Abstract
Mammalian red blood cells, unlike those of other vertebrates, must withstand the rigors of circulation in the absence of new protein synthesis. Key to this is plasma membrane elasticity deriving from the protein spectrin, which forms a network on the cytoplasmic face. Spectrin is a tetramer (αβ)2, made up of αβ dimers linked head to head. We show here that one component of erythrocyte spectrin, αI, is encoded by a gene unique to mammals. Phylogenetic analysis suggests that the other α-spectrin gene (αII) common to all vertebrates was duplicated after the emergence of amphibia, and that the resulting αI gene was preserved only in mammals. The activities of αI and αII spectrins differ in the context of the human red cell membrane. An αI-spectrin fragment containing the site of head-to-head interaction with the β-chain binds more weakly than the corresponding αII fragment to this site. The latter competes so strongly with endogenous αI as to cause destabilization of membranes at 100-fold lower concentration than the αI fragment. The efficacies of αI/αII chimeras indicate that the partial structural repeat, which binds to the complementaryβ-spectrin element, and the adjacent complete repeat together determine the strength of the dimer–dimer interaction on the membrane. Alignment of all available α-spectrin N-terminal sequences reveals three blocks of sequence unique to αI. Furthermore, human αII-spectrin is closer to fruitfly α-spectrin than to human αI-spectrin, consistent with adaptation of αI to new functions. We conclude that αI-spectrin represents a neofunctionalized spectrin adapted to the rapid make and break of tetramers.
Keywords: cytoskeleton, membrane, red blood cell, triple-helix
Red blood cells transport oxygen from respiratory organs to tissues in all vertebrates, but both their overall size and organelle content differ among vertebrate classes. Mammalian erythrocytes are distinguished from those of other vertebrates by their small size and lack of nuclei and other organelles (1). In general, mammalian red cells are smaller than those of birds, which are smaller than those of reptiles (2, 3). The small size and simple structure of mammalian red cells are widely considered to represent an adaptation to homeothermy, because smaller cells are thought to be better-adapted to rapid circulation (3).
To understand the evolutionary origin of mammalian red blood cells, we need to investigate the adaptations that have allowed them to dispense with a nucleus and survive in circulation in the absence of major repair mechanisms. Over the last two decades, it has become clear that the mammalian red cell membrane is exquisitely adapted to survival by an elastic network associated with its cytoplasmic face (4, 5). The protein spectrin is the major filamentous component of the lattice. It is an elongated molecule made up of two types of subunit, α and β. The α and β chains form dimers by lateral association, and pairs of dimers self-associate to make a tetramer.
The elongated shape of spectrin comes from 16–20 consecutive homologous repeating units (6) that are left-handed antiparallel triple-helical coiled coils (7, 8). The tetramer is generated by formation of a complete triple-helical repeating unit from partial repeats (9): a single helix from the N terminus of the α-chain (equivalent to the third helix, helix C, of a full repeat) unites with two helices, equivalent to helices A and B from the β-chain. The association results in a noncovalent triple-helical segment. The first full triple helix adjacent to the partial repeat modulates the affinity of interaction (10, 11).
The sequence of the human genome indicates there are multiple spectrin genes, two α and five β (12). Human red cell spectrin is a product of the SPTA1 (αI-spectrin) and SPTB (βI-spectrin) genes. The other genes are expressed outside red cells; the most wide-spread are products of SPTAN1 (αII-spectrin) and SPTBN1 (βII-spectrin).
Some biochemical differences between αI/αII and βI/βII spectrin are now evident. Thus αI- and βI-spectrins form tetramers with much lower affinity than αII- and βII-spectrins (11). Furthermore, although all of the chains bind phosphatidylserine, the locations of the binding sites differ between erythroid and nonerythroid polypeptides (13).
Now that multiple animal genome sequences are available, we can address the evolutionary origin of the different spectrin genes and their relation to functional aspects of the protein. In this report, we take advantage both of genomic sequences and the properties of cloned fragments of the proteins for this purpose. We report that the gene encoding αI-spectrin is unique to mammals, and that other vertebrates possess only a single common α-spectrin gene, equivalent to the human gene for αII-spectrin. Furthermore, analysis of the activities of fragments of αI- and αII-spectrin in competing with endogenous αI-spectrin in intact red cell membranes reveals adaptation of αI-spectrin as a polypeptide specialized for rapid making and breaking of tetramers in response to shear-induced deformation of the cell.
Results
αI-Spectrin Is a Mammalian Adaptation. We interrogated the ENSEMBL and University of California at Santa Cruz genome databases to identify all α-spectrin genes and the Uniprot and European Molecular Biology Laboratory/GenBank databases to identify their products. Tables 1 and 2, which are published as supporting information on the PNAS web site, lists the sequences used here.
