Abstract
The number of red blood cells is normally tightly regulated by a classic homeostatic mechanism based on oxygen sensing in the kidney. Decreased oxygen delivery resulting from anemia induces the production of erythropoietin, which increases red cell production and hence oxygen delivery. Investigations of erythropoietin regulation identified the transcription factor hypoxia-inducible factor (HIF). HIF is now recognized as being a key regulator of genes that function in a comprehensive range of processes besides erythropoiesis, including energy metabolism and angiogenesis. HIF itself is regulated through the α-subunit, which is hydroxylated in the presence of oxygen by a family of three prolyl hydroxylase domain proteins (PHDs)/HIF prolyl hydroxylases/egg-laying-defective nine enzymes. Hydroxylation allows capture by the von Hippel–Lindau tumor suppressor gene product, ubiquitination, and destruction by the proteasome. Here we describe an inherited mutation in a mammalian PHD enzyme. We show that this mutation in PHD2 results in a marked decrease in enzyme activity and is associated with familial erythrocytosis, identifying a previously unrecognized cause of this condition. Our findings indicate that PHD2 is critical for normal regulation of HIF in humans.
Keywords: EGLN1, hypoxia-inducible factor, Epo, VHL, proteasome
Familial erythrocytosis is rare, but identification of the underlying defects has the potential to give important insights into the pathway by which humans regulate erythropoietin (Epo) production and, more broadly, how cells sense and respond to low oxygen tension. The framework for oxygen sensing in this pathway has been provided by recent dramatic advances in our understanding of the molecular mechanisms by which proteins transduce changes in oxygen tension into alterations in Epo gene transcription. The salient features of this pathway are as follows. Under normoxic conditions, the prolyl hydroxylases (PHDs) site-specifically hydroxylate the α-subunits of hypoxia-inducible factor (HIF) (1–7). This hydroxylation allows modification-dependent binding by the von Hippel–Lindau tumor suppressor protein (VHL), the substrate recognition subunit of an E3 ubiquitin ligase complex, resulting in the constitutive degradation of HIF and maintenance of very low levels of HIF protein (8–12). Under hypoxic conditions, PHD-catalyzed hydroxylation, which employs molecular oxygen as an obligatory cosubstrate, is diminished, thereby allowing HIF to escape VHL-mediated proteasomal degradation. HIF levels, therefore, rise, and HIF then binds to a transcriptional enhancer element located 3′ of the Epo gene (13–15). Thus, HIF can regulate Epo transcription, and hence production, in a manner exquisitely sensitive to oxygen concentration.
This distinctly unusual signal transduction mechanism plays a far broader role in hypoxic adaptation than was initially realized because HIF regulates a large number of genes involved in cellular and systemic responses in addition to Epo. Protein products of HIF gene targets include those involved in glucose uptake, such as Glut1 and Glut3; glycolysis; pH regulation, such as carbonic anhydrase IX; and angiogenesis, such as VEGF and Flt-1 (16, 17). Hence, the identification of genetic alterations in the HIF pathway has important implications for understanding human physiology that extend beyond insights into the control of Epo production.
Autosomal recessive Chuvash polycythemia (18–20) was recently shown to arise from a mutation in VHL that impairs its ability to polyubiquitinate HIF, resulting in excess red cell production with what is typically an inappropriately elevated Epo level (18). Because a significant cohort of familial erythrocytosis cases does not display VHL mutations (21), we hypothesized that a defect in other aspects of the oxygen-sensing pathway might account for some of these cases. Here, we report one such family and identify a mutation in the active site of one of the three PHD isoforms that segregates with the phenotype. Functional studies reveal that this mutation results in a marked defect in PHD activity and provide evidence that this mutation is the cause of the erythrocytosis in this family.
Results
A family with erythrocytosis presented as follows. The father came to medical attention at the age of 45 years with erythrocytosis (Hb 18.0 g/dl) and intermittent claudication and was treated with repeated venesection. He was a smoker and died at the age of 61 from esophageal carcinoma. The mother is hematologically normal. The daughter and son subsequently presented at ages 26 and 30, respectively, with Hb of 18.0 and 17.5 g/dl. Given the elevated Hb, one would expect depressed Epo levels. In fact, they were within the normal range (Table 1), suggesting dysregulation of the Epo axis and thereby favoring a secondary as opposed to primary erythrocytosis. Although these values are lower than those typically seen in patients with Chuvash polycythemia, it is also relevant to note that there is considerable variation in the Epo levels reported in these patients, with some displaying normal Epo levels (19, 21–24). On subsequent testing of the daughter, the Hb had fallen to 14.8 g/dl [hematocrit (Hct) 0.43], with normal white cell and platelet counts; however, there was a concomitant history of menorrhagia. The daughter has regularly required venesection to maintain a Hct below 0.50 and had an episode of superficial thrombophlebitis 4 years after presentation.
Table 1. Hematological parameters in the PHD-2-associated erythrocytosis family.
| Parameter | Father | Daughter* | Son* | Mother |
|---|---|---|---|---|
| Hb, g/dl | 18.0 | 18.0 | 17.5 | 14.5 |
| RBC × 1012 per liter | 6.4 | N/A | 6.1 | 4.5 |
| Hct | 0.53 | N/A | 0.52 | 0.41 |
| WBC × 109 per liter | 5.6 | 11 | 5.7 | 4.6 |
| Platelets × 109 per liter | 266 | 268 | 244 | 191 |
| Epo, mU/ml (normal range 5-25 mU/ml) | N/A | 6.3 | 6.4 | N/A |
RBC, red blood cell; Hct, hematocrit; WBC, white blood cell; N/A, not available; mU, milliunits.
