC1a, -SC1b, and -SC2 genes, and picky flies express significantly less PGRP-SC1a. The picky defects can be rescued by expression of PGRP-SC1a, indicating that PGRP-SC1a is important for phagocytosis and activation of AMP responses. These results differ from the prevailing model of catalytic PGRPs as scavengers and suggest that in vivo, PGRP-SC1a is required for initiating immune pathways. By comparing a wild-type PGRP-SC1a with a cysteine–serine mutant PGRP-SC1a for rescue of picky phenotypes, we find that the catalytic activity is essential for phagocytosis of live S. aureus, but not for activation of the Toll pathway.
Results
picky, a Mutation Affecting Phagocytosis of S. aureus and the Induction of Antibacterial Responses. To identify genes important for phagocytosis, we screened a collection of ethylmethane sulfonate-induced adult viable mutants (25) using an in vivo phagocytosis assay (1). To determine what a mutant might look like, we first examined the PGRP-LC (ird7) and PGRP-SA (seml) mutants. PGRP-LC was reported to be a recognition receptor for phagocytosis of E. coli in S2 cells (9). However, this study contradicted another report citing that ird7 blood cells can phagocytose bacteria (4). We found that ird7 mutants were able to phagocytose both E. coli and S. aureus particles (Fig. 1). This finding suggested that phagocytosis of Gram– bacteria may require other receptors in addition to PGRP-LC in vivo. In contrast, we found that seml mutants were specifically impaired in their ability to phagocytose S. aureus but not E. coli (Fig. 1). We found that 94% (29 of 31 flies) of the seml mutant flies were able to phagocytose E. coli, but only 25% (17 of 69 flies) were able to phagocytose S. aureus. This finding suggests that PGRP-SA may be important for recognition of Gram+ bacteria for phagocytosis in addition to its role in activating the Toll pathway. In contrast, flies with mutations affecting other Toll pathway components, spatzle (the Toll ligand) and Dif (an NF-κB) were still able to phagocytose S. aureus (Fig. 5, which is published as supporting information on the PNAS web site), indicating that the phagocytosis defect is not a secondary effect from loss of Toll signaling.
Fig. 1.
picky mutants fail to phagocytose S. aureus but are able to phagocytose E. coli. Wild-type (wt) phagocytosis of fluorescein-conjugated E. coli (Ec) and rhodamine-conjugated S. aureus (Sa) particles are shown. Arrows indicate phagocytes under the cuticle on the dorsal anterior side of the abdomen that have phagocytosed bacterial particles. Also shown are the responses for E. coli and S. aureus phagocytosis in ird7, seml, and picky mutant flies. Both seml and picky mutants are unable to phagocytose S. aureus particles.
From our pilot screen, we identified one mutation that we named picky eaterZ2–4761 (picky) because the mutants fail to phagocytose S. aureus particles, but can still phagocytose E. coli and Saccharomyces cerevisiae zymosan particles (Fig. 1; data not shown). Only 25% of picky mutant flies showed any phagocytosis response to S. aureus, but 83% of those same flies were still able to phagocytose E. coli. To investigate whether the recognition involved the difference in peptidoglycan between Gram+ and Gram– bacteria, we examined phagocytosis of a B. subtilis–GFP strain (27). picky mutants were able to phagocytose B. subtilis (data not shown). This finding suggests that the picky gene product is important for the specific recognition of the Lys-type peptidoglycans, typical of most Gram+ bacteria, but not found in Bacillus spp.
We next examined whether the picky flies were susceptible to S. aureus or E. coli infection by assessing survival over a 30-h period (Fig. 2A). Compared with the isogenic parental flies, picky flies were significantly more susceptible to S. aureus infection (P << 0.001) but not to E. coli or PBS alone (P = 0.60 and P = 0.69, respectively). For comparison with a mutant known to be susceptible to S. aureus (23), the susceptibility of seml to S. aureus infection was also examined (Fig. 6A, which is published as supporting information on the PNAS web site). seml flies were significantly more susceptible than the parental flies (P << 0.001) but not significantly different from picky (P = 0.19).