The genes for human, rat, and mouse αI- and αII-spectrins are all annotated in ENSEMBL, and their coding sequences are verifiable by comparison with cDNA/protein sequences. Cow and macaque spectrin genes were readily identified by blast in the preliminary assemblies of their genomes available in pre-ENSEMBL. Although some elephant, sheep, and pig genome and/or EST sequences for both αI- and αII-spectrin are available, there is incomplete coverage of any substantial region, so these are excluded from our analyses. In addition to the sequences from placental (eutherian) mammals, we identified both genes in the marsupial (metatherian) Monodelphis domestica (gray short-tailed opossum) by searching the available genomic scaffolds. Only small amounts of genomic sequence from the monotreme (prototherian) Ornithorhynchus anatinus (platypus) are available, but in the current Whole Genome Shotgun sequence traces are fragments of two α-spectrin genes. We conclude that both genes are represented in the genomes of all three subclasses of the mammals.
Sequences similar to αII-spectrin were evident in all vertebrates in one gene per genome, except in the case of the bony fish Tetraodon nigroviridis. This organism has two α-spectrin genes (ENSEMBL genes GSTENG00004619001 and GSTENG00004620001), but this is unsurprising, because a whole genome duplication event is known in the ray-finned fish lineage (14). GSTENG00004619001 encodes a truncated α-spectrin that is probably a pseudogene. The sequence is very closely related to GSTENG00004620001 and does not represent an αI-spectrin (data not shown).
In each invertebrate genome, an α-spectrin similar to the known Drosophila melanogaster and Caenorhabditis elegans genes was readily detected. The platyhelminth Taenia cellulosae is also represented in the range of sequences in Uniprot. The origins of α-spectrin appear to extend back at least as far as the cnidarian hydra. Several ESTs from Hydra magnipapillata were detected, as well as some genomic sequence traces that appear to be from α-spectrin, as judged by blastx comparison to the whole Uniprot database. Additional ESTs were identified in the hagfish Eptatretus burgeri, the scallop Argopecten irradians, the ascidian Molgula tectiformis, the planarian Dugesia japonica, and the schistosomes Schistosoma mansoni and Schistosoma japonica (data not shown). These sequences emphasize the ubiquity of spectrin in the animal kingdom.
However, although we could detect the gene for αI-spectrin in all available mammal databases, we could not find it in any other animal. We searched exhaustively for further αI-spectrins using a combination of blat and blast searches of genomic sequences available through the genome portals. We also interrogated the EST databases and the raw genomic sequence trace archives by blast. We constructed hidden Markov model sequence profiles for the distinctive structural regions of the α-spectrins (the tetramerforming site, the SH3 domain, and the calmodulin-like domain) and used hmmsearch to analyze the predicted proteins in the Uniprot and ENSEMBL peptide databases. In no case could we detect a second α-spectrin gene in nonmammalian organisms.
These results lead to the conclusion that the αI-spectrin gene is a mammalian adaptation, and further that α-spectrin is ubiquitous in animals but lacking in other organisms.
Relationship Among the αI, αII, and Invertebrate α-Spectrin Genes. To understand more of the relationship among the spectrin genes and their proteins, we compared the sequences of the coding regions of the genes and their protein products. In doing this, however, we were limited by the incompleteness of some of the genomic data currently available. Because the spectrin genes are quite large (the ORF contains 7.5 kb and spans ≈80 kb in the human genome), there are gaps in many genomic sequences. Moreover, an informative cDNA from T. cellulosae is a partial sequence covering just the first 1.2 kb of the ORF. Therefore, it was not possible to align the full coding sequences of all of the genes. We were able, nevertheless, to align the complete sequences of a well defined and functionally important region, namely the N-terminal sequence that binds β-spectrin. The aligned N-terminal protein sequences (from the start of the β-interactive site through the first full repeat) are shown in Fig. 1. Note the similarities within groups of α-spectrins and in particular the similarities between the αI-spectrins and the clustering of mammalian αII-spectrins with other vertebrate α-spectrins. There are few positions at which the sequences consistently differ from each other, but we note three short blocks of sequence in αI-spectrin that diverge from all others. These are highlighted in red in Fig. 1. These regions are candidate mediators of isoform-specific activities of α-spectrin.
Fig. 1.
Alignment of the amino acid sequences of the N-terminal regions of α-spectrins. Shown are the alignment of a structurally well defined and functionally essential region of α-spectrin, namely the N-terminal helix that binds β-spectrin and the first full triple-helical repeat. The secondary structure elements are assigned by alignment with the NMR structure of human αI-spectrin (Protein Data Bank ID code 1OWA).