Hb concentration shown for these individuals is that at presentation; other values were obtained upon subsequent examination.
The son, who also had paresthesia, has had consistently elevated Hb levels, reaching as high as 19.1 g/dl (Hct 0.54). The year of presentation, the son's red cell mass was normal and the plasma volume was reduced; several months later, there was a normal plasma volume, and on this occasion, the red cell mass approached the cutoff for an abnormal result. Upon the most recent testing (6 years after presentation), the laboratory tests performed on the son revealed the following: Hb, 17.8 g/dl; white cell count, 4.14 × 109 per liter; platelets, 191 × 109 per liter; Hct, 0.53; red cell count, 6.11 × 1012 per liter; mean cell volume, 84.5 fl; mean cell Hb, 29.1 pg; and mean cell Hb concentration, 34.5 g/dl. Differential: neutrophils, 54%; lymphocytes, 28%; monocytes, 17%; eosinophils, 2%; and basophils, 0%. Iron status results: iron, 15.5 μmol/liter (normal range: 10–30 μmol/liter); transferrin, 2.16 g/liter (normal range: 1.7–2.86 g/liter); and ferritin, 192.8 μg/liter (normal range: 18–280 μg/liter). Results from testing a second sample were similar (e.g., Hb and Hct values of 17.6 g/dl and 0.52, respectively). The level of Epo was also measured and was 8.6 mU/ml (normal range: 5–25 mU/ml), showing a persistently inappropriate level given the Hct and Hb values.
Investigations of both siblings failed to reveal evidence for known primary or secondary causes of erythrocytosis and included a normal oxygen saturation, p50, bone marrow aspirate, and trephine biopsy. Molecular screening for Epo receptor and VHL gene mutations did not reveal any abnormality in either sibling. There was no evidence of renal dysfunction, with both siblings being normotensive, with no significant proteinuria on dipstick urinalysis and plasma creatinine in the normal range. Ultrasound scans revealed an absent left kidney and enlarged right kidney in the son. Fluorescein retinal angiography was normal with no evidence of hemangioblastoma. CNS and abdominal MRI (including T1 and T2 weighted sequences and pre- and postdynamic contrast sequences) was performed on the son and daughter to seek CNS hemangioblastomas, endolymphatic sac tumors, adrenal tumors, and tumors or cysts in the pancreas, kidney, or liver. The only abnormalities detected were a known minor scoliosis and absent left kidney in the son.
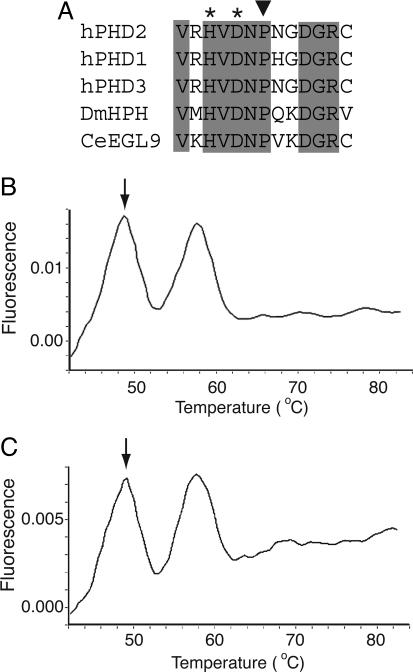
Sequencing of DNA obtained from the two children and a tumor sample from the deceased father revealed in all cases a heterozygous C to G change at base 950 of the coding sequence of PHD2 (data not shown), resulting in a predicted proline to arginine change at codon 317, an evolutionarily conserved residue (Fig. 1A). This residue is in close proximity in the primary sequence to two of the three residues in PHD2 responsible for coordinating the ferrous atom at the active site. Additional confirmation of this mutation was provided by melting curve analysis employing a hybridization probe specific to the normal PHD2 sequence (Fig. 1 B and C). The change was not detected in the hematologically normal mother (Table 1) or in 200 normal control samples (data not shown). Sequencing of the entire coding sequence for PHD1, exons 1–3 of PHD3, and exon 12 of HIF-1α failed to reveal any additional mutations.
Fig. 1.
The 950 C → G mutation affects a residue in the active site of PHD2. (A) Comparison of amino acid sequence from residues 311–323 (human PHD2 numbering) in human HIF prolyl hydroxylases as well as those from Drosophila melanogaster (DmHPH) and Caenorhabditis elegans (CeEGL9). Shading indicates completely conserved residues, asterisks indicate iron-chelating residues, and the inverted triangle indicates Pro-317 of human PHD2, which is predicted to be changed to Arg by the 950 C → G mutation. (B and C) Detection of 950 C → G PHD2 mutation by a hybridization probe. By using a fluorescently labeled hybridization probe specific to the normal sequence, the two heterozygous erythrocytosis siblings were screened with the Roche Diagnostics LightCycler. The annealing temperature of the probe was 58°C when hybridized to the normal sequence. The mismatch due to the presence of the 950 C → G PHD2 mutation reduced the annealing temperature to 49°C (arrows). Both siblings exhibited two peaks, indicating the presence of both the normal and mutant sequences.