Fig. 2.
pickyZ4761 mutants are more susceptible to S. aureus infection and fail to induce Drosomycin.(A) picky flies are more sensitive to S. aureus infection than the isogenic parental (iso) wild-type stock. Shown are survival curves over a 30-h period for wild-type and picky flies injected with S. aureus, E. coli, or PBS. Survival after S. aureus injection differed significantly between wild-type and picky (P << 0.001). Additionally, survival of S. aureus injection was very significantly less than survival of PBS injection for picky but not for wild type (P << 0.001 and P = 0.40, respectively). There was no statistically significant difference in survival after E. coli injection between wild type and picky (P = 0.60), or between survival after E. coli or PBS for wild-type or picky adults (P = 0.40 and P = 0.38 respectively). (B) picky mutants are impaired in the induction of Drosomycin but not Diptericin. Shown are bar graphs comparing the wild type with picky with expression of Diptericin and Drosomycin in response to S. aureus (+S) or E. coli (+E) at 2 h compared with uninfected (–). The graphs are normalized to the parental wild-type uninfected (–), and error bars show standard deviation.
Because picky flies appeared to be impaired in the recognition of S. aureus for phagocytosis and survival, it was possible that the mutation might also affect activation of the Toll or Imd pathways. The induction of Drosomycin and Diptericin AMP genes is often used to assess activation of the Toll/Gram+ and Imd/Gram– signaling pathways, respectively (1, 4–6, 9, 28–35). The AMP responses in picky were compared with those of the known Toll pathway components, seml and Dif, and to an Imd pathway component, ird7 (Fig. 6B). picky flies showed no induction of Drosomycin in response to S. aureus, E. coli, or Micrococcus luteus (Figs. 2B and 6B) at either 2 or 24 h. All of the S. aureus-infected picky flies were dead at 24 h, so the Drosomycin expression in this sample was not assessed. seml and Dif both show some induction of Drosomycin at 2 and 24 h. In comparison, picky has a more consistent and dramatic effect on Drosomycin expression and likely encodes an essential component of the Toll pathway. In contrast, the expression of Diptericin in response to bacterial infection in picky mutants was higher than that seen in wild type (Fig. 2B). Therefore, the defect in picky appears to be selective for the Toll pathway. To determine where in the Toll pathway picky might lie, we crossed the picky mutation into a Toll10b gain of function mutant. picky was not able to suppress the constitutive expression of Drosomycin in a Toll10b mutant (Fig. 6C). This epistasis analysis places picky either upstream of Toll, which would be consistent with a role in bacterial recognition or in a parallel pathway.
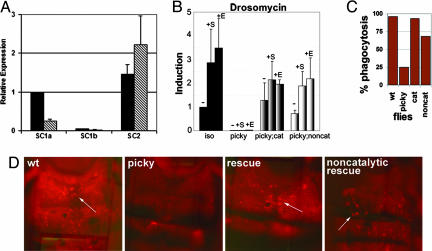
picky Mutants Are Rescued with Transgenic Expression of PGRP-SC1a. Because the phenotype of picky flies suggested that recognition occurring at the peptidoglycan level was affected, the PGRP genes were obvious candidate genes to test. The picky mutation was analyzed in trans to overlapping deficiency chromosomes in the region of the three PGRP genes, PGRP-SC1a, -SC1b, and -SC2, on the second chromosome. Two deficiencies failed to complement picky for S. aureus phagocytosis: Df H3E1 that deletes a genomic interval 44D1–4 to 44F12 and Df H3D3 that deletes 44D1–4 to 44F4–5. picky does complement Df 44Ce, which deletes 44C1–2 to 44E1–4, and Df H3C1 deletes 43F to 44D3–8 (data not shown). picky maps to the 44E1–4; 44F4–5 interval, which likely contains the PGRP gene cluster at 44E2. The ORFs of these three genes showed no sequence changes in the picky mutant. We examined the expression levels of 16 genes in the 100 kb surrounding the PGRP-SC1/2 locus and found that only the PGRP-SC1a and PGRP-SC1b expression levels were decreased by >50% in the picky mutants (Fig. 3A and Fig. 7, which is published as supporting information on the PNAS web site). PGRP-SC1a and PGRP-SC1b are identical in amino acid sequence; PGRP-SC2 is 68% identical to PGRP-SC1a in amino acid sequence. Specific probes designed to distinguish between the three genes with quantitative real-time PCR were used to analyze their levels of gene expression in mutant vs. parental flies. PGRP-SC1a expression levels were significantly reduced (25% of wild-type levels). PGRP-SC1b was expressed at half the level in the mutant compared with wild type. A comparison of the relative expression levels of PGRP-SC1a and PGRP-SC1b in wild-type flies indicated that PGRP-SC1a was the predominant message in parental adult flies and contributed to >95% of the total PGRP-SC1a and -SC1b RNA. PGRP-SC2 was expressed in the mutant at wild-type levels. The primary site of expression of PGRP-SC1a/b had been shown to be the gut (17), raising one question as to whether it could be found in the hemolymph to function in phagocytosis and AMP responses. All three PGRP transcripts could be detected in the adult gut, hemocytes, and fat body (Fig. 8, which is published as supporting information on the PNAS web site); however, the expression of PGRP-SC1a in the gut is 15-fold higher than that seen in the hemocytes or fat body. In sum, these results indicated that the picky mutation is likely a regulatory mutation affecting the expression of PGRP-SC1a.