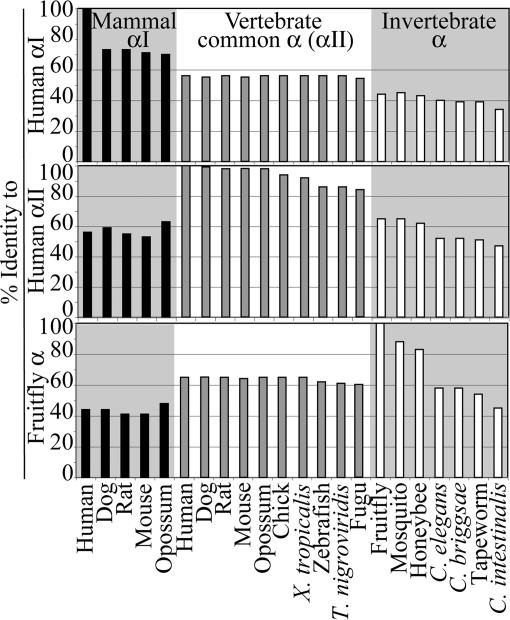
To quantify the relationships between the different spectrins, Fig. 2 and Table 3, which is published as supporting information on the PNAS web site, show the sequence identity (as judged by blastp) between the different proteins, using different N-terminal sequences as reference points. Note that the proteins cluster into obvious groups in this analysis, the αI, αII/vertebrate α, insect, and worm groups in particular. These data confirm the impression given in Fig. 1 that mammalian αII-spectrin is a representative of a common vertebrate α-spectrin group. Interestingly, the representatives of the common vertebrate α-spectrin all have similar sequence identities to human αI-spectrin (e.g., human αII-spectrin and zebrafish α-spectrin are 56% identical).
Fig. 2.
Sequence identities between αI-, αII-, and invertebrate α-spectrins. The sequences of the N-terminal regions of human αI-, αII-, or fruitfly α-spectrin were compared with other sequences, as indicated.
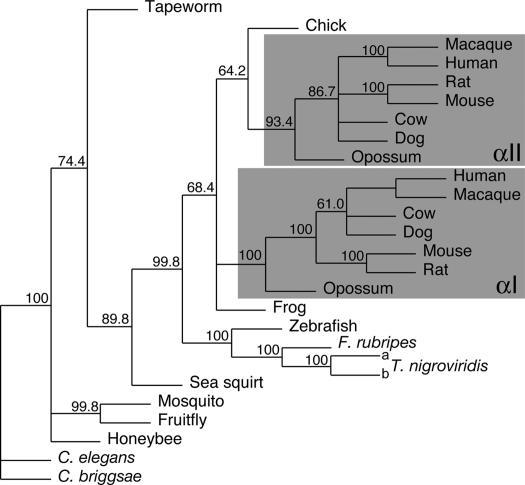
To learn more of their relationship, we turned to a phylogenetic approach. We aligned the cDNA sequences encoding polypeptides from the start of the β-interactive site through the third full repeat (see Table 1). This alignment was analyzed by the maximum-likelihood method by using phyml. Fig. 3 shows a consensus unrooted tree generated by the phylip program consense from 500 bootstrapped trees generated by phyml. Note that there is a clear separate clustering of the mammalian αI- and αII-spectrins. The two T. nigroviridis genes cluster together with the other fish α-spectrins, confirming that neither is an αI-spectrin.
Fig. 3.
Maximum likelihood phylogenetic tree of α-spectrins. cDNA sequences encoding the region from the N-terminal helix through the second full repeat were aligned and analyzed by maximum likelihood with the aid of phyml. Five hundred bootstrapped replicates were examined, and a consensus tree was built from these by using phylip consense. Numbers shown at forks indicate the percentage occurrence among the trees of the group consisting of the species to the right of that fork. The groups of organisms containing both αI- and αII-spectrin are shaded. The organisms and cDNA sequences used are as listed in Table 1.
Fig. 3 indicates that the αI-spectrins separated from the common vertebrate α-spectrin at a point defined by a node between the frog Xenopus tropicalis and chicken Gallus gallus. The node between fish and other vertebrates is robust (99.8% bootstrap value), indicating that the duplication event giving rise to αI-spectrin is unlikely to have occurred in fish. In none of our analyses, whether by using other maximum-likelihood, parsimony, or distance-matrix methods, was the node placed above G. gallus (not shown). It seems most likely that the vertebrate α-spectrin gene underwent duplication after vertebrates began to inhabit land, but the resulting αI-spectrin gene was lost in the lineage leading to G. gallus.
αII-Spectrin Interacts with Human Red Cell Membranes with High Affinity. We have previously demonstrated that purified fragments of αII- and βII-spectrin interact in solution with very high affinity compared with αI and βI (11). Also, αII binds with much higher affinity than αI to βI (11). We have further found that a fragment of αI-spectrin can compete for binding to βI-spectrin in intact red cell ghosts, thereby disrupting the endogenous tetramers and destabilizing the membrane (15).