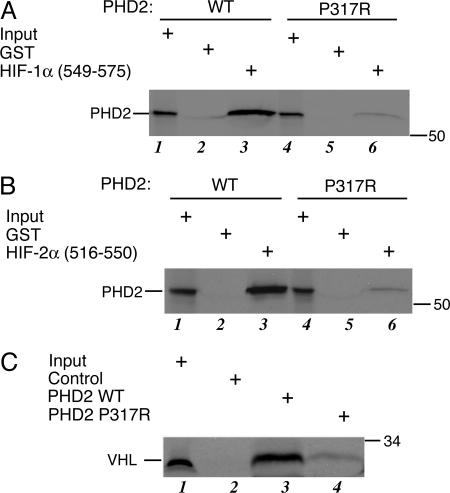
To investigate the functional consequences of this mutation, we first examined the binding of wild-type or mutant PHD2 to HIF-1α-(549–575), which contains the primary hydroxylacceptor proline, Pro-564. Toward this end, we prepared 35S-labeled, in vitro-translated wild-type or P317R PHD2, incubated these proteins with GST-HIF-1α-(549–575) immobilized on glutathione (GSH)-agarose, and then washed the agarose and eluted the proteins. As shown in Fig. 2A, mutant PHD2 binds substantially more weakly to HIF-1α-(549–575) than does wild-type PHD2 (compare lanes 3 and 6). Similar results were observed with a HIF-2α peptide [HIF-2α-(516–550)] that contains the hydroxylacceptor Pro-531 (Fig. 2B, compare lanes 3 and 6). As is evident, the mutation substantially decreases, although does not abolish, binding activity. To determine whether the mutation affects prolyl hydroxylase activity, we prepared in vitro-translated wild-type and P317R PHD2, which we incubated with GST-HIF-1α (549–575) in the presence of 2-oxoglutarate and ascorbic acid. We subsequently assessed the extent of HIF hydroxylation by measuring the binding of 35S-labeled VHL to the HIF product. As shown in Fig. 2C, mutated PHD2 displays dramatically less HIF hydroxylase activity than does the wild type (compare lanes 3 and 4). Importantly, and consistent with the binding assays, the P317R mutation results in a substantial loss of function.
Fig. 2.
PHD2 P317R is defective in HIF binding and HIF hydroxylase activity. (A) 35S-labeled, in vitro-translated wild-type or P317R Flag-PHD2 was incubated with 40 μg of either GST or GST-HIF-1α-(549–575) immobilized on GSH-agarose. The agarose was washed, bound proteins were eluted, and the eluates were subjected to SDS/PAGE and autoradiography. Input represented 10% of the total. The relative recovery of wild-type PHD2 from three replicates was 100 ± 8.7 (arbitrary units), whereas that of P317R PHD2 was 2.5 ± 1.3 (P < 0.001). (B) 35S-labeled, in vitro-translated wild-type or P317R Flag-PHD2 was incubated with 60 μg of either GST or GST-HIF-2α-(516–550) immobilized on GSH-agarose. The agarose was washed, the bound proteins were eluted, and the eluates were subjected to SDS/PAGE and autoradiography. Input represented 10% of the total. The relative recovery of wild-type PHD2 from three replicates was 100 ± 19 (arbitrary units), whereas that of P317R PHD2 was 4.2 ± 0.42 (P = 0.001). (C) In vitro-translated wild-type or P317R Flag-PHD2, or mock in vitro translation reaction, was incubated with 20 μg of GST-HIF-1α-(549–575) immobilized on 10 μl of GSH-agarose in the presence of 2-oxoglutarate, ascorbic acid, and FeCl2. The agarose was then washed, and the degree of HIF hydroxylation was assessed by subsequent incubation with 35S-labeled, in vitro-translated VHL. Input represented 5% of the total. Under the conditions of the assay, the recovery of 35S-labeled, in vitro-translated VHL when wild-type PHD2 was used in three independent experiments was 100 ± 10.2 (arbitrary units), whereas that when P317R PHD2 was used was 8.8 ± 3.4 (P < 0.001).
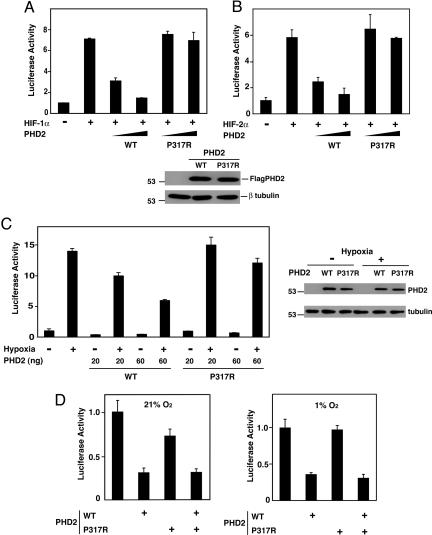
We next examined the functional consequences of this mutation in reporter gene assays. Accordingly, we coexpressed wild-type or P317R PHD2 with HIF-1α along with a reporter gene linked to a hypoxia response element (HRE) in HEK293 cells. Overexpression of wild-type PHD2 suppresses HIF-1α-induced activation of this reporter gene (Fig. 3A, third and fourth columns). We consistently found that P317R PHD2 was less effective than wild-type PHD2 in suppressing such activity (for example, compare the fourth and sixth columns of Fig. 3A). Similar results were obtained when HIF-2α, instead of HIF-1α, was used to activate the HRE reporter gene (Fig. 3B, compare the third and fifth columns). In separate experiments, we also activated the HRE reporter by exposing cells to reduced levels of oxygen and found the mutant PHD2 to be less effective in suppressing hypoxia-induced activation of the reporter (Fig. 3C).