Fig. 3.
picky mutant phenotype is due to loss of PGRP-SC1a. (A) QRT-PCR using SYBR-green to detect expression levels of PGRP-SC1a, PGRP-SC1b, and PGRP-SC2 in wild-type (black bars) and picky (gray bars) flies. The samples were normalized to the wild-type PGRP-SC1a transcript level for cross comparison. Error bars indicate standard error (n = 6). (B–D) Both the catalytic and the cysteine–serine noncatalytic PGRP-SC1a transgenic rescue flies are able to rescue induction of Drosomycin in response to S. aureus (+S) or E. coli (+E) as compared with uninfected (–) (B) and phagocytosis of S. aureus particles (C and D). The bar graph shows the percentage of animals showing phagocytosis of S. aureus particles.
To verify that the picky immune defects were due to a loss of PGRP-SC1a function, we used P element-mediated transformation to express PGRP-SC1a under the control of a UAS promoter in the mutant flies. To determine the role of the catalytic activity, a wild-type PGRP-SC1a and a catalytically dead mutant PGRP-SC1a (PGRP-SC1am) were made and compared for their ability to rescue the picky phenotypes. To generate the catalytically dead PGRP-SC1a, a mutation changing the active site cysteine-168 to a serine was introduced. This mutation should generate a PGRP-SC1a that can still bind, but can no longer cleave peptidoglycan (18). Both transgenes were able to rescue the picky AMP phenotype and the initial phagocytosis defect observed in the picky mutant (Fig. 3). Three independent PGRP-SC1a transgenic lines were tested for rescue, and 93% of the flies (27 of 29) examined recovered the ability to phagocytose S. aureus particles (Fig. 3C). With expression of the cysteineserine mutant PGRP-SC1am, 68% (19 of 28) of the flies examined showed phagocytosis of S. aureus particles, but the intensity of fluorescence observed in the phagocytes was much less than seen in wild type or with the catalytic rescue. The ability of the transgenes to rescue both AMP responses and phagocytosis defect confirmed that loss of PGRP-SC1a in picky flies was indeed the cause of the observed immune phenotypes. These results also suggested that the catalytic activity of PGRP-SC1a may be dispensable for these two immune functions.
PGRP-SC1a Catalytic Activity Is Essential for Recognition and Clearance of S. aureus. To uncover the role of the catalytic activity in PGRP-SC1a function, it made sense to examine the host response to live bacteria, instead of microbial particles. S. aureus injection is lethal to wild-type flies, but there are significant differences in susceptibility between parental and picky mutant flies within the first 24 h (Fig. 2 A). picky mutants showed a dramatic die-off within the first 12 h, whereas wild-type flies took 24 h to reach the same percent death. The catalytic PGRP-SC1a rescue line is able to rescue survival, but not to wild-type levels (Fig. 4A). The expression of the transgene with the UAS-GAL4 system may not fully replicate wild-type regulation of PGRP-SC1a. In contrast, the cysteine–serine mutant PGRP-SC1am is not able to rescue to a significant degree (P > 0.01) and shows susceptibility similar to the picky mutants. This result indicates that despite the activation of the Toll pathway and phagocytosis of S. aureus particles in the cysteine–serine mutant rescue flies, this is not sufficient for rescue of survival to S. aureus infection.
Fig. 4.