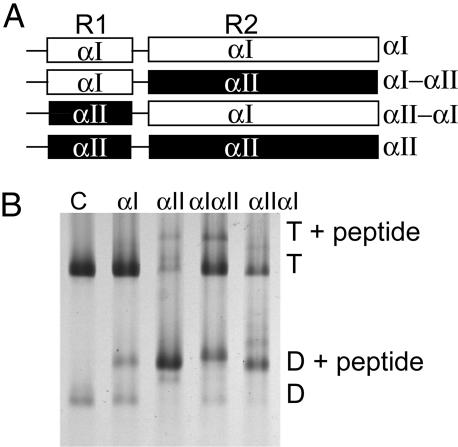
Because αI- and αII-spectrins differ in their β-spectrin-binding capacities, we inferred that addition of heterologous αII-spectrin fragment to red cell membranes should afford a functional comparison of the common vertebrate α-spectrin with αI-spectrin in the context of the intact membrane. We therefore examined binding of α-spectrin fragments to red cell ghosts, taking advantage of our well characterized αI-, αII-, and chimeric αIαII- and αIIαI-spectrin fragments (Fig. 4A and ref. 11). In these experiments, the peptides were sealed into ghosts in isotonic buffer; the ghosts were incubated at 37°C for 40 min to allow binding, then relysed in hypotonic buffer, and the spectrin (with any bound peptides) extracted in very low ionic strength buffer.
Fig. 4.
Binding of αI- and αII-spectrin fragments to spectrin dimer self-association sites in ghost membranes at 37°C. (A) Schematic diagram of α-spectrin fragments. R1 is the partial triple helical repeat that binds to β.R2 is the first full triple helix. αIαII and αIIαI are hybrids in which R1 and R2 are swapped between the proteins. (B) Spectrin extracted from the resealed ghosts was analyzed by electrophoresis in 5% nondenaturing gels. Incorporation at 100 μM total concentration of each fragment was demonstrated by the presence of a new band, migrating above the spectrin dimer (D) and tetramer (T). Lane 1, control spectrin with no peptide introduced into the ghosts; lane 2, αI fragment incorporated; lane 3, αII fragment incorporation; lane 4, αIαII; lane 5 αIIαI.
The extract was analyzed by nondenaturing gel electrophoresis (Fig. 4B). Note that in the absence of peptide, at least 90% of the spectrin is recovered as tetramer. In the presence of the αI-spectrin peptide, there is marked incorporation of the fragment into the dimer fraction, and the total proportion of dimer increases as noted (15). However, in the presence of the αII-spectrin peptide, tetramers are almost completely dissociated. The major band on the gel corresponds to spectrin dimer, with bound αII-spectrin peptide. It follows that at 37°C, in the absence of any applied shear, the spectrin tetramers exist in association-dissociation equilibrium, making dimer ends available to the complementary peptide. The hybrid peptides (αIIαI and αIαII) show intermediate levels of incorporation.
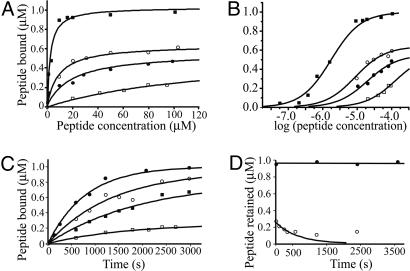
Fig. 5 A and B show the concentration dependence of incorporation of fragments into spectrin in the membrane. The affinities at 37°C differed by ≈2 orders of magnitude, with apparent association constants of 5 × 105 and 8 × 103 M–1 for αII- and αI-spectrin respectively, whereas the hybrid peptides, αIIαI and αIαII, gave association constants of 1 × 105 and 5 × 104 M–1, respectively. The binding profiles can also be fitted with equal precision and elimination of the discrepancies in apparent saturation levels by a negative-cooperativity model, whereby, for instance, the binding of a peptide to one of the dimers of a spectrin tetramer inhibits the entry of a second peptide. For the chimeric peptides, this leads to thermodynamic association constants of 2.1 × 105 and 6.0 × 104 M–1, respectively, for the first site (differing from the microscopic binding constants by a factor of 2) and association constants for the second site ≈2 orders of magnitude lower. It is clear, regardless of the model, that both the N-terminal helix and the adjoining complete repeat influence the affinity of the peptide for spectrin on the membrane, the former exerting the greater effect.
Fig. 5.
Binding isotherms and kinetics of interaction for α-spectrin fragments to spectrin in intact membranes. (A and B) Binding of α-spectrin fragments to membranes. Open squares, αI; solid squares, αII; open circles, αIIαI; solid circles, αIαII. The curves are calculated, assuming a set of independent identical sites in each case. B shows the data plotted on a log scale. (C) Association rates for αI (open squares), αII (filled circles), αIαII (filled squares), and αIIαI (open circles) peptides; (D) dissociation rates of αI (open circles) and αII (closed circles) peptides from membranes. The curve is calculated from the initial rate (see text).
The time courses of these binding processes are shown in Fig. 5C. Because of the large excess of free peptide in the system, binding can be expected to obey pseudofirst-order kinetics, as indeed observed. True second-order rate constants at 37°C were 11 and 0.9 Ms–1 for assimilation of the αI and αII peptides, respectively, onto the membrane. Dissociation rates measured at 37°C revealed an unexpected phenomenon; whereas an off-rate constant of 1.6 × 10–3 s–1 was recorded for the loss of the αI peptide from the membrane, there was no measurable dissociation of the αII peptide over periods of at least 1 hr (Fig. 5D). This implies an off-rate constant of less than ≈10–5·s–1. A possible explanation is that the αII-spectrin peptide binds not only to β-spectrin but also to membrane phospholipids, because it contains a phosphatidylserine-binding site missing from the αI-spectrin peptide (13).