Fig. 3.
P317R PHD2 is defective in inhibiting HRE reporter gene activity. (A) HEK293 cells were cotransfected with 100 ng of (eHRE)3-Luc, which contains the Epo HRE, 100 ng of pRL-TK, 200 ng of either pcDNA3 or pcDNA3-HA-HIF-1α, and 0, 5, or 15 ng of pcDNA3-Flag-PHD2 (wild type or P317R). The total DNA dose was adjusted to 415 ng with pcDNA3. Eighteen hours after transfection, the cells were harvested and assayed for luciferase activity. Error bars represent standard deviations. In separate experiments, HEK293 cells were transfected with 1 μg of wild-type or P317R pcDNA3-Flag-PHD2, and 18 h later, 20-μg extracts were analyzed by Western blotting with anti-Flag or anti-β-tubulin antibodies. The positions of PHD2, β-tubulin, and a molecular weight marker are indicated. (B) HEK293 cells were cotransfected with 100 ng of (eHRE)3-Luc, 100 ng of pRL-TK, 300 ng of either pcDNA3 or pSV-Sport-HA-hHIF-2α, and 0, 2, or 6 ng of pcDNA3-Flag-PHD2 (wild type or P317R). The total DNA dose was adjusted to 506 ng with pcDNA3. Eighteen hours after transfection, the cells were harvested and assayed for luciferase activity. (C) HEK293 cells were cotransfected with 50 ng of (eHRE)3-Luc, 100 ng of pRL-TK, and either 0, 20, or 60 ng of pcDNA3-Flag-PHD2 (wild type or P317R). The total DNA dose was adjusted to 210 ng with pcDNA3. Twenty-four hours after transfection, some cells were subjected to 1% O2 for an additional 18 h. All cells were then harvested and assayed for luciferase activity. In separate experiments, HEK293 cells were transfected with 0.5 μg of wild-type or P317R pcDNA3-Flag-PHD2, and 24 h later, some cells were subjected to 1% O2 for an additional 18 h. Cellular extracts (10 μg) were analyzed by Western blotting with anti-Flag or anti-β-tubulin antibodies. (D) HEK293 cells were cotransfected with 50 ng of (eHRE)3-Luc, 100 ng of pRL-TK, and either 0 or 100 ng of wild-type or P317R pcDNA3-Flag-PHD2. The total DNA dose was equalized with pcDNA3. (Left) Cells were harvested 42 h after transfection and assayed for luciferase activity. (Right) Results from a similar experiment, except that at 24 h after transfection, cells were subjected to 1% O2 for 18 h. With all luciferase assays, activities are normalized to that of a Renilla luciferase internal transfection control. Shown are results that are representative of two or three independent experiments performed in duplicate.
Taken together, the binding, enzymatic, and reporter gene data all point toward a marked loss of PHD2 function arising from the P317R mutation. One possibility this finding raises is that the P317R mutant might act as a dominant negative inhibitor. To examine this possibility, we transfected the HRE reporter gene with an expression vector for wild-type PHD2 in the absence or presence of one producing P317R PHD2. As expected, overexpression of wild-type PHD2 inhibits basal HRE reporter gene activity under normoxic conditions (Fig. 3D Left, first and second columns). Significantly, overexpression of P317R PHD2 fails to reverse wild-type PHD2-induced inhibition of HRE activity (fourth column), and perhaps more importantly, does not activate the HRE reporter by itself (third column). Similar results were observed under hypoxic conditions (Fig. 3D Right). We conclude that the P317R PHD2 displays substantial loss of function without acting as a dominant negative inhibitor.
Discussion
The present findings provide evidence that PHD2 plays a critical role in regulating HIF levels in Epo-producing cells in humans. More broadly, these findings confirm previous studies in cultured cells implicating PHD2 as the primary PHD isoform that regulates HIF under physiologic conditions (25, 26). These studies showed that short interfering RNA knockdown of PHD2 stabilizes HIF-1α and HIF-2α in a number of different human cell lines under standard culture conditions in which cells are incubated at ambient levels of oxygen. PHD2 is also the isoform that appears to be the most abundant in reticulocyte lysates as well as in most transformed human cell lines (7, 26) and is the most closely related ancestrally to the HIF prolyl hydroxylase from either Drosophila melanogaster or Caenorhabditis elegans (27). Because the P317R mutation described in the present report affects PHD2 activity toward both HIF-1α and HIF-2α, the data do not indicate which HIF is more important in Epo gene regulation. In fact, there is evidence that implicates both (28–33). The data, in addition, do not rule out the possibility that other PHD isoforms may, under certain circumstances, also contribute to normoxic stabilization of HIF (26). Indeed, RNA interference studies have shown that PHD1 contributes to normoxic stabilization of HIF-1α in estrogen-stimulated breast carcinoma BT-474 cells and that both PHD1 and PHD3 contribute to normoxic stabilization of HIF-2α in breast carcinoma MCF7 cells (26).