PGRP-SC1a catalytic activity is necessary to survive S. aureus infection. (A) Shown are survival curves comparing the responses of wild type, picky, catalytic rescue, and noncatalytic rescue flies with S. aureus infection. The catalytic rescue showed a significant increase in survival, as compared with picky mutants (P << 0.01), but remained below the survival levels of wild-type adults. The noncatalytic rescue did not show very significantly increased survival as compared with picky (P > 0.01). Injection of PBS in the two rescue lines was not significantly different from PBS injections in wild type and picky (all P >> 0.01) (data not shown). (B) picky mutants and the noncatalytic rescue flies are impaired in their ability to clear S. aureus–GFP bacteria from the hemocoel. The ability to collect S. aureus–GFP into blood cells corresponds with ability to survive infection. Shown are wild type at 12 h (the 24 h time point looks the same), picky at 12 h (the 24 h time point does not survive), and catalytic and noncatalytic rescue flies at 24 h after S. aureus-GFP infection.
To try and identify a difference between the wild-type PGRP-SC1a and the cysteine–serine mutant PGRP-SC1am rescue that accounts for differences in survival, S. aureus-GFP (36) infection was monitored in the wild-type, picky, and rescue lines over the course of a day. The two rescue lines differed in their ability to clear an S. aureus-GFP infection that correlated well with their ability to survive infection (Fig. 4B). In wild-type flies, most of the S. aureus-GFP fluorescence was seen in the blood cells 12 and 24 h after infection. The persistence of fluorescence at 24 h suggested that some S. aureus were able to survive in wild-type phagocytes and may be the cause of the eventual lethality. This clearance assay is similar to the phagocytosis assay in that fluorescent bacteria is seen in the phagocytes, but it differs in that there is no quenching of extracellular fluorescence, so the visualization of S. aureus-GFP in the phagocytes is due to a noticeable relocalization of GFP to those cells. Also, the phagocytosis assay indicates whether any bacteria can be found in the phagocytes after 30 min, whereas this clearance assay gives a sense of the flies' ability to clear or collect all of the S. aureus into phagocytes over the course of a day. In picky flies, the GFP fluorescence remains diffuse throughout the abdomen at 12 h after infection. The phenotype was not assessed at 24 h because all of the picky flies were dead at this time point. The inability of picky flies to noticeably collect the S. aureus-GFP in the phagocytes is consistent with their inability to phagocytose S. aureus particles. All of the wild-type (catalytic) rescue flies were able to gather the S. aureus-GFP into the phagocytes after 24 h, indicating rescue of the picky phenotype. In contrast, none of the cysteine–serine mutant (noncatalytic) rescue flies were able to collect the S. aureus-GFP into the phagocytes, even after 24 h. Overall, this clearance assay proved to be quite robust; of the 30–45 flies representing each class that were examined, all of the flies within a given class showed identical responses to that shown in Fig. 4B. These results indicate that the catalytic activity of PGRP-SC1a is absolutely required for efficient phagocytosis of live S. aureus over time. The clearance results correlate well with the survival curves, indicating that the ability to collect S. aureus into the phagocytes is essential for staving off an early death.
Addition of Free Peptidoglycan Can Substitute for the PGRP-SC1a Catalytic Activity. We were curious as to how the presence of free peptidoglycan, either shed from a growing bacterium or digested from the bacterial cell wall by a catalytic PGRP like PGRP-SC1a, might affect the host phagocytosis response. To address this question, the effect of conditioned bacterial growth media on the ability of the picky mutant and transgenic rescue flies to phagocytose S. aureus-GFP was examined. A mid-log culture of S. aureus left in LB growth media should contain shed peptidoglycan, whereas S. aureus grown in LB, then washed and resuspended in PBS, will not. picky mutants were unable to phagocytose either preparation of S. aureus-GFP bacteria within 30 min, whereas wild type and the catalytic rescue were able to phagocytose both (data not shown). Of interest, the cysteine–serine mutant rescue flies were unable to phagocytose S. aureus in PBS, but were able to phagocytose S. aureus in growth media. This result indicates that the catalytic activity is absolutely necessary for recognition and phagocytosis of live S. aureus bacteria in PBS. To determine whether the effect is due to free peptidoglycan, PBS-washed bacteria were supplemented with increasing amounts of S. aureus peptidoglycan. This was sufficient to restore phagocytosis in the cysteine–serine mutant rescue line (Fig. 9, which is published as supporting information on the PNAS web site) but not in the picky mutant (data not shown). Of note, the addition of exogenous peptidoglycan was also sufficient to enable the cysteine–serine (noncatalytic) rescue flies to rescue the picky S. aureus survival defect (Fig. 9). Again, this rescue was not to wild-type levels, but was significantly improved relative to the picky mutant (P < 0.01) and was similar to that seen with the catalytic rescue line (P = 0.32). Together, these results suggest that the cleavage of peptidoglycan can be substituted by the addition of free peptidoglycan and that PGRP-SC1a needs this free peptidoglycan to efficiently recognize and phagocytose S. aureus.