Dissociation of the αI peptide was also incomplete, with an undissociable residue in excess of the very low calculated equilibrium level of binding. We cannot exclude slow binding to a hitherto unrecognized population of sites in the membrane or sequestration of protein in lipid vesicles generated by destabilized membrane.
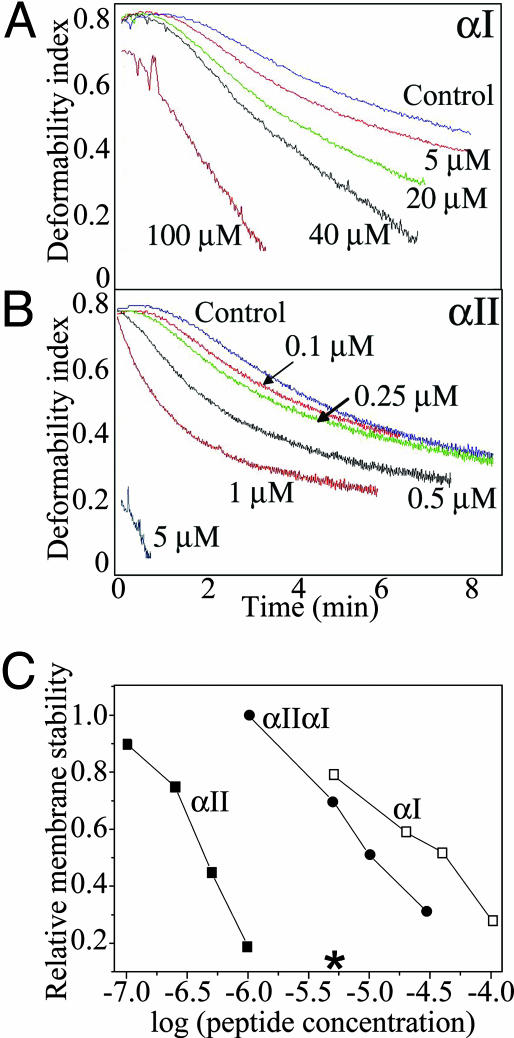
Effect of Peptide Incorporation on Membrane Stability. The concentration-dependent effects of the α-spectrin N-terminal peptides on the mechanical stability of the membrane were assessed by shearing in the ektacytometer at room temperature (16, 17). Fig. 6A shows that, as before (15), membrane stability was markedly reduced by incorporation of the αI peptide, as reflected by a faster rate of decay of the deformability index. The degree of destabilization followed the extent of incorporation of the peptide. The αII peptide, which, as indicated, binds more strongly to spectrin in the membrane, exerted a strikingly more powerful destabilizing effect (Fig. 6B): a perceptible perturbation of the stability was observed at peptide concentrations as low as 0.1 μM, and by 5 μM, the membrane rapidly disintegrated under low shear. At the same concentration, the αI-spectrin peptide showed only a minimal effect. The hybrid peptide, αIIαI, exhibited an intermediate effect on membrane stability (Fig. 6C).
Fig. 6.
Effect of peptide incorporation on membrane stability. Membrane mechanical stability of the resealed ghosts was measured by ektacytometry. Membrane stability is expressed in terms of the rate of decline in deformability index (DI). Faster decay (DI curve shifted to the left) reflects destabilization of the membrane. A and B show the effects of incorporation of the αI and αII peptides, respectively; (C) correlation between relative membrane stability (the time taken to reach 50% loss of deformability) and peptide concentrations; open squares, αI; filled squares, αII; filled circles, αIIαI. *, A concentration of αII peptide that caused essentially instantaneous fragmentation.
Discussion
Birds and mammals are widely supposed to have found two different solutions to the problem of rapid blood cell circulation. Birds generally have smaller genomes than other terrestrial vertebrates, and this reduction in genomic load allows smaller nuclei and smaller cell size. By contrast, mammals evolved minimal red cells, from which the nucleus and other organelles were eliminated. The mammalian solution carries with it a demand for an exceptionally resilient membrane that can survive in circulation in the absence of major biosynthetic repair mechanisms. The observations described here afford insight into the molecular adaptations underlying the evolution of mammalian red cells.