Pro-317 of PHD2 is two residues C-terminal to the iron-chelating residue Asp-315. Although the three-dimensional structure of PHD2 has not yet been reported, it should be noted that the residue in the hydroxylase factor inhibiting HIF (FIH), which is two residues C-terminal to the iron-chelating aspartic acid residue, Gln-203, is in the vicinity of, or indeed, a contact residue with a cysteinyl residue in HIF-1α that is in the –3 position with respect to the hydroxylacceptor asparagine (34). This observation therefore raises the possibility that Pro-317 may be a substrate-binding residue in PHD2. However, whether Pro-317 functions in this manner or serves some other role, such as contributing to the structural integrity of the iron-binding site, must await additional studies.
The current studies, together with previous studies on Chuvash polycythemia (18), indicate that multiple defects in the oxygen-sensing pathway leading to HIF activation can lead to inappropriate up-regulation of HIF and Epo production, leading to erythrocytosis. The present case has features of a secondary erythrocytosis, but in the absence of erythroid progenitor culture assays, we cannot rule out the possibility that there may be features of primary disease. Indeed, Chuvash polycythemia has features of both (18). Interestingly, the present disorder is inherited in an autosomal dominant fashion, in contrast to Chuvash polycythemia, which is an autosomal recessive disorder. The most likely explanation is that both result in a hypomorphic phenotype; the former being due to near-haploinsufficiency for PHD2, and the latter being due to two copies of a hypomorphic allele. It is also possible that PHD2-dependent, VHL-independent, or conversely PHD2-independent, VHL-dependent pathways could contribute to the phenotype. This possibility would imply that other proteins unrelated to HIF regulation interact selectively with PHD2 or VHL, and indeed a number of examples of proteins interacting with the latter have already been reported (35–38).
The elevated Hb and Hct values observed in the siblings would satisfy the World Health Organization Hb criteria for polycythemia vera, but we recognize that the affected individuals described here do not have this disease. The red cell mass was measured in only one member of the kindred, in whom the case for regular venesection was considered borderline. Several months later, after a venesection, a repeat study demonstrated a normal plasma volume and a red cell mass approaching the upper limit of the quoted normal range. The family therefore shows a degree of phenotypic variation, which may reflect how a partial loss of function in the Epo pathway can lead to a small but significant alteration in red cell production and which may also reflect the subtle nature of the underlying defect. This family additionally illustrates difficulties in the diagnostic approach to erythrocytosis and interpretation of red cell mass studies.
We did not observe loss of heterozygosity in the esophageal carcinoma sample obtained from the father, suggesting that this carcinoma was not related to the PHD2 mutation. However, it will be of interest to examine the relationship of this mutation to some of the other clinical features observed in these patients, such as thrombophlebitis and claudication, as well as to examine PHD2 in other tumor contexts. In regard to latter, somatic mutations in PHD2 have recently been reported in endometrial cancer (39). We considered the possibility that the patients might harbor tumors similar to those seen in von Hippel–Lindau disease because these patients benefit from early detection and treatment. We therefore screened them for retinal and CNS hemangioblastoma, endolymphatic sac tumor, renal cysts and tumors, and pheochromocytomas. No evidence of any tumors was found.
Overall, it appears likely that near-haploinsufficiency for PHD2 in the cells that produce Epo is the mechanism leading to erythrocytosis, implying that normal PHD2 concentrations in these cells are not greatly in excess of those required to hydroxylate HIF in normoxia. Interestingly, we did not observe changes in HIF target genes in Epstein–Barr virus-transformed lymphocytes from these patients (data not shown), suggesting that different cell types may differ in the sensitivity of the HIF pathway to reductions in PHD2 activity. On the basis of our findings, one might predict that pharmacological PHD2 inhibition could be used to increase Epo levels relatively selectively. However, enthusiasm for this therapeutic approach must be tempered by the realization that such an inhibitor could potentially activate broader HIF-mediated changes in gene expression, resulting in other consequences such as increased angiogenesis. Conversely, our studies highlight erythrocytosis as a potential side effect if diseases involving ischemia were to be treated with compounds that target PHD2.
Materials and Methods
Patients. Idiopathic erythrocytosis patients with a family history were selected from our registry (19, 40). Ethical approval was obtained from the Queen's University Research Ethics Committee, and written, informed consent was obtained from all individuals. Panels of normal control DNA samples were purchased from the European Collection of Cell Cultures (ECACC) (Porton Down, Salisbury, England). The samples were prepared from U.K. Caucasian blood donors with informed consent for research use. Patient Epo levels were measured at King's College Hospital by an automated sandwich antibody assay (Nichols Advantage Chemiluminescence Immunometric assay; Nichols Institute Diagnostics, Middlesex, U.K.).
PHD Mutational Analysis. Using PCR-direct sequencing, we examined PHD1 (exons 1–5), PHD2 (exons 1–4), PHD3 (exons 1–3), and HIF-1α (exon 12). PHD2 exon 2 was also examined in DNA obtained from a fixed, paraffin-embedded sample of the esophageal carcinoma from the father. A group of 200 normal control samples was screened for the C950G PHD2 mutation by using a fluorescently labeled hybridization probe specific to the normal PHD2 sequence, and melting curve analysis was performed by using the Roche Diagnostics LightCycler according to the manufacturer's protocols. Protocols and oligonucleotide sequences are available upon request.