Discussion
The peptidoglycan recognition protein family is important for allowing the Drosophila immune response to distinguish between Gram+ and Gram– bacteria for the activation of specific AMP signaling pathways (4–10). Our data indicate that phagocytosis also relies on PGRPs to distinguish between Gram+ and Gram– bacterial peptidoglycan. We report that a catalytic PGRP, PGRP-SC1a, is essential for the phagocytosis of S. aureus and the activation of AMP responses. The fact that PGRP-SC1a mutants have a striking phagocytosis defect indicates that it is absolutely required and that other PGRPs are not able to substitute for its loss of function. PGRP-SC1a differs from the existing PGRP mutants in that it has catalytic activity that is essential for efficient phagocytosis and ultimately limiting the infection process. Hence, noncatalytic PGRPs, such as PGRP-SA, may not be able to substitute for PGRP-SC1a function. Because PGRP-SC1a is likely present in the hemolymph, it may be acting as an opsonin to bind bacteria, with the PGRP-bacterial complex then being recognized by transmembrane phagocytosis receptors that complete the uptake into the phagocyte. An alternative model is that PGRP-SC1a may generate peptidoglycan cleavage products that function as immune modulators to stimulate phagocytic activity.
Our result showing that ird7 mutants can phagocytose E. coli in vivo differs from a previous report that found that RNAi of PGRP-LC caused a defect in the phagocytosis of E. coli in S2 cells (9). This discrepancy in results may reflect inherent differences in the assays. The S2 cell phagocytosis assay is able to detect quantitative differences in phagocytosis using fluorescence-activated sorting, but does not directly distinguish between bacteria at the surface and internalized bacteria (21). The in vivo assay by our method is not as quantitative but is able to test mutations in the animal where the phagocytes are in their natural cellular environment and detects only internalized bacteria. Our experiments suggest that Drosophila adults may require other receptors in addition to PGRP-LC for the cellular recognition of Gram– bacteria in vivo. The fact that PGRP-LC mutants can still phagocytose E. coli, but fail to activate AMP responses (4–7, 9, 10), also indicates that the recognition events activating these two responses can be distinct.
For S. aureus, PGRP-SC1a is necessary for recognition of bacteria for both the phagocytosis and AMP responses. However, for E. coli, PGRP-SC1a is similar to PGRP-LC in that it is not necessary for recognition for phagocytosis, but is necessary for the activation of an AMP response. This finding raises the interesting issue of the role PGRPs play in recognizing Gram– bacteria. In Gram– bacteria, the peptidoglycan is buried under an outer membrane, so it has been proposed that the small amounts of PGN that are shed may be sufficient to trigger the AMP response (7). Because the cell wall PGN is not easily accessible, it makes sense that the PGRPs are not playing a direct role in recognition for phagocytosis. An alternate possibility is that the exposure of bacterial peptidoglycan occurs after the initial uptake into phagocytes, and at that later point, is then recognized by PGRP-LC and PGRP-SC1a to trigger AMP responses. There is precedence for intracellular recognition in the phagosome in mammalian immune responses, with studies indicating that mammalian short PGRPs function in the phagosome to inhibit bacterial growth (12, 15, 37) and the observation that Toll-like receptor recognition of bacteria in the phagosome can influence the rate of phagosome maturation (38). A related possibility is that some basal level of activation of PGRP-SC1a and the Toll pathway may be required for any expression of Drosomycin. picky mutants show much lower expression of Drosomycin than wild type in the absence of infection, so perhaps an induction may not be detectable.
We also found that seml mutants are defective in the phagocytosis of S. aureus but not E. coli. Recent reports have indicated that the processing of peptidoglycan into monomers, lactyltetrapeptides, or muropeptides, can yield potent activators of the AMP response for both the Imd (6) and the Toll pathway (39). It was further suggested for the Toll pathway (39) that processing of peptidoglycan may be a prerequisite step upstream of PGRP-SA/seml recognition and activation of the Toll AMP pathway. This model would be consistent with catalytic PGRPs, such as PGRP-SC1a, playing a supporting role in the processing of peptidoglycan for initiating recognition events.