Genomic data indicate that α-spectrin genes were not multiplied in genomes during the vertebrate radiation. This contrasts with other components of the membrane skeleton. Thus, ankyrin, the adaptor that links spectrin to transmembrane proteins, is encoded by a single gene in invertebrates (two in insects) (18, 19). All vertebrates have at least three copies of ankyrin (known fish genomes having six), and there is little evidence of specific mammalian adaptation in the ANK1 gene, which encodes mammalian red cell ankyrin. Similar considerations apply to β-spectrin and protein 4.1; there are four conventional β-spectrin and four 4.1 genes in all vertebrates, some of them duplicated in ray-finned fish (data not shown). In this context, it is striking that α-spectrin is found as a single copy in all available vertebrate genomes, other than those of mammals and the fish T. nigroviridis. In the latter, the whole genome duplication event in the ray-finned fish lineage (14) probably gave rise to the second gene, but it is truncated and presumably inert. The other available fish genomes contain only a single α-spectrin gene, implying that in all of the fish, retention of a second functional α-spectrin was not advantageous. Our phylogenetic analysis (Fig. 3) indicates that the α-spectrin gene was duplicated once again after the advent of terrestrial vertebrates, but the duplicated gene is not retained in chicken. One possibility is that, given that avian erythrocytes retain nuclei, a second α-spectrin was not required and was lost as the genomes compacted.
We explored the functional consequences of the adaptation of mammalian αI-spectrin for the membrane of an enucleate red cell. The different rates of incorporation of the αI and αII peptides and of their hybrids show that binding of the peptide is the rate-limiting step in its incorporation into the membrane cytoskeleton (Fig. 5). Thus we may suppose that the opening and closing of the tetramers in situ is fast. The relative affinities of the different constructs for membrane-bound spectrin (Fig. 5) and their activities in destabilizing the red cell membrane (Fig. 6) mirror the relative affinities for binding βI-spectrin in vitro (11). The chimeras are particularly revealing, because they show that both the single helix at the α-chain N terminus and the adjacent complete triple-helical repeat determine the strength of the interactions.
Alignment of the sequences of all available α-spectrin N-terminal regions reveals sequence differences that correlate with function (Fig. 1). Most notably, a block of 19 residues that covers the end of helix B of the first full triple-helical repeat and the helix B-C loop is well conserved in mammalian αI-spectrin but differs substantially from the consensus of all other α-spectrins. This sequence is a likely candidate for mediating the functional differences between αI- and αII-spectrins. It contains more charged residues in αI- than in αII-spectrin and is overall more hydrophilic. An important topic for future investigation will be to establish the relationship of this block to affinity for β-spectrin and for phosphatidylserine.
Our results offer a rationalization for the evolution of erythrocyte spectrin as a labile tetramer, rather than a single continuous elastic element. In this respect, we view αI-spectrin as neofunctionalized. There is no reason to suppose that after duplication, the two genes for α-spectrin became subfunctionalized, i.e., that their total functional capacity is equivalent to that of the preduplicated gene. On the basis of sequence comparison (Figs. 1 and 2 and Table 3) and phylogeny (Fig. 3), mammalian αII-spectrin is clearly a representative of a single common vertebrate α-spectrin. αI-spectrin has been so changed by mutation as to adapt to a new function, namely, to provide a flexible shear-resistant and elastic membrane-cytoskeleton of a mammal-specific enucleate red blood cell. This conclusion is supported by the consideration of global amino acid sequence identity; human αII-spectrin is 55% identical (72% similar) to human αI-spectrin in a Needleman–Wunsch (20) alignment of the whole proteins; human αII-spectrin is 64% identical (79% similar) and human αI-spectrin 46% identical (67% similar) to D. melanogaster α-spectrin. Human αII-spectrin, then, has diverged further from human αI-spectrin than from fruitfly spectrin.
Our results also suggest that the stable nonerythroid spectrin tetramer, in contrast to its highly labile erythroid homolog, may bestow increased membrane mechanical integrity on nonerythroid cells. Experiments in the invertebrate C. elegans and D. melanogaster reveal that spectrin is required for stabilizing cell junctions against the forces of animal movement (21–23). It seems most likely that analogous functions are preserved and extended in the common vertebrate α-spectrin; thus, for example, the need for the spectrin-binding domain of an ankyrin isoform, ankG, for stability and biogenesis of human epithelial cell lateral membranes (24) argues for an essential function of spectrin in this system also.
The much lower extent of reversible dissociation of nonerythroid spectrin tetramers may also limit the “gated” diffusion in intact membranes of free-floating transmembrane proteins in nonerythroid cells. In the red cell, conversely, the more frequent opening and closing of the (αIβI)2 tetramers facilitates diffusion (25–27).
Materials and Methods
Materials. Human venous blood was donated with informed consent by healthy volunteers. Glutathione-Sepharose 4B and pGEX vectors were purchased from Amersham Pharmacia Biotech, Dextran T40 from Amersham Pharmacia Biotech, proteinase inhibitor mixture set II from Calbiochem, SDS/PAGE and electrophoresis reagents from Bio-Rad, and GelCode Blue Reagent from Pierce. Top Pfu polymerase and BL21(DE3) bacteria were from Stratagene and T4 ligase and restriction enzymes were from New England BioLabs. All other chemicals were reagent grade from commercial sources.