Plasmids and Proteins. pcDNA3-Flag-PHD2 P317R was prepared in two steps. First, pcDNA3-PHD2-C P317R was prepared by QuikChange mutagenesis (Stratagene) using pcDNA3-PHD2-C (41) as a template and the following two oligonucleotides: 5′-GTACGTCATGTTGATAATCGAAATGGAGATGGAAGATG-3′ and 5′-CATCTTCCATCTCCATTTCGATTATCAACATGACGTAC-3′. The 0.6-kb HindIII/XhoI fragment of pcDNA3-Flag-PHD2 was then subcloned into the HindIII/XhoI site of pcDNA3-PHD2-C P317R to yield pcDNA3-Flag-PHD2 P317R. The entire PHD2 coding region was sequenced to confirm its authenticity.
pGEX-HIF-2α-(516–550) was constructed by subcloning into the EcoRI/XhoI site of pGEX-5X-1 (Amersham Pharmacia) appropriate oligonucleotides encoding the indicated sequence. pSV-Sport-HA-hHIF-2α was constructed by subcloning the 0.06-kb KpnI/XhoI fragment from pcDNA3-HA into the KpnI/SalI site of pSport-MOP2 (42) (a gift of Christopher Bradfield, University of Wisconsin, Madison). The sources of pGEX-HIF-1α-(549–575), pcDNA3-Flag-VHL, pcDNA3-HA-HIF-1α, (eHRE)3-Luc, and the HEK293 cells used in the reporter gene assays have been described in refs. 3, 43, and 44.
GST-HIF-1α-(549–575), GST-HIF-2α-(516–550), and GST proteins were purified from Escherichia coli transformed with the appropriate plasmids by affinity chromatography on GSH-agarose (45). 35S-labeled, in vitro-translated Flag-PHD2 or Flag-PHD2 P317R was prepared by using either rabbit reticulocyte lysate TnT Quick or wheat germ TnT kits (Promega) and pcDNA3-Flag-PHD2 or pcDNA3-Flag-PHD2 P317R as templates, respectively. FeCl2 (50 μM) was included in the reaction mixture. 35S-labeled, in vitro-translated Flag-VHL was prepared by using a rabbit reticulocyte lysate TnT Quick kit and pcDNA3-Flag-VHL as a template.
Assays. For GST pulldown assays, GST-HIF-1α-(549–575), GST-HIF2-α-(516–550), or GST prebound to 10 μl of GSH-agarose was incubated with rocking for 1 h at 4°C with 5 μl of in vitro-translated, 35S-labeled Flag-PHD2 or Flag-PHD2 P317R prepared in reticulocyte lysates in 500 μl of buffer A (20 mM Hepes, pH 7.9/100 mM KCl/1 mM DTT) supplemented with 0.2% Nonidet P-40 (Roche Applied Science, Indianapolis), 0.5 mg/ml BSA, and 10 μM FeCl2. The resins were washed three times with buffer A supplemented with 0.2% Nonidet P-40 and 10 μM FeCl2 and eluted with 2× SDS loading buffer followed by heating at 100°C for 3 min, and the eluates were subjected to SDS/PAGE and autoradiography. Recovery was quantitated by using a Molecular Dynamics Storm 860 PhosphorImager.
The prolyl hydroxylase assay was performed in two steps. First, equivalent amounts (as determined by PhosphorImager analysis) of 35S-labeled Flag-PHD2 or Flag-PHD2 P317R prepared in wheat germ extracts were incubated with GST-HIF-1α-(549–575) immobilized on GSH-agarose in 25 μl of buffer A supplemented with 50 μM FeCl2, 1 mM 2-oxoglutarate, 5 mM ascorbate, and 0.3 μg/μl bovine liver catalase (Sigma) for 1 h at 30°C. The resins were washed and then incubated with 10 μl of 35S-labeled, in vitro-translated Flag-VHL in 500 μl of buffer A supplemented with 0.2% Nonidet P-40 and 0.2% BSA for 1 h at 4° C with rocking. The resins were washed three times with buffer A supplemented with 0.2% Nonidet P-40, eluted with 2× SDS/PAGE loading buffer, and heated at 100°C for 3 min, and the eluates were then subjected to SDS/PAGE and autoradiography.
For luciferase assays, HEK293 cells were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Transient transfections were performed by using FuGENE 6 (Roche Diagnostics) at a 2:1 FuGENE to DNA ratio and with cells at 60–70% confluency. Luciferase activities were measured by using a Dual-Luciferase Reporter Assay System (Promega) and an LB9507 luminometer (Wallac, Gaithersburg, MD). Cells were exposed to hypoxic conditions (1% O2/5% CO2/94% N2) in a modular incubator (Billups-Rothenberg, Del Mar, CA). For immunoblotting, cellular lysates were prepared as described in ref. 43, except that dithiothreitol was omitted, and protein concentrations of extract were determined by using a Bio-Rad DC protein assay kit. Anti-Flag (M2, Sigma) and anti-β-tubulin (D-10, Santa Cruz Biotechnology) monoclonal antibodies were used as described in ref. 43.
Acknowledgments
We thank all of the clinicians who have referred erythrocytosis patients from the U.K. and Ireland to our registry and provided patient samples. We are grateful to Dr. Christopher Bradfield for the gift of pSport-MOP2. We also thank Ms. Wanhua Lu for technical assistance and Mr. Jerome Brooks for providing laboratory data on the patients and for helpful comments on the manuscript. This work was supported by the Northern Ireland Leukaemia Research Fund and National Institutes of Health Grant R01 CA090261 (to F.S.L.).