As many PGRPs are clearly required for activation of immune effector responses, the issue arises as to the specificity of PGRPs for their substrates. Work from several groups indicates that alternative splice variants (in PGRP-LC) (4, 6, 10) or small differences in amino acid sequences (from PGRP-LB and human PGRP-Iα structural analyses) (40, 41) may result in the ability of different PGRPs to have distinct recognition potential. It will be interesting to explore the range of bacterial types recognized by the 17 Drosophila PGRP proteins and to determine how subtle differences in recognition may be reflected by specific amino acid sequences. There may also be genetic interactions or antagonism in their function, so using the genetic tools available in Drosophila may be necessary for ultimately determining both their unique and redundant roles.
This work demonstrates the utility of a forward genetic approach to understanding the recognition of pathogen types for phagocytosis and activation of AMP responses. It is hoped that identification of genes important for recognition of bacteria in Drosophila should increase our understanding of how the process works in our own immune system.
Materials and Methods
Phagocytosis Assay and Genetic Screen. The assay was done as described (1). Adult flies were injected in the abdomen with fluorescently labeled microbial particles or bacteria expressing GFP. After 30 min, trypan blue was injected; if hemocytes take up the fluorescent particles, the fluorescence can be visualized through the cuticle on the dorsal side of the abdomen. For the genetic screen, we injected a 4 mg/ml mixture of rhodamine-labeled S. aureus particles and fluorescein-labeled E. coli particles (Molecular Probes). Injecting a mixture provides an internal control because most of our mutants are able to phagocytose one particle type but not the other. The phagocytosis assay is not quantitative with our method, although we can get a sense whether the fluorescent signal is weaker than normal or there is a decrease in the number of cells showing phagocytosis. Because of the range in the fluorescent signal or numbers of phagocytes showing fluorescence in wild-type, we use a stringent criteria and score lines with any evident fluorescence in phagocytes as positive for phagocytosis. In a potential phagocytosis mutant, we may see anywhere between 0 to 25% of the ≥10 flies examined as still being able to phagocytose some bacteria. For phagocytosis mutants, the assay was repeated several times (in ≈30–40 animals total) to verify the phenotype. For phenotypic analysis of the picky mutant, the isogenic parental cn bw strain that was mutagenized to generate the mutant (25) was used as the wild-type control.
Reverse Transcription and Quantitative Real-Time PCR. For AMP expression, for the 2-h time point, adult flies were injected with either an overnight culture of E. coli, M. luteus, or S. aureus resuspended in PBS. For the 24-h time point, adult flies were injected with a 1:100 dilution of a mid-log culture (OD600 0.5–0.6) of E. coli, M. luteus, or S. aureus that was washed and resuspended in PBS. RNA was isolated from adult flies by homogenizing flies in STAT-60 buffer according to the manufacturer's protocol (Isotex Diagnostics). The RNA was digested with RNase-free DNase, then subjected to reverse transcription using Superscript II (Invitrogen) and quantitative real-time PCR using either LUX probes (Invitrogen) or SYBR-green (Applied Biosystems) on an ABI 5700 following the manufacturers' protocols. The specific primers used can be found in Supporting Text, which is published as supporting information on the PNAS web site. The experiments were done in triplicate and the error bars represent standard deviation.
Generation of Transgenic Rescue Flies. The cloning and mutagenesis strategy to generate UAS-PGRP-SC1a and the UAS-PGRP-SC1am (noncatalytic version) are detailed in Supporting Text. Once transformant flies were identified, the transgenic rescue chromosomes were crossed into the picky mutant background by using standard genetic crosses. These flies were then crossed to flies carrying picky and a T155-GAL4 driver that expresses GAL4 in the lymph gland, fat body, and other tissues (26).
Survival Experiments. Adult flies were aged to at least 5 days old and injected in the abdomen with either PBS as a control or a 1:100 dilution of a mid-log culture (OD600 0.5–0.6) of either E. coli or S. aureus, washed and resuspended in PBS by using a Pneumatic PicoPump PV820 (World Precision Instruments) for injections. Survival after injection was assessed every 6 h over a 30-h period. Sixty flies were injected for each experiment, and the experiments were done in triplicate. A log-rank test was used to compare survival curves, and P values <0.01 were deemed significant.