Bioinformatics. Protein sequences were retrieved from the UniProt (SwissProt/TrEmbl) Knowledgebase, cDNA sequences from European Molecular Biology Laboratory (EMBL)/GenBank, and genomic sequences from Ensembl or University of California at Santa Cruz (UCSC) genome portals. blast (28) analyses were done with either online servers at European Bioinformatics Institute, National Center for Biotechnology Information, and EMBL or with a local installation. blat (29) comparisons to genomic sequences were done with the server at UCSC. It was sometimes necessary to refine automated cDNA predictions from genomic sequence; for this purpose, relevant protein, cDNA, or EST sequences were identified by blast and matched to genomic sequence by wise2 (protein) or the emboss (30) program est2genome (nucleotide) for display and annotation in artemis (31). The hmmer package (32) was used for hidden Markov model (HMM) analysis. Multiple sequence alignments were made by using the GCG (33) program pileup or t-coffee (34). For phylogenetic analyses, the phylip package (35) and phyml (36) were used. For pairwise global alignment, the emboss (30) program needle was used. The structure of the N-terminal region of αI-spectrin [Protein Data Bank (PDB) ID code 1OWA] (37) was retrieved from PDB.
Preparation of Recombinant Spectrin Fragments. The cDNA encoding the N-terminal region of human erythrocyte α-spectrin (αI-spectrin), comprising residues 1–154 (αI 1–154) was expressed from the vector pGEX-2T by using the construct described by Nicolas et al. (10). The cDNA encoding the N-terminal region of nonerythroid αII-spectrin comprising residues 1–149 (αII 1–149) was expressed from the vector pGEX-2T, using the construct described by Bignone and Baines (11). Two hybrid proteins were also used. In these, the N-terminal helix and first full repeat from one protein are exchanged with those of the other; thus peptide αIαII contains the N-terminal helix of αI and first full repeat of αII, whereas peptide αIIαI comprises the N-terminal helix of αII and first full repeat of αI (11). All proteins were purified by affinity chromatography on glutathione-Sepharose.
Introduction of Spectrin Fragments into Erythrocyte Ghosts. Red cells were isolated from freshly drawn blood by centrifugation and washed three times with Tris-buffered isotonic saline (10 mM Tris, pH 7.4/0.15 M potassium chloride). The cells were lysed with 35 volumes of ice-cold hypotonic buffer A (5 mM Tris/5 mM potassium chloride, pH 7.4). The resulting ghosts were collected by centrifugation and washed once in cold lysis buffer. Isotonicity was restored by adding 0.1 volume of 1.5 M potassium chloride/50 mM Tris, pH 7.4, and the ghosts (5 × 109 per ml) were resealed by incubating at 37°C for 40 min with the required concentrations of the spectrin fragments.
Extraction and Analysis of Spectrin from Resealed Ghosts. The resealed ghosts were washed with isotonic buffer and relysed with 30 volumes of ice-cold Buffer A and washed three times with the same buffer. To extract the spectrin, the ghosts were suspended in 0.25 mM sodium phosphate, pH 7.4, and incubated overnight at 4°C. The spectrin was collected by centrifugation (21,000 × g) and examined by gel electrophoresis in the native state in a Tris-Bicine buffer system, run in the cold (38). The spectrin tetramer, dimer, and their complexes with the peptide fragment were well resolved, and the absence of a zone corresponding to the free fragment or of a trail of stained material showed that no dissociation had occurred during electrophoresis. The gels, stained with GelCode blue, were evaluated by densitometry. Equilibrium binding data were analyzed as described in Supporting Text, which is published as supporting information on the PNAS web site.
Dissociation of Incorporated α-Spectrin Peptide from Resealed Ghosts. Sealed ghosts were cooled on ice and washed free of unbound peptides by centrifugation in the cold. They were relysed by exposure to hypotonic buffer (40 volumes per volume of packed ghosts) in the cold and again washed three times. Ghosts were then resuspended in 10 volumes of isotonic buffer and incubated at 37°C for up to 40 min. Samples were taken at intervals and examined by gel electrophoresis. The concentration of residual dimer–peptide complex was estimated as before. Kinetic binding data were analyzed as described in Supporting Text.
Measurement of Membrane Stability. The resealed ghosts were suspended in 40% dextran, and membrane mechanical stability was quantified by ektacytometry, as described (16). The measure of membrane stability was taken as the rate of decrease of deformability index at a constant applied shear stress of 750 dynes·cm–2 (1 dyne = 10 μN).