Conflict of interest statement: P.H.M. is a director and consultant of and holds equity in ReOx Ltd., which aims to develop therapeutic inhibitors of the HIF hydroxylase enzymes.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Epo, erythropoietin; GSH, glutathione; Hct, hematocrit; mU, milliunits; HIF, hypoxia-inducible factor; PHD, prolyl hydroxylase domain protein; VHL, von Hippel–Lindau tumor suppressor protein; HRE, hypoxia response element.
References
- 1.Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J. M., Lane, W. S. & Kaelin, W. G., Jr. (2001) Science 292, 464–468. [DOI] [PubMed] [Google Scholar]
- 2.Jaakkola, P., Mole, D. R., Tian, Y. M., Wilson, M. I., Gielbert, J., Gaskell, S. J., Kriegsheim Av, A., Hebestreit, H. F., Mukherji, M., Schofield, C. J., et al. (2001) Science 292, 468–472. [DOI] [PubMed] [Google Scholar]
- 3.Yu, F., White, S. B., Zhao, Q. & Lee, F. S. (2001) Proc. Natl. Acad. Sci. USA 98, 9630–9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masson, N., Willam, C., Maxwell, P. H., Pugh, C. W. & Ratcliffe, P. J. (2001) EMBO J. 20, 5197–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epstein, A. C., Gleadle, J. M., McNeill, L. A., Hewitson, K. S., O'Rourke, J., Mole, D. R., Mukherji, M., Metzen, E., Wilson, M. I., Dhanda, A., et al. (2001) Cell 107, 43–54. [DOI] [PubMed] [Google Scholar]
- 6.Bruick, R. K. & McKnight, S. L. (2001) Science 294, 1337–1340. [DOI] [PubMed] [Google Scholar]
- 7.Ivan, M., Haberberger, T., Gervasi, D. C., Michelson, K. S., Gunzler, V., Kondo, K., Yang, H., Sorokina, I., Conaway, R. C., Conaway, J. W. & Kaelin, W. G., Jr. (2002) Proc. Natl. Acad. Sci. USA 99, 13459–13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maxwell, P. H., Wiesener, M. S., Chang, G. W., Clifford, S. C., Vaux, E. C., Cockman, M. E., Wykoff, C. C., Pugh, C. W., Maher, E. R. & Ratcliffe, P. J. (1999) Nature 399, 271–275. [DOI] [PubMed] [Google Scholar]
- 9.Ohh, M., Park, C. W., Ivan, M., Hoffman, M. A., Kim, T. Y., Huang, L. E., Pavletich, N., Chau, V. & Kaelin, W. G. (2000) Nat. Cell Biol. 2, 423–427. [DOI] [PubMed] [Google Scholar]
- 10.Kamura, T., Sato, S., Iwai, K., Czyzyk-Krzeska, M., Conaway, R. C. & Conaway, J. W. (2000) Proc. Natl. Acad. Sci. USA 97, 10430–10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cockman, M. E., Masson, N., Mole, D. R., Jaakkola, P., Chang, G. W., Clifford, S. C., Maher, E. R., Pugh, C. W., Ratcliffe, P. J. & Maxwell, P. H. (2000) J. Biol. Chem. 275, 25733–25741. [DOI] [PubMed] [Google Scholar]
- 12.Tanimoto, K., Makino, Y., Pereira, T. & Poellinger, L. (2000) EMBO J. 19, 4298–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semenza, G. L., Nejfelt, M. K., Chi, S. M. & Antonarakis, S. E. (1991) Proc. Natl. Acad. Sci. USA 88, 5680–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pugh, C. W., Tan, C. C., Jones, R. W. & Ratcliffe, P. J. (1991) Proc. Natl. Acad. Sci. USA 88, 10553–10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck, I., Ramirez, S., Weinmann, R. & Caro, J. (1991) J. Biol. Chem. 266, 15563–15566. [PubMed] [Google Scholar]
- 16.Semenza, G. L. (2003) Nat. Rev. Cancer 3, 721–732. [DOI] [PubMed] [Google Scholar]
- 17.Pugh, C. W. & Ratcliffe, P. J. (2003) Nat. Med. 9, 677–684. [DOI] [PubMed] [Google Scholar]
- 18.Ang, S. O., Chen, H., Hirota, K., Gordeuk, V. R., Jelinek, J., Guan, Y., Liu, E., Sergueeva, A. I., Miasnikova, G. Y., Mole, D., et al. (2002) Nat. Genet 32, 614–621. [DOI] [PubMed] [Google Scholar]
- 19.Percy, M. J., McMullin, M. F., Jowitt, S. N., Potter, M., Treacy, M., Watson, W. H. & Lappin, T. R. (2003) Blood 102, 1097–1099. [DOI] [PubMed] [Google Scholar]
- 20.Pastore, Y. D., Jelinek, J., Ang, S., Guan, Y., Liu, E., Jedlickova, K., Krishnamurti, L. & Prchal, J. T. (2003) Blood 101, 1591–1595. [DOI] [PubMed] [Google Scholar]