Supplementary Material
Acknowledgments
We thank A. Fitzgerald for initial work on the phagocytosis screen; M. Erdinc and K. Randle for technical assistance; E. Koundakjian, M. Cahill, and C. Zuker for the EMS collection of adult viable lines; D. Schneider for advice on cellular assays; A. Cheung (Dartmouth, Hanover, NH) for S. aureus–GFP; D. Rudner (Harvard Medical School, Boston) for B. subtilis–GFP; D. Ferrandon, K. Anderson, and the Bloomington Drosophila Stock Center for fly stocks; Duke University Model Systems Genomics Unit for P-element transformation services; and E. Baehrecke, D. Mosser, N. Silverman, and Wu laboratory members for helpful comments on the manuscript. The work was supported in part by National Institutes of Health Grant GM62316, start-up funds from University of Maryland Biotechnology Institute, and a Howard Hughes Medical Institute Undergraduate Research Fellowship (to L.S.G.).
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AMP, antimicrobial peptide; PGRP, peptidoglycan recognition protein.
References
- 1.Elrod-Erickson, M., Mishra, S. & Schneider, D. (2000) Curr. Biol. 10, 781–784. [DOI] [PubMed] [Google Scholar]
- 2.Dziarski, R. (2004) Mol. Immunol. 40, 877–886. [DOI] [PubMed] [Google Scholar]
- 3.Steiner, H. (2004) Immunol. Rev. 198, 83–96. [DOI] [PubMed] [Google Scholar]
- 4.Choe, K. M., Werner, T., Stoven, S., Hultmark, D. & Anderson, K. V. (2002) Science 296, 359–362. [DOI] [PubMed] [Google Scholar]
- 5.Gottar, M., Gobert, V., Michel, T., Belvin, M., Duyk, G., Hoffmann, J. A., Ferrandon, D. & Royet, J. (2002) Nature 416, 640–644. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko, T., Goldman, W. E., Mellroth, P., Steiner, H., Fukase, K., Kusumoto, S., Harley, W., Fox, A., Golenbock, D. & Silverman, N. (2004) Immunity 20, 637–649. [DOI] [PubMed] [Google Scholar]
- 7.Leulier, F., Parquet, C., Pili-Floury, S., Ryu, J. H., Caroff, M., Lee, W. J., Mengin-Lecreulx, D. & Lemaitre, B. (2003) Nat. Immunol. 4, 478–484. [DOI] [PubMed] [Google Scholar]
- 8.Michel, T., Reichhart, J. M., Hoffmann, J. A. & Royet, J. (2001) Nature 414, 756–759. [DOI] [PubMed] [Google Scholar]
- 9.Ramet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B. & Ezekowitz, R. A. (2002) Nature 416, 644–648. [DOI] [PubMed] [Google Scholar]
- 10.Werner, T., Borge-Renberg, K., Mellroth, P., Steiner, H. & Hultmark, D. (2003) J. Biol. Chem. 278, 26319–26322. [DOI] [PubMed] [Google Scholar]
- 11.Kang, D., Liu, G., Lundstrom, A., Gelius, E. & Steiner, H. (1998) Proc. Natl. Acad. Sci. USA 95, 10078–10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, C., Gelius, E., Liu, G., Steiner, H. & Dziarski, R. (2000) J. Biol. Chem. 275, 24490–24499. [DOI] [PubMed] [Google Scholar]
- 13.Ochiai, M. & Ashida, M. (1999) J. Biol. Chem. 274, 11854–11858. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida, H., Kinoshita, K. & Ashida, M. (1996) J. Biol. Chem. 271, 13854–13860. [DOI] [PubMed] [Google Scholar]
- 15.Dziarski, R., Platt, K. A., Gelius, E., Steiner, H. & Gupta, D. (2003) Blood 102, 689–697. [DOI] [PubMed] [Google Scholar]
- 16.Xu, M., Wang, Z. & Locksley, R. M. (2004) Mol. Cell. Biol. 24, 7949–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werner, T., Liu, G., Kang, D., Ekengren, S., Steiner, H. & Hultmark, D. (2000) Proc. Natl. Acad. Sci. USA 97, 13772–13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellroth, P., Karlsson, J. & Steiner, H. (2003) J. Biol. Chem. 278, 7059–7064. [DOI] [PubMed] [Google Scholar]