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants DK 26263, HL 31579, and DK 32094. A.J.B. acknowledges a Leverhulme Trust Research Fellowship and Project Grant 96/18062 from the Biotechnology and Biological Sciences Research Council.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gulliver, G. (1875) Proc. Zool. Soc. London 1875, 474–495. [Google Scholar]
- 2.Hawkey, C. M., Bennett, P. M., Gascoyne, S. C., Hart, M. G. & Kirkwood, J. K. (1991) Br. J. Haematol. 77, 392–397. [DOI] [PubMed] [Google Scholar]
- 3.Gregory, T. R. (2001) Blood Cells Mol. Dis. 27, 830–843. [DOI] [PubMed] [Google Scholar]
- 4.Mohandas, N. & Evans, E. (1994) Annu. Rev. Biophys. Biomol. Struct. 23, 787–818. [DOI] [PubMed] [Google Scholar]
- 5.Discher, D. E. & Carl, P. (2001) Cell Mol. Biol. Lett. 6, 593–606. [PubMed] [Google Scholar]
- 6.Speicher, D. W. & Marchesi, V. T. (1984) Nature 311, 177–180. [DOI] [PubMed] [Google Scholar]
- 7.Yan, Y., Winograd, E., Viel, A., Cronin, T., Harrison, S. C. & Branton, D. (1993) Science 262, 2027–2030. [DOI] [PubMed] [Google Scholar]
- 8.Pascual, J., Pfuhl, M., Walther, D., Saraste, M. & Nilges, M. (1997) J. Mol. Biol. 273, 740–751. [DOI] [PubMed] [Google Scholar]
- 9.Kotula, L., DeSilva, T. M., Speicher, D. W. & Curtis, P. J. (1993) J. Biol. Chem. 268, 14788–14793. [PubMed] [Google Scholar]
- 10.Nicolas, G., Pedroni, S., Fournier, C., Gautero, H., Craescu, C., Dhermy, D. & Lecomte, M. C. (1998) Biochem. J. 332, 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bignone, P. A. & Baines, A. J. (2003) Biochem. J. 374, 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett, V. & Baines, A. J. (2001) Physiol. Rev. 81, 1353–1392. [DOI] [PubMed] [Google Scholar]
- 13.An, X., Guo, X., Gratzer, W. & Mohandas, N. (2005) Biochem. Biophys. Res. Commun. 327, 794–800. [DOI] [PubMed] [Google Scholar]
- 14.Van de Peer, Y. (2004) Genome Biol. 5, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An, X., Lecomte, M. C., Chasis, J. A., Mohandas, N. & Gratzer, W. (2002) J. Biol. Chem. 277, 31796–31800. [DOI] [PubMed] [Google Scholar]
- 16.Chasis, J. A. & Mohandas, N. (1986) J. Cell Biol. 103, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohandas, N., Clark, M. R., Health, B. P., Rossi, M., Wolfe, L. C., Lux, S. E. & Shohet, S. B. (1982) Blood 59, 768–774. [PubMed] [Google Scholar]
- 18.Hortsch, M., Paisley, K. L., Tian, M. Z., Qian, M., Bouley, M. & Chandler, R. (2002) Mol. Cell Neurosci. 20, 43–55. [DOI] [PubMed] [Google Scholar]
- 19.Hopitzan, A. A., Baines, A. J. & Kordeli, E. (2006) Mol. Biol. Evol. 23, 46–55. [DOI] [PubMed] [Google Scholar]
- 20.Needleman, S. B. & Wunsch, C. D. (1970) J. Mol. Biol. 48, 443–453. [DOI] [PubMed] [Google Scholar]
- 21.Hammarlund, M., Davis, W. S. & Jorgensen, E. M. (2000) J. Cell Biol. 149, 931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moorthy, S., Chen, L. & Bennett, V. (2000) J. Cell Biol. 149, 915–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng, H., Lee, J. K., Goldstein, L. S. & Branton, D. (1995) J. Cell Biol. 128, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kizhatil, K. & Bennett, V. (2004) J. Biol. Chem. 279, 16706–16714. [DOI] [PubMed] [Google Scholar]
- 25.Golan, D. E. & Veatch, W. (1980) Proc. Natl. Acad. Sci. USA 77, 2537–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koppel, D. E., Sheetz, M. P. & Schindler, M. (1981) Proc. Natl. Acad. Sci. USA 78, 3576–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomishige, M., Sako, Y. & Kusumi, A. (1998) J. Cell Biol. 142, 989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 29.Kent, W. J. (2002) Genome Res. 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice, P., Longden, I. & Bleasby, A. (2000) Trends Genet. 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 31.Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A. & Barrell, B. (2000) Bioinformatics 16, 944–945. [DOI] [PubMed] [Google Scholar]
- 32.Eddy, S. R. (1998) Bioinformatics 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 33.Genetics Computer Group (1996) wisconsin package (Genetics Computer Group, Madison, WI).
- 34.Notredame, C., Higgins, D. G. & Heringa, J. (2000) J. Mol. Biol. 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 35.Felsenstein, J. (1993) phylip (Phylogeny Inference Package) (Dept. of Genetics, University of Washington, Seattle), Ver. 35c.
- 36.Guindon, S., Lethiec, F., Duroux, P. & Gascuel, O. (2005) Nucleic Acids Res. 33, W557–W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, S., Caffrey, M. S., Johnson, M. E. & Fung, L. W. (2003) J. Biol. Chem. 278, 21837–21844. [DOI] [PubMed] [Google Scholar]
- 38.Shahbakhti, F. & Gratzer, W. B. (1986) Biochemistry 25, 5969–5975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.