- 21.Gordeuk, V. R., Stockton, D. W. & Prchal, J. T. (2005) Haematologica 90, 109–116. [PubMed] [Google Scholar]
- 22.Percy, M. J., Beard, M. E., Carter, C. & Thein, S. L. (2003) Br. J. Haematol. 123, 371–372. [DOI] [PubMed] [Google Scholar]
- 23.Cario, H., Schwarz, K., Jorch, N., Kyank, U., Petrides, P. E., Schneider, D. T., Uhle, R., Debatin, K. M. & Kohne, E. (2005) Haematologica 90, 19–24. [PubMed] [Google Scholar]
- 24.Gordeuk, V. R., Sergueeva, A. I., Miasnikova, G. Y., Okhotin, D., Voloshin, Y., Choyke, P. L., Butman, J. A., Jedlickova, K., Prchal, J. T. & Polyakova, L. A. (2004) Blood 103, 3924–3932. [DOI] [PubMed] [Google Scholar]
- 25.Berra, E., Benizri, E., Ginouves, A., Volmat, V., Roux, D. & Pouyssegur, J. (2003) EMBO J. 22, 4082–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Appelhoff, R. J., Tian, Y. M., Raval, R. R., Turley, H., Harris, A. L., Pugh, C. W., Ratcliffe, P. J. & Gleadle, J. M. (2004) J. Biol. Chem. 279, 38458–38465. [DOI] [PubMed] [Google Scholar]
- 27.Taylor, M. S. (2001) Gene 275, 125–132. [DOI] [PubMed] [Google Scholar]
- 28.Yu, A. Y., Shimoda, L. A., Iyer, N. V., Huso, D. L., Sun, X., McWilliams, R., Beaty, T., Sham, J. S., Wiener, C. M., Sylvester, J. T. & Semenza, G. L. (1999) J. Clin. Invest. 103, 691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai, Z., Manalo, D. J., Wei, G., Rodriguez, E. R., Fox-Talbot, K., Lu, H., Zweier, J. L. & Semenza, G. L. (2003) Circulation 108, 79–85. [DOI] [PubMed] [Google Scholar]
- 30.Scortegagna, M., Morris, M. A., Oktay, Y., Bennett, M. & Garcia, J. A. (2003) Blood 102, 1634–1640. [DOI] [PubMed] [Google Scholar]
- 31.Warnecke, C., Zaborowska, Z., Kurreck, J., Erdmann, V. A., Frei, U., Wiesener, M. & Eckardt, K. U. (2004) FASEB J. 18, 1462–1464. [DOI] [PubMed] [Google Scholar]
- 32.Scortegagna, M., Ding, K., Zhang, Q., Oktay, Y., Bennett, M. J., Bennett, M., Shelton, J. M., Richardson, J. A., Moe, O. & Garcia, J. A. (2005) Blood 105, 3133–3140. [DOI] [PubMed] [Google Scholar]
- 33.Rankin, E. B., Higgins, D. F., Walisser, J. A., Johnson, R. S., Bradfield, C. A. & Haase, V. H. (2005) Mol. Cell. Biol. 25, 3163–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elkins, J. M., Hewitson, K. S., McNeill, L. A., Seibel, J. F., Schlemminger, I., Pugh, C. W., Ratcliffe, P. J. & Schofield, C. J. (2003) J. Biol. Chem. 278, 1802–1806. [DOI] [PubMed] [Google Scholar]
- 35.Okuda, H., Saitoh, K., Hirai, S., Iwai, K., Takaki, Y., Baba, M., Minato, N., Ohno, S. & Shuin, T. (2001) J. Biol. Chem. 276, 43611–43617. [DOI] [PubMed] [Google Scholar]
- 36.Corn, P. G., McDonald, E. R., III, Herman, J. G. & El-Deiry, W. S. (2003) Nat. Genet 35, 229–237. [DOI] [PubMed] [Google Scholar]
- 37.Li, Z., Wang, D., Na, X., Schoen, S. R., Messing, E. M. & Wu, G. (2003) EMBO J. 22, 1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, Z., Wang, D., Messing, E. M. & Wu, G. (2005) EMBO Rep. 6, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato, H., Inoue, T., Asanoma, K., Nishimura, C., Matsuda, T. & Wake, N. (2005) Int. J. Cancer 118, 1144–1153. [DOI] [PubMed] [Google Scholar]
- 40.Percy, M. J., Mooney, S. M., McMullin, M. F., Flores, A., Lappin, T. R. & Lee, F. S. (2003) Mol. Cancer 2, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang, J., Zhao, Q., Mooney, S. M. & Lee, F. S. (2002) J. Biol. Chem. 277, 39792–39800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hogenesch, J. B., Chan, W. K., Jackiw, V. H., Brown, R. C., Gu, Y. Z., Pray-Grant, M., Perdew, G. H. & Bradfield, C. A. (1997) J. Biol. Chem. 272, 8581–8593. [DOI] [PubMed] [Google Scholar]
- 43.Yu, F., White, S. B., Zhao, Q. & Lee, F. S. (2001) Cancer Res. 61, 4136–4142. [PubMed] [Google Scholar]
- 44.Zhao, Q. & Lee, F. S. (2003) Biochemistry 42, 3627–3634. [DOI] [PubMed] [Google Scholar]
- 45.Smith, D. B. & Johnson, K. S. (1988) Gene 67, 31–40. [DOI] [PubMed] [Google Scholar]