- 19.Chang, C. I., Pili-Floury, S., Herve, M., Parquet, C., Chelliah, Y., Lemaitre, B., Mengin-Lecreulx, D. & Deisenhofer, J. (2004) PLoS Biol. 2, E277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takehana, A., Katsuyama, T., Yano, T., Oshima, Y., Takada, H., Aigaki, T. & Kurata, S. (2002) Proc. Natl. Acad. Sci. USA 99, 13705–13710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramet, M., Pearson, A., Manfruelli, P., Li, X., Koziel, H., Gobel, V., Chung, E., Krieger, M. & Ezekowitz, R. A. (2001) Immunity 15, 1027–1038. [DOI] [PubMed] [Google Scholar]
- 22.Schleifer, K. H. & Kandler, O. (1972) Bacteriol. Rev. 36, 407–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bischoff, V., Vignal, C., Boneca, I. G., Michel, T., Hoffmann, J. A. & Royet, J. (2004) Nat. Immunol. 5, 1175–1180. [DOI] [PubMed] [Google Scholar]
- 24.Takehana, A., Yano, T., Mita, S., Kotani, A., Oshima, Y. & Kurata, S. (2004) EMBO J. 23, 4690–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koundakjian, E. J., Cowan, D. M., Hardy, R. W. & Becker, A. H. (2004) Genetics 167, 203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison, D. A., Binari, R., Nahreini, T. S., Gilman, M. & Perrimon, N. (1995) EMBO J. 14, 2857–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita, M. & Losick, R. (2002) Mol. Microbiol. 43, 27–38. [DOI] [PubMed] [Google Scholar]
- 28.Georgel, P., Naitza, S., Kappler, C., Ferrandon, D., Zachary, D., Swimmer, C., Kopczynski, C., Duyk, G., Reichhart, J. M. & Hoffmann, J. A. (2001) Dev. Cell 1, 503–514. [DOI] [PubMed] [Google Scholar]
- 29.Hedengren, M., Asling, B., Dushay, M. S., Ando, I., Ekengren, S., Wihlborg, M. & Hultmark, D. (1999) Mol. Cell 4, 827–837. [DOI] [PubMed] [Google Scholar]
- 30.Khush, R. S., Cornwell, W. D., Uram, J. N. & Lemaitre, B. (2002) Curr. Biol. 12, 1728–1737. [DOI] [PubMed] [Google Scholar]
- 31.Leulier, F., Rodriguez, A., Khush, R. S., Abrams, J. M. & Lemaitre, B. (2000) EMBO Rep. 1, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, Y., Wu, L. P. & Anderson, K. V. (2001) Genes Dev. 15, 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rutschmann, S., Jung, A. C., Zhou, R., Silverman, N., Hoffmann, J. A. & Ferrandon, D. (2000) Nat. Immunol. 1, 342–347. [DOI] [PubMed] [Google Scholar]
- 34.Silverman, N., Zhou, R., Stoven, S., Pandey, N., Hultmark, D. & Maniatis, T. (2000) Genes Dev. 14, 2461–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vidal, S., Khush, R. S., Leulier, F., Tzou, P., Nakamura, M. & Lemaitre, B. (2001) Genes Dev. 15, 1900–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kahl, B. C., Goulian, M., van Wamel, W., Herrmann, M., Simon, S. M., Kaplan, G., Peters, G. & Cheung, A. L. (2000) Infect. Immun. 68, 5385–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tydell, C. C., Yount, N., Tran, D., Yuan, J. & Selsted, M. E. (2002) J. Biol. Chem. 277, 19658–19664. [DOI] [PubMed] [Google Scholar]
- 38.Blander, J. M. & Medzhitov, R. (2004) Science 304, 1014–1018. [DOI] [PubMed] [Google Scholar]
- 39.Filipe, S. R., Tomasz, A. & Ligoxygakis, P. (2005) EMBO Rep. 6, 327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan, R., Roychowdhury, A., Ember, B., Kumar, S., Boons, G. J. & Mariuzza, R. A. (2004) Proc. Natl. Acad. Sci. USA 101, 17168–17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, M. S., Byun, M. & Oh, B. H. (2003) Nat. Immunol. 4, 787–793